Abstract
For the generation of energy, the important human pathogen Streptococcus pneumoniae relies on host-derived sugars, including β-glucoside analogs. The catabolism of these nutrients involves the action of 6-phospho-β-glucosidase to convert them into usable monosaccharaides. In this study, we characterized a 6-phospho-β-glucosidase (BglA3) encoded by SPD_0247. We found that this enzyme has a cell membrane localization and is active only against a phosphorylated substrate. A mutated pneumococcal ΔSPD0247 strain had reduced 6-phospho-glucosidase activity and was attenuated in growth on cellobiose and hyaluronic acid compared to the growth of wild-type D39. ΔSPD0247-infected mice survived significantly longer than the wild-type-infected cohort, and the colony counts of the mutant were lower than those of the wild type in the lungs. The expression of SPD_0247 in S. pneumoniae harvested from infected tissues was significantly increased relative to its expression in vitro on glucose. Additionally, ΔSPD0247 is severely impaired in its attachment to an abiotic surface. These results indicate the importance of β-glucoside metabolism in pneumococcal survival and virulence.
INTRODUCTION
Streptococcus pneumoniae is a frequent occupant of the human nasopharynx, where it resides without causing symptoms (1). Conversely, the bacterium also is a major human pathogen, being a leading cause of bacterial pneumonia, otitis media, meningitis, and septicemia (1). The increasing trend of antibiotic resistance and the shortcomings of existing vaccines mean that a better understanding of the pathogenesis of pneumococcal diseases is required.
Almost one-third of the transporters in the pneumococcal genome are dedicated to sugars (2), stressing the important role that these carbon sources play in the ability of pneumococci to survive in the host. The pneumococcus has been shown to ferment 32 different sugars in vitro, including various hexoses, α- and β-galactosides, and glucosides, as well as polysaccharides (3, 4). However, the concentrations of readily available simple carbohydrates in the respiratory tract are low (5); thus, the pneumococcus relies on complex host glycans for growth in vivo (4). Central to this is the ability to sequentially deglycosylate host glycoproteins (6).
Mammalian extracellular matrix is rich in glycosaminoglycans (GAGs; e.g., hyaluronic acid), which contain β-linked disaccharide repeating units (7, 8). The degradation of GAGs leads to the generation of structural analogues of cellobiose or N,N′-diacetylchitobiose [(GlcNAc)2] and other β-linked disaccharides (9). Disaccharides such as these can be transported into the bacterial cell through phosphoenolpyruvate-dependent phosphotransferase systems (PTS), which transport and phosphorylate sugars simultaneously (10). The phosphorylated disaccharides then are hydrolyzed by cytoplasmic phospho-β-glucosidases (EC 3.2.1.86) or phospho-β-galactosidases (EC 3.2.1.85) that usually do not have hydrolytic activity toward nonphosphorylated substrates. The resulting glucose and glucose 6-phosphate are further metabolized by the glycolytic pathway (11).
β-Glucoside metabolism has been linked to bacterial survival in vivo and virulence. For example, Listeria monocytogenes expresses β-glucoside permease in vivo (12), and Streptococcus gordonii genes involved in β-glucoside metabolism are upregulated in vivo on infected heart valves during experimental endocarditis and in vitro during biofilm formation on saliva-coated hydroxyapatite (7). However, the full extent of pneumococcal β-glucoside metabolism and its relation to virulence require further study.
It was demonstrated that most pneumococcal strains are able to utilize hyaluronic acid as a sole source of carbon, and growth on hyaluronic acid is dependent on hyl, ugl, and pts, coding for hyaluronidase, glucuronyl hydrolase, and the PTS, respectively (13). In addition, the celBCD region has been shown to be important for cellobiose metabolism (14). The activity of phospho-β-glucosidases or phospho-β-galactosidases is important for β-glucoside utilization, and the pneumococcus has six genes annotated as either 6-phosphogalactosidase or 6-phospho-β-glucosidase (SPD_0247, SPD_277, SPD_0427, SPD_0503, SPD_1046, and SPD_1830) (15). The presence of multiple copies of phosphoglycosyl hydrolases indicates that the pneumococcus encounters β-glucosides in the host. Among these, BglA2 (SPD_0503) has been structurally characterized (16), and all except SPD_0247 are associated with sugar transporters implicated in the transport of various sugars, including β-glucosides and lactose. While most of the phosphoglycosyl hydrolases have been linked to the utilization of sugars, the function of the putative orphan protein encoded by SPD_0247 is not known. Such proteins are crucial for nutrient metabolism by many mucosal pathogens. In this study, we characterized a pneumococcal enzyme with 6-phospho-β-glucosidase activity (referred to as BglA3) encoded by SPD_0247 and investigated its role in β-glucoside metabolism, biofilm formation, and virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptococcus pneumoniae serotype 2 strain D39 was used in this study. Pneumococci were grown in brain heart infusion (BHI) broth or on blood agar plates supplemented with 5% (vol/vol) defibrinated horse blood under microaerophilic conditions at 37°C. Where appropriate, the medium was supplemented with spectinomycin (100 μg/ml) or kanamycin (250 μg/ml).
Sicard's defined medium (17) also was used for bacterial growth. When needed, the medium was supplemented either with glucose or cellobiose (each at 22 mM) or with hyaluronic acid (5 mg/ml), concentrations at which they are known to support pneumococcal growth (13). Pneumococci were grown in chemically defined medium with the desired carbon source until the optical density at 600 nm (OD600) had reached approximately 0.3 to 0.4 to prepare starter cultures. Five-microliter aliquots of starter cultures then were transferred into wells containing 195 μl medium. Growth was monitored by measuring the optical density at 600 nm using a Multiskan GO microplate spectrophotometer (Thermo Scientific). Growth rates (μ) were calculated through linear regressions of the plots of ln(OD600) versus time during the exponential growth phase (18).
Construction of mutants.
In vitro mariner mutagenesis was used to mutate SPD_0247 as described previously (18, 19). The chromosomal region to be mutated was amplified with the primers listed in Table 1 and incubated with Himar 1 transposase (20) and plasmid pR412, which contains the mariner minitransposon conferring spectinomycin resistance (21). The in vitro-mutagenized DNA then was transformed into the pneumococcus using competence-stimulating peptide (22). The insertion of the resistance cassette was confirmed by PCR using transposon-specific primer MP127 or MP128, with appropriate chromosomal primers, and by sequencing. The resulting mutant was designated ΔSPD0247.
TABLE 1.
Oligonucleotide primers used in this study
| Primer IDa | Primer sequenceb (5′–3′) | Target |
|---|---|---|
| SPD0247-F | GAAGCAAGCTTGATGGGAGC | SPD_0247 in D39 |
| SPD0247-R | CCGATACCAGTTGTTCCCTC | SPD_0247 in D39 |
| SPD0247NcoI | CGCCATGGCGTGCTTGAAACGCTTG | SPD_0247 in D39 |
| SPD0247SphI | GACGCATGCGGATAATCAGTCAGAGTC | SPD_0247 in D39 |
| SPD0709-RTF | TCGTGTGGCTGCCAAGCGTG | SPD_0709 in D39 |
| SPD0709-RTR | GGCTGATCCACCAGCTGAGTC | SPD_0709 in D39 |
| MP 127 | CCGGGGACTTATCAGCCAACC | pR412 (21) |
| MP 128 | TACTAGCGACGCCATCTATGTG | pR412 (21) |
| MAL-F | GCTTGAAAAGGAGTATACTT | pCEP (23) |
| PCEP-R | AGGAGACATTCCTTCCGTATC | pCEP (23) |
Primer identifiers (ID) with an F or R tag were used for amplification of gene targets for mutational work, while primer ID with an RTF or RTR tag were utilized for gene expression analysis.
NcoI and SphI recognition sites are italicized.
Complementation of ΔSPD0247.
To confirm that the mutation of SPD_0247 introduced no polar effects, ΔSPD0247 was complemented with an intact copy of the gene using pCEP, a nonreplicative plasmid that allows controlled gene expression following ectopic integration into the chromosome (23). The entire SPD_0247 sequence (1,380 bp) along with its upstream sequence was amplified with SPD_0247NcoI and SPD_0247SphI primers (Table 1). The amplicons were digested with NcoI and SphI and were ligated into similarly digested pCEP. An aliquot of ligation mixture was transferred into Stellar competent Escherichia coli cells (Clontech, Saint-Germain-en-Laye, France). The presence of an insert was determined by colony PCR using MalF and pCEPR primers, whose recognition sites are localized immediately up- and downstream of the cloning site. Recombinant plasmid then was transformed into ΔSPD247 as described above. The transformants were selected on blood agar plates supplemented with spectinomycin and kanamycin. The complemented strain was designated ΔSPD0247Comp.
RNA extraction from bacterial cells and purification.
The extraction of RNA was done using the TRIzol method on mid-log-phase cultures, as described previously (24). Before use, the RNA was treated with amplification-grade DNase I (Qiagen) and subsequently purified with an RNeasy minikit (Qiagen).
Extraction of pneumococcal RNA from infected tissues.
Outbred 9-week-old female MF1 mice (Harlan Olac, Bicester, United Kingdom) were intranasally infected with 50 μl phosphate-buffered saline (PBS) containing approximately 1 × 106 passaged D39 pneumococci (18, 25). When mice became severely lethargic, they were anesthetized and the blood was collected by cardiac puncture. The lungs and nasopharynx of the sacrificed mice were removed and homogenized on ice in sterile PBS. Pneumococci were separated from host cells by centrifugation at 900 × g for 6 min at 4°C. Supernatants subsequently were centrifuged at 15,500 × g for 2 min at 4°C, and the bacterial pellet was stored at −80°C until further processing. Prior to pelleting, a sample of homogenate was plated onto blood agar for the quantification of pneumococci and to exclude the presence of contaminating microflora. RNA extraction and purification were done as described previously (18).
Quantitative reverse transcription-PCR (RT-PCR).
First-strand cDNA synthesis was performed on approximately 1 μg of DNase-treated total RNA, immediately after isolation, using 200 U of SuperScript II reverse transcriptase (Invitrogen, Paisley, United Kingdom) at 42°C for 55 min and random hexamers (18, 25). cDNA (15 ng) was amplified in a 20-μl reaction volume that contained 1× SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and 3 pmol of each primer (indicated with an RTF or RTR tag in Table 1). The transcription levels of specific genes were normalized to the transcription of gyrB, which was amplified in parallel with SPD_0709F and SPD_0709R primers. The results were analyzed by the comparative threshold cycle (CT) method (26).
Cloning, expression, and purification of 6-phospho-β-glucosidase.
SPD_0247 was amplified by PCR with SPD_0247IFF and SPD0247IFR primers (Table 1). The amplicons were mixed with prelinearized In-Fusion Ready pEcoli-Nterm 6XHN vector and In-Fusion dry-down mixture (Clontech, Saint-Germain-en-Laye, France). The recombinant constructs were transformed into Fusion-Blue competent E. coli, and a transformant was selected for sequencing to rule out any mutation. The construct DNA then was transformed into E. coli BL21(DE3) (Novagen, Nottingham, United Kingdom) for recombinant protein expression. The protein purification used immobilized metal affinit chromatography (IMAC) resin and nondenaturing conditions as instructed by the manufacturer (Clontech). The identity of the protein was verified by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometric analysis of tryptic digests of the products by PNACL at the University of Leicester.
Substrate determination and kinetic characterization of the recombinant protein.
Purified recombinant enzyme was tested against various synthetic chromogenic and fluorogenic substrates for determination of the substrate specificity of the enzyme. This was done by incubating 10 μl of 10 mM substrate, 35 μl Tris-HCl (pH 7.5) buffer, 45 μl distilled water, and 10 μl of enzyme (2.4 μg). Once the specific substrate was determined, the enzyme was characterized kinetically using different concentrations of protein and substrate, p-nitrophenol (pNP)-6-phospho-β-glucoside, ranging from 0 to 2.0 mM. The reactions were monitored for 1 h at 37°C on a Bio-Rad plate reader model 680, with monitoring of the assay every minute and registering the absorbance at 415 nm. Km and Vmax were assessed at the pH optimum of the enzyme using the Lineweaver-Burk method (27). Lactate dehydrogenase (28) and neuraminidase (29) activities in cellular fractions were determined as previously described.
Preparation of antisera against recombinant BglA3.
Ten-week-old female MFI outbred mice (Harlan Olac) were injected with 25 μg of recombinant protein, 33 μl of Imject alum adjuvant (Perbio Science, Cramlington, United Kingdom), and 67 μl of PBS. The control group was administered only adjuvant and PBS. Injections were repeated three times at 2-week intervals. At 2 weeks after the last injection, mice were anesthetized with 5% (vol/vol) fluothane (Astra Zeneca, Macclesfield, United Kingdom) over oxygen (1.5 to 2 liters/min), and blood was collected by cardiac puncture. The blood was left at room temperature for 1 h to clot; the serum was recovered by centrifugation at 5,000 × g for 10 min and stored at −80°C until needed.
Cellular localization of BglA3.
The pneumococcal culture pellet was resuspended in 10 mM Tris-HCl–1 mM EDTA buffer (pH 8.0) containing 25% (wt/vol) sucrose and 12.5 mg/ml lysozyme. The suspension was incubated at 37°C, and the cell lysate was centrifuged at 3,000 × g for 5 min. The supernatant containing the cell wall was kept. The pellet then was resuspended in 500 μl of PBS, sonicated, and centrifuged at 20,000 × g for 10 min. The supernatant and the pellet were kept for the analysis of cytoplasmic and membrane proteins, respectively. The quality of fractionation was determined by assessing activities of lactate dehydrogenase, which is known to have an intracellular localization (30), and neuraminidase, which has cell wall localization (31). Western hybridization followed procedures we described previously (18).
In vivo virulence studies.
To determine the virulence of pneumococcal strains, 10-week-old female MFI outbred mice (Harlan Olac) were lightly anesthetized, and 50 μl containing approximately 5 × 105 CFU/ml S. pneumoniae in PBS was administered dropwise into the nostrils (18, 25). The inoculum dose was confirmed by viable counting on blood agar plates. Mice were monitored for disease signs (progressively starry coat, hunched posture, and lethargy) for 7 days (18, 25). The mice that reached the severely lethargic stage were considered to have reached the endpoint of the assay and were humanely sacrificed. The time to this point was defined as the survival time. Mice that were alive 7 days after infection were deemed to have survived the infection. Survival times were analyzed by the Mann-Whitney U test.
To determine the development of bacteremia in each mouse, approximately 20 μl of venous blood was collected at predetermined time points after infection, and viable counts were determined as described above. The growth of pneumococci in the nasopharynx and lungs also was determined. For this, at predetermined time intervals following intranasal infection, set groups of mice were deeply anesthetized and humanely sacrificed by cervical dislocation. The lungs and nasopharynx were transferred separately into PBS and homogenized. Viable counts in homogenates were determined as described above. Data were analyzed by analysis of variance followed by the Bonferroni posttest. P values of <0.05 were considered statistically significant.
Investigation of involvement of BglA3 in biofilm formation.
Pneumococcal cultures in BHI medium were incubated at 37°C in flat-bottom microtiter plates (Nunc) in 200 μl containing approximately 1 × 106 CFU/ml of pneumococci. After overnight growth, there was no difference in the numbers of pneumococci, between 8.8 × 108 and 9.1 1 × 108 CFU/ml. On the following day, the cells in suspension that had not adhered were collected, serially diluted, and plated for enumeration. The wells were washed three times with BHI, 100 μl of fresh BHI was added to the wells, and the plate was sonicated for 3 s. The number of attached pneumococci was quantified by plating serial dilutions of sonicated homogenate. The percentage of adhered cells was normalized against the nonadherent planktonic cells for each strain. The data then were analyzed using the Mann-Whitney U test.
RESULTS
Characterization of BglA3.
SPD_0247 encodes a putative protein annotated as a 6-phospho-β-glucosidase (14). This putative protein (BglA3) has 459 amino acids and a predicted molecular mass of 53.2 kDa. In the D39 strain there are five BglA3 paralogs displaying 31 to 47% similarity, namely, SPD_0277, SPD_0427, SPD_0503, SPD_1046, and SPD_1830 (15). Recently an enzyme with 6-phospho-β-glucosidase activity (BglA2; SPD_0503) in S. pneumoniae was characterized structurally (16). Interestingly, this protein has only 31% identity, while BglJ (sgo_1759) of Streptococcus gordonii has 88% amino acid sequence identity to BglA3.
The substrate specificity for pneumococcal BglA3 (SPD_0247) is not known. Therefore, we expressed and purified recombinant BglA3 and tested it against nine synthetic substrates: O-nitrophenyl (ONP)-β-d-galactopyranoside, 4-methylumbelliferyl-N-acetyl-α-d-glucosaminide, 4-methylumbelliferyl-β-d-glucopyranoside, methylumbelliferyl-α-d-glucopyranoside, O-nitrophenyl-α-d-galactopyranoside, pNP-α-d-glucoside-6-phosphate, pNP-α-d-mannose-6-phosphate, pNP-α-d-galactose-6-phosphate, and pNP-β-d-glucose-6-phosphate. The results showed that purified protein was active only against the synthetic substrate pNP-β-d-glucose-6-phosphate. The enzyme had a specific activity of 3.37 ± 0.2 μU/min/μg of protein (n = 9) against this substrate. There was no detectable activity against the α-linked glucoside substrates. There also was no detectable activity against 4-methylumbelliferyl-β-d-glucopyranoside and 4-methylumbelliferyl-α-d-glucopyranoside, indicating that the presence of a phosphate group is an essential requirement for substrate recognition. The data suggest that this enzyme is indeed a 6-phospho-β-glucosidase. The kinetic parameters also were determined as a Km of 1,416 ± 319 μM and Vmax of 0.795 ± 0.09 μmol · min−1 · mg−1 (n = 9).
Subcellular localization of BglA3.
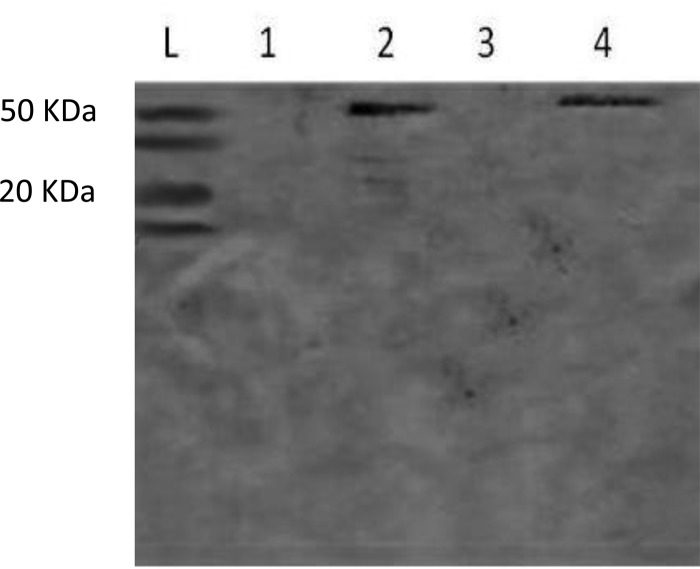
It was found that the protein BglA3 is membrane associated, since the antibodies raised against this protein reacted only with the extract containing the membrane fractions (Fig. 1, lane 2) but not with the cell wall (lane 1) or with the cytoplasmic fractions (lane 3). The band seen in lane 4 corresponds to the positive control, D39 whole-cell extract. Moreover, the antibodies did not react with the whole-cell lysate of the mutant in Western blot analysis, showing the specificity of antibody (data not shown). In this regard, it also is worth noting that in silico analysis found five putative glucosidases of various molecular sizes, 53 to 55 kDa. If the antibodies were cross-reactive with these, the bands would have been seen in the blot shown for the D39 extract; however, no additional bands were seen (Fig. 1). Given that the enzyme is active against the phosphorylated substrates and that hyaluronic acid disaccharides are imported into the cell (13), we predict that the enzyme is localized inside the cell. The fractionation quality was assessed by determining activities for lactate dehydrogenase, known for having an intracellular location (30), and neuraminidase, known to have cell surface localization (31). The results showed that the cytoplasmic fraction contained a much higher level of lactate dehydrogenase activity (357.5 ± 21 mU/mg) than the cell membrane (20.7 ± 11.4 mU/mg) and cell wall (9.4 ± 3.6 mU/mg) fractions. In addition, the highest neuraminidase activity was detected in the cell wall (43.9 ± 3.8 mU/mg), compared to the cytoplasmic (4.9 ± 1.9 mU/mg) and membrane (1.8 ± 0.3 mU/mg) fractions. These results show that the fractionation had been successful.
FIG 1.
Determination of 6-phospho-β-glucosidase subcellular localization by immunoblotting in cellular fractions of pneumococcal strain D39. Lane 1, cell wall extract; lane 2, membrane extract; lane 3, cytoplasmic extract; lane 4, whole-cell extract; lane L, protein size marker.
Characterization of ΔSPD0247 by growth studies.
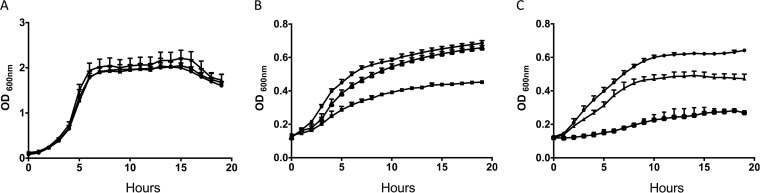
To determine the functional role of BglA3, a loss-of-function mutant, ΔSPD0247, was created. The mutant was tested for its ability to grow in defined medium in the presence of cellobiose or hyaluronic acid. The mutant grew as well as the wild type in rich medium, BHI (data not shown), and in defined medium with glucose (Fig. 2A). However, compared to that of the wild type, ΔSPD0247 was impaired in its ability to grow in Sicard's defined medium without glucose but supplemented either with cellobiose (Fig. 2B) or hyaluronic acid (Fig. 2C). ΔSPD0247 grown in defined medium supplemented with cellobiose had a doubling time of 5.09 ± 0.08 h (n = 4), whereas D39 had a doubling time of 3.89 ± 0.05 h (n = 4) (P < 0.001). On the other hand, the genetically complemented strain ΔSPD0247Comp had a growth profile (doubling time of 3.84 ± 0.06 h; P > 0.05) similar to that of the wild type on cellobiose, indicating that the mutation did not cause a polar effect.
FIG 2.
Growth of pneumococcal strains in Sicard's defined medium either with glucose (A) or without glucose but supplemented with either cellobiose (22 mM) (B) or hyaluronic acid (5 mg/ml) (C) as the sole carbon source. The mean values from 4 independent experiments with the standard error of the means have been plotted. ●, D39; ▲, ΔSPD0247Comp; ■, ΔSPD0247.
A decreased ability to grow on hyaluronic acid also was observed with the mutant (Fig. 2C). Wild-type D39 had a doubling time of 5.52 ± 0.14 h (n = 4), whereas ΔSPD0247 had a doubling time of 9.40 ± 0.08 h (n = 4) (P < 0.001). Once again, a restoration of the phenotype was achieved when the mutant was complemented: a doubling time of 5.88 ± 0.18 h (n = 4) was observed (P > 0.05). Although on hyaluronic acid ΔSPD0247Comp had the same rate of growth as the wild type, it had a lower growth yield. This indicates that bglA3 expression is affected by the genome topology, since in the complemented strain the genome location of an intact copy of bglA3 is different from that in the wild type.
Virulence studies.
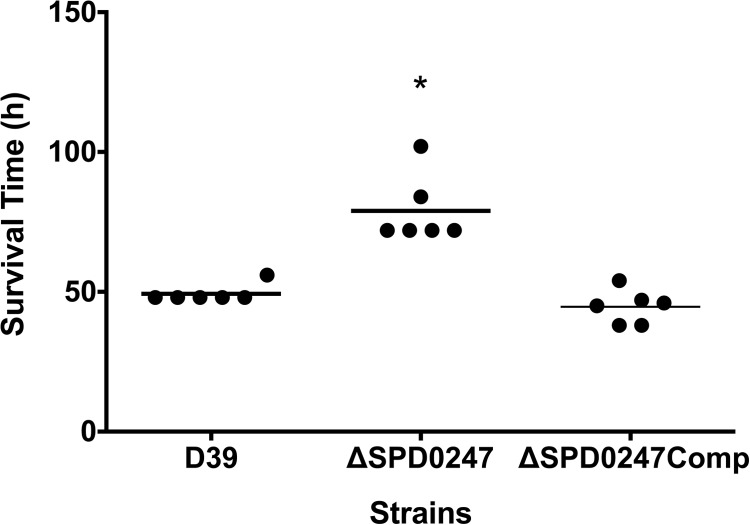
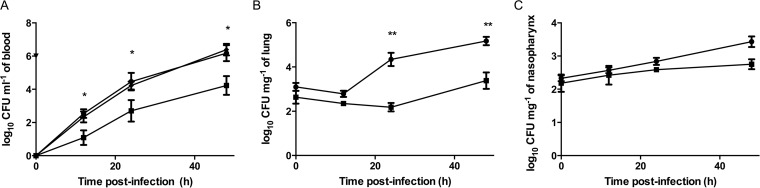
The contribution of BglA3 to pneumococcal virulence was investigated. The median survival time of mice infected intranasally with ΔSPD0247 (79 h ± 12 h; n = 6) was significantly longer than those for the cohorts infected with D39 (49 h ± 3 h; n = 6) and ΔSPD0247Comp (45.5 h ± 8 h; n = 6) (P < 0.01) (Fig. 3). To determine the contribution of BglA3 to pneumococcal growth in different tissues, the lungs, nasopharynx, and blood were collected from groups of mice at predetermined time points after intranasal infection (Fig. 4A, B, and C). There was a significant difference in colony counts in the blood for wild-type D39 and the ΔSPD0247 mutant at 12 h [log10(2.54 ± 0.26) (n = 20) and log10(1.09 ± 0.44) (n = 10), respectively], 24 h [log10(4.46 ± 0.53) (n = 20) and log10(2.70 ± 0.65) (n = 10), respectively], and 48 h postinfection [log10(6.18 ± 0.48) (n = 20) and log10(4.23 ± 1.1) (n = 10), respectively] (P < 0.05). Moreover, the colony counts of ΔSPD0247Comp at 12, 24, and 48 h postinfection [log10(2.34 ± 0.33), log10(4.23 ± 0.24), and log10(6.39 ± 0.35) (n = 10), respectively] were significantly higher than those of the mutant, ruling out the polar effect of the mutation. In the lungs at 24 h [log10(2.18 ± 0.19); n = 10] and 48 h [log10(3.38 ± 0.37); n = 10] postinfection, the colony counts of the mutant were significantly lower than those of the wild type [log10(4.34 ± 0.3) and log10(5.17 ± 0.19) (n = 20) for 24 h and 48 h, respectively] (P < 0.001), but there was no significant difference in the lungs at 12 h postinfection (P > 0.05) (Fig. 4B). ΔSPD0247 colonized the nasopharynx as well as the wild type, with no significant differences throughout the experiment (Fig. 4C) (P > 0.05).
FIG 3.
Survival of mice infected intranasally with 5 × 105 CFU/ml S. pneumoniae D39, isogenic mutant strain ΔSPD0247, or ΔSPD0247Comp. For each group, 6 mice were used. Each dot represents the survival time of an individual mouse. The horizontal bar indicates the median survival time. *, P < 0.05 (significant difference between the mutant and the D39 and ΔSPD0247Comp strains).
FIG 4.
Number of pneumococci recovered from blood (A), the lungs (B), and nasopharynx (C) after intranasal infection. Note that ΔSPD0247Comp was tested only in the blood. Each mouse received approximately 5 × 105 CFU/ml. For each time point, 10 mice for ΔSPD0247 (■) and ΔSPD0247Comp (◆) and 15 to 20 mice for D39 (●) were used. Vertical bars indicate the standard errors of the means. *, P < 0.05; **, P < 0.01 (significant difference between the ΔSPD0247 mutant and D39).
Gene expression analysis.
SPD_0247 mRNA levels were measured in D39 cells recovered from infected mouse tissues (lungs, blood, and nasopharynx). Compared to growth in Sicard's defined medium supplemented with glucose, the expression of SPD_0247 was greater in the nasopharynx (8.3- ± 0.4-fold; n = 3) and the lungs (17- ± 0.7-fold; n = 3). There was no statistically significant difference in the expression of SPD_0247 in blood compared to the expression in vitro.
Contribution of BglA3 to biofilm formation.
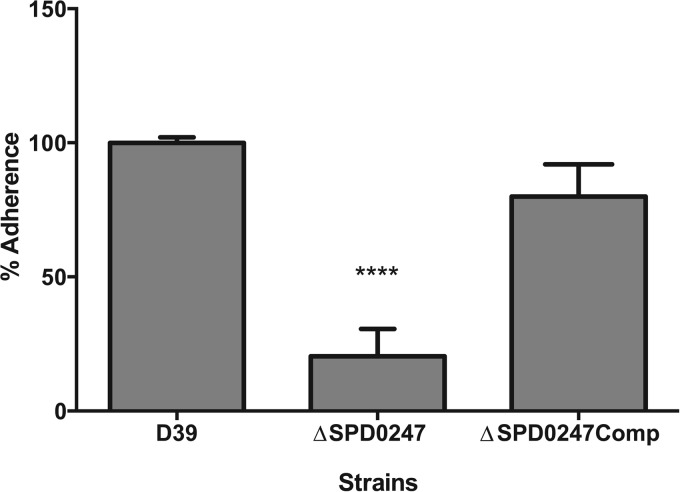
The biofilm-forming ability of the pneumococcal strains was assessed because it had been demonstrated that genes associated with β-glucoside metabolism may regulate adhesion and biofilm formation in Streptococcus gordonii (7). The adherence capability of mutant ΔSPD0247 also was assessed by comparing the proportion of adherent bacteria to those of nonadherent cells. It was observed that the mutant had a significantly decreased ability to adhere to flat-bottom microtiter plates (20% ± 10%; n = 5) compared to the wild type (100% ± 2%; n = 5) (P < 0.05) (Fig. 5). Genetic complementation of the mutant restored its biofilm-forming ability.
FIG 5.
Biofilm formation of wild-type D39, ΔSPD0247, and ΔSPD0247Comp. The percentage of cells adhered to microtiter plates was calculated relative to that of the wild type after normalizing the adherent cells to the nonadherent planktonic cells for each strain (n = 5). ****, P < 0.0001 (significant difference between the mutant and the D39 and ΔSPD0247Comp strains).
DISCUSSION
The pneumococcus relies on carbohydrates for the generation of energy. The microbe encounters diverse host environments in which the sugar content varies (4). Thus, mechanisms allowing flexibility in the use of different sugar substrates provide a survival advantage to the pneumococcus. Indeed, the pneumococcus is known to have the ability to ferment 32 different sugars (3), and a large proportion of genome-coding capacity is devoted to sugar transporters (2). However, there is an incomplete understanding of the mechanisms enabling pneumococcus to utilize host-derived sugars.
β-Glucoside analogs are plentiful in the respiratory tract, and their utilization requires 6-phospho-β-glucosidase activity (7). In this study, the enzymatic and functional role of BglA3 encoded by SPD_0247 was investigated. It was found that the presence of a phosphate group is a requirement for enzyme-substrate recognition, and that the enzyme plays a role in the metabolism of phosphorylated substrates. In this study, chromophoric substrates have been used. The kinetics and the reaction mechanisms obtained with the chromophoric substrates can be different from those of the natural substrates; however, synthetic substrates provide a simple, rapid, and precise assay for enzyme activity and are widely used in enzymology (32). It is not always possible to obtain natural substrates, and it is known that natural substrates can have enzyme inhibitory effects as a result of steric hindrance caused by the larger molecules of natural substrates, which complicates enzyme kinetic measurements. Certain differences can be seen depending on the source of substrate; therefore, caution must be taken when comparing enzyme kinetics obtained with different sources of substrate (32).
The activity of this enzyme is required for efficient utilization of hyaluronic acid and cellobiose, suggesting the importance of the enzyme in pneumococcal survival in the host. BglA3 also makes a significant contribution to invasive pneumococcal disease. The number of ΔSPD0247 organisms in the lungs was significantly lower than that of the wild type throughout the infection. It is very likely that this enzyme contributes to virulence in the lungs, and its contribution to virulence is due to its effect on nutrient metabolism. BglA3's contribution to virulence in the lungs also was supported by the fact that the expression of SPD_0247 went up significantly in wild-type bacteria recovered from the nasopharynx and the lungs compared to growth on glucose in vitro. Both the wild type and the mutant had disseminated into blood at 12 h postinfection. Although the number of ΔSPD0247 organisms in the blood was lower than that of the wild type throughout the infection, once in the blood, the mutant's growth rate was similar to that of the wild type. The difference in blood counts could have arisen due to impaired dissemination of the mutant pneumococci into the blood. Moreover, the role of BglA3 in the utilization of β-glucosides in blood cannot be ruled out (31).
The mutation of genes in β-glucoside uptake systems of S. mutans adversely affected in vitro adhesion and/or growth rate in liquid medium (33). Similar results were observed in this study. A recent study demonstrated that the mutation of components of the hyaluronic acid utilization locus, consisting of hyaluranate lyase, a PTS transporter for the hyaluronic acid disaccharide, and the unsaturated glucuronyl hydrolase, abrogated hyaluronic acid utilization and adversely effected pneumococcal colonization (13). BglA3 is an intracellular enzyme, and these enzymes usually are not often associated with biofilm formation. Furthermore, recombinant enzyme provided externally did not restore the adherence ability of ΔSPD0247, supporting the proposed intracellular role for BglA3 (data not shown). It may be speculated that the metabolic pathway in which this enzyme is playing a role has an effect on cell surface composition.
Although the BglA3-deficient strain is attenuated in growth on both cellobiose and hyaluronic acid, the attenuation on hyaluranic acid was more pronounced than that on cellobiose, indicating that BglA3 has a crucial role in hyaluranic acid metabolism. The preference of different 6-phospho-β-glucosidases for different substrates also has been shown in S. mutans. One of the three β-glucosidases present in S. mutans, i.e., amygdalin, gentiobiose, and salicin, was required for the fermentation of cellobiose, whereas the other two had no effect on the fermentation of these four β-glucosides (33), indicating that the substrate specificity of β-glucosidases varies.
A total abolishment of the growth of ΔSPD0247 on cellobiose or hyaluronic acid was not expected, as there are other loci responsible for β-glucoside metabolism. One of the best-characterized β-glucoside operons of S. pneumoniae is the SPD_0279-83 lactose-type PTS (TC_4.A.3) (34). This operon is responsible for the uptake of the β-glucosides cellobiose, gentiobiose, arbutin, amygdalin, and esculin (34). In addition, there are other loci implicated in β-glucoside utilization, such as SPD_0502-3 and SPD_1831-3 (4, 14), which contain a gene coding for β-glucosidase. Previously, it was shown that the strains mutated in 6-phospho-β-glucosidase genes, in SPD_0277, SPD_0502, or both, could grow as well as the wild-type strain on cellobiose, showing that it is possible to compensate for the lack of one or multiple 6-phospho-β-glucosidases.
SPD_0247 is associated with neither a regulatory nor a transporter gene. This observation leads to the possibility that BglA3 is processing sugars transported by other loci, such as celBCD as well as SPD_0293 and SPD_0295-7 loci, which are implicated in cellobiose and hyaluronic acid transport, respectively (3, 14, 35). BglA3's involvement in the cleavage of sugars transported by multiple loci offers an explanation for the in vivo attenuation of ΔSPD0247 despite the presence of multiple β-glucoside operons in S. pneumoniae.
In this study, BglA3 was studied in the D39 strain; therefore, we cannot generalize the data to other pneumococcal strains. However, it is plausible that this enzyme also plays an important role in the biology of other pneumococcal strains, because similar to other phosphoglycosyl hydrolases, SPD_0247 is part of a pneumococcal core genome (36). Pneumococcal fitness depends largely on its ability to acquire and utilize host glycoconjugates. Detailed knowledge of pneumococcal sugar metabolism may allow the identification of targets for new anti-infectives against this important pathogen (37). The acquisition of this knowledge is our future aim.
ACKNOWLEDGMENT
We are grateful to Jack Thompson, National Institutes of Health, Bethesda, MD, for kindly providing some of the substrates used in this study.
REFERENCES
- 1.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 2.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 3.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King SJ. 2010. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol 25:15–24. doi: 10.1111/j.2041-1014.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- 5.Philips BJ, Meguer JX, Redman J, Baker EH. 2003. Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intensive Care Med 29:2204–2210. doi: 10.1007/s00134-003-1961-2. [DOI] [PubMed] [Google Scholar]
- 6.King SJ, Hippe KR, Weiser JN. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol 59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 7.Kilic AO, Tao L, Zhang Y, Lei Y, Khammanivong A, Herzberg MC. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J Bacteriol 186:4246–4253. doi: 10.1128/JB.186.13.4246-4253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masuda H. 1984. The preparation of heparan sulfate from the mitral valve of the human heart. Int J Biochem 16:99–103. doi: 10.1016/0020-711X(84)90057-0. [DOI] [PubMed] [Google Scholar]
- 9.Pritchard DG, Lin B, Willingham TR, Baker JR. 1994. Characterization of the group B streptococcal hyaluronate lyase. Arch Biochem Biophys 315:431–437. doi: 10.1006/abbi.1994.1521. [DOI] [PubMed] [Google Scholar]
- 10.McKessar SJ, Hakenbeck R. 2007. The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J Bacteriol 189:1342–1350. doi: 10.1128/JB.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocaign-Bousquet M, Garrigues C, Loubiere P, Lindley ND. 1996. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Van Leeuwenhoek 70:253–267. doi: 10.1007/BF00395936. [DOI] [PubMed] [Google Scholar]
- 12.Gahan CG, Hill C. 2000. The use of listeriolysin to identify in vivo induced genes in the gram-positive intracellular pathogen Listeria monocytogenes. Mol Microbiol 36:498–507. doi: 10.1046/j.1365-2958.2000.01869.x. [DOI] [PubMed] [Google Scholar]
- 13.Marion C, Stewart JM, Tazi MF, Burnaugh AM, Linke CM, Woodiga SA, King SJ. 2012. Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect Immun 80:1390–1398. doi: 10.1128/IAI.05756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shafeeq S, Kloosterman TG, Kuipers OP. 2011. CelR-mediated activation of the cellobiose-utilization gene cluster in Streptococcus pneumoniae. Microbiology 157:2854–2861. doi: 10.1099/mic.0.051359-0. [DOI] [PubMed] [Google Scholar]
- 15.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu WL, Jiang YL, Pikis A, Cheng W, Bai XH, Ren YM, Thompson J, Zhou CZ, Chen Y. 2013. Structural insights into the substrate specificity of a 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4. J Biol Chem 288:14949–14958. doi: 10.1074/jbc.M113.454751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sicard AM. 1964. A new synthetic medium for Diplococcus pneumoniae, and its use for the study of reciprocal transformations at the amiA locus. Genetics 50:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terra VS, Homer KA, Rao SG, Andrew PW, Yesilkaya H. 2010. Characterization of novel beta-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect Immun 78:348–357. doi: 10.1128/IAI.00721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yesilkaya H, Spissu F, Carvalho SM, Terra VS, Homer KA, Benisty R, Porat N, Neves AR, Andrew PW. 2009. Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect Immun 77:5418–5427. doi: 10.1128/IAI.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampe DJ, Churchill ME, Robertson HM. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 21.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol 38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 22.Alloing G, Granadel C, Morrison DA, Claverys JP. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol 21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 23.Guiral S, Henard V, Laaberki MH, Granadel C, Prudhomme M, Martin B, Claverys JP. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152:343–349. doi: 10.1099/mic.0.28433-0. [DOI] [PubMed] [Google Scholar]
- 24.Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett 278:231–235. doi: 10.1111/j.1574-6968.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- 25.Hajaj B, Yesilkaya H, Benisty R, David M, Andrew PW, Porat N. 2012. Thiol peroxidase is an important component of Streptococcus pneumoniae in oxygenated environments. Infect Immun 80:4333–4343. doi: 10.1128/IAI.00126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta CT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Hans Lineweaver DB. 1934. The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666. doi: 10.1021/ja01318a036. [DOI] [Google Scholar]
- 28.Gaspar P, Al-Bayati FA, Andrew PW, Neves AR, Yesilkaya H. 2014. Lactate dehydrogenase is the key enzyme for pneumococcal pyruvate metabolism and pneumococcal survival in blood. Infect Immun 82:5099–5109. doi: 10.1128/IAI.02005-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yesilkaya H, Soma-Haddrick S, Crennell SJ, Andrew PW. 2006. Identification of amino acids essential for catalytic activity of pneumococcal neuraminidase A. Res Microbiol 157:569–574. doi: 10.1016/j.resmic.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 30.O'Sullivan L, Morgan SM, Ross RP, Hill C. 2002. Elevated enzyme release from lactococcal starter cultures on exposure to the lantibiotic lacticin 481, produced by Lactococcus lactis DPC5552. J Dairy Sci 85:2130–2140. doi: 10.3168/jds.S0022-0302(02)74291-4. [DOI] [PubMed] [Google Scholar]
- 31.Camara M, Boulnois GJ, Andrew PW, Mitchell TJ. 1994. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect Immun 62:3688–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kakade ML, Simons N, Liener IE. 1969. An evaluation of natural vs. synthetic substrates for measuring the antitryptic activity of soybean samples. Cereal Chem 46:8. [Google Scholar]
- 33.Old LA, Lowes S, Russell RR. 2006. Genomic variation in Streptococcus mutans: deletions affecting the multiple pathways of beta-glucoside metabolism. Oral Microbiol Immunol 21:21–27. doi: 10.1111/j.1399-302X.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 34.Boianelli A, Bidossi A, Gualdi L, Mulas L, Mocenni C, Pozzi G, Vicino A, Oggioni MR. 2012. A non-linear deterministic model for regulation of diauxic lag on cellobiose by the pneumococcal multidomain transcriptional regulator CelR. PLoS One 7:e47393. doi: 10.1371/journal.pone.0047393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marion C, Burnaugh AM, Woodiga SA, King SJ. 2011. Sialic acid transport contributes to pneumococcal colonization. Infect Immun 79:1262–1269. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tettelin H, Hollingshead SK. 2004. Comparative genomics of Streptococcus pneumoniae: intrastrain diversity and genome plasticity, p 15–29. In Tuomanen E, Mitchell T, Morrison D, Spratt B (ed), The pneumococcus. ASM, Washington, DC. [Google Scholar]
- 37.Buckwalter CM, King SJ. 2012. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol 20:517–522. doi: 10.1016/j.tim.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]