Abstract
The ability of the subgingival microbial community to induce an inappropriate inflammatory response ultimately results in the destruction of bone and gingival tissue. In this study, subgingival plaque samples from both healthy and diseased sites in the same individual were obtained from adults with chronic periodontitis and screened for their ability to either activate Toll-like receptor 2 (TLR2) or TLR4 and to antagonize TLR4-specific activation by agonist, Fusobacterium nucleatum LPS. Subgingival plaque from diseased sites strongly activated TLR4, whereas matched plaque samples obtained from healthy sites were significantly more variable, with some samples displaying strong TLR4 antagonism, while others were strong TLR4 agonists when combined with F. nucleatum LPS. Similar results were observed when TLR4 dependent E-selectin expression by endothelial cells was determined. These results are the first to demonstrate TLR4 antagonism from human plaque samples and demonstrate that healthy but not diseased sites display a wide variation in TLR4 agonist and antagonist behavior. The results have identified a novel characteristic of clinically healthy sites and warrant further study on the contribution of TLR4 antagonism in the progression of a healthy periodontal site to a diseased one.
INTRODUCTION
Periodontal disease is characterized by marked inflammation and destruction of bone and gingival tissue. Although the disease can be classified into different subtypes (1), bacterially induced periodontitis in adults is often a chronic inflammatory condition in which pathogenic plaque biofilm accumulates and adheres to the tooth surface above and below the gingiva. These supra- and subgingival plaque biofilms not only differ in location, but also in microbial composition and in relation to the development of periodontal diseases (2). Although suspected periodontal pathogens may be detected in supragingival plaque from diseased sites, the biofilm below the gingiva ultimately interacts with the periodontium and resides in a distinct environment, limited by space and host immune protection but enriched with nutrients from gingival crevicular fluid (3).
Consequently, the subgingival plaque biofilm also includes bacterial antigens, which directly engage the innate immune system at the site of infection. One of these antigens, lipopolysaccharide (LPS), is a well-characterized ligand specific to innate immune receptor, Toll-like receptor 4 (TLR4). LPS is located in the outer membrane of Gram-negative bacteria and structural differences can potentiate different activities on TLR4 signaling (4, 5). For example, Fusobacterium nucleatum LPS can potentiate a relatively strong TLR4 agonistic response due to its bisphosphorylated, hexaacylated lipid A moiety, the endotoxic portion of LPS which interacts directly with the TLR4 signaling complex (6). On the other hand, other periodontal bacteria, such as Porphyromonas gingivalis may modulate its LPS structural composition by removing phosphate residues and acyl chains on its lipid A backbone. These LPS structures antagonize TLR4 activation when mixed with strong agonist Escherichia coli LPS (7). Furthermore, the Gram-positive bacterial cell wall component, lipoteichoic acid, a known TLR2 activator, can also act as a TLR4 antagonist by interacting with coreceptor CD14 (8). Therefore, the subgingival oral microbial community has the potential to modulate TLR4 activity by the relative expression of TLR4 agonists and antagonists. In addition, the modulation of TLR4 activity is also dependent on the expression levels of TLR4 and MD-2 (9). Consequently, the potential for modulation of TLR4 activity as a component of periodontal homeostasis (10) exists both from the subgingival microbial community, as well as from the host as manifested in the expression levels of key TLR4 activation pathway components found in the local periodontal environment (11).
Therefore, in this study, TLR4 activation, as well as inhibition, was determined for subgingival plaque samples obtained from clinically healthy and diseased sites where both the microbial composition and expression of TLR4 pathway components are known to be altered (11). In addition, TLR2 activation was examined to determine if periodontal health status affected activation of this key inflammatory mediator. It was found, consistent with the inflammatory nature of periodontitis, that diseased plaque samples potently activated both TLR2 and TLR4 and that these activities were associated with increasing disease. These data demonstrate a strong proinflammatory state in response to a dysbiotic microbial community in disease. In contrast, plaque sampled from healthy sites exhibited both TLR4 activation and antagonism. TLR4 antagonism from human clinical samples is novel and suggests that TLR4 modulation may contribute to periodontal health homeostatic mechanisms.
MATERIALS AND METHODS
Study population.
Systemically healthy, untreated patients (9 males and 6 females; age range, 43 to 61 years) with generalized chronic periodontitis were recruited in this study while seeking dental treatment in the School of Dentistry, Ege University, İzmir, Turkey. The study was conducted in full accordance with ethical principles, including the World Medical Association's Declaration of Helsinki, as revised in 2000. The study protocol was explained, and written informed consent was received from each individual before clinical periodontal examinations and subgingival plaque sampling. Medical and dental histories were obtained and smoking habits were recorded. Individuals with medical disorders, such as diabetes mellitus and immunological disorders, those who had antibiotic or periodontal treatment in the last 6 months, and smokers were excluded from the study.
Individuals with chronic periodontitis were diagnosed in accordance with the clinical criteria stated in the consensus report of the World Workshop in Periodontitis (12). These individuals had ≥4 teeth in each jaw with a probing depth (PD) of ≥5 mm, clinical attachment level (CAL) of ≥4 mm, and ≥50% alveolar bone loss at least in two quadrants. Assessment of the extent and severity of alveolar bone loss was done radiographically. Bitewing radiographs were evaluated for interproximal bone loss from the cemento-enamel junction of the tooth to the bone crest. These individuals also had bleeding on probing (BOP) at >80% of the proximal sites.
Subgingival plaque sample collection.
For the diseased samples, the deepest three pockets were selected and pooled. Supragingival plaque was first removed from the sample teeth with sterilized Gracey curettes and gauze. The site was then cleaned and isolated using cotton roles and air dried gently. Another sterilized Gracey curette was inserted to the deepest part of the pocket and removed applying a slight force toward the root surface. The tip of the curette was then inserted in the microcentrifuge tube containing 0.5 ml of distilled water and shaken until the plaque was removed from the curette.
For the healthy subgingival plaque samples in the same patient, three healthy sites with a PD of <3 mm and no sign of inflammation and bleeding on probing were chosen and pooled in a single microcentrifuge tube containing 0.5 ml of distilled water. The samples were frozen and stored at −40°C until the sample collection period was completed.
Clinical periodontal measurements.
Subsequent to saliva and serum sampling, clinical periodontal recordings, including plaque index (PI), PD, CAL, and BOP (recorded as “+” or “−”) were performed at six sites (mesiobuccal, midbuccal, distobuccal, mesiolingual, midlingual, and distolingual locations) on each tooth present, except the third molars, using a Williams periodontal probe (Hu-Friedy, Chicago, IL). CAL was assessed from the cemento-enamel junction to the base of the probable pocket. BOP (deemed positive if it occurred within 15 s after periodontal probing) was recorded dichotomously by visual examination. All measurements were performed by two precalibrated examiners (P.G. and N.N.). Interexaminer and intraexaminer calibration was analyzed using the Kappa-Cohen test. The initial intraexaminer kappa values were 0.96 (PD) and 0.86 (CAL) for P.G. and 0.93 (PD) and 0.79 (CAL) for N.N. The interexaminer values were 0.92 (PD) and 0.75 (CAL).
Plaque sample processing.
Lyophilized plaque samples were weighed with an analytical microscale and then resuspended in 1× phosphate-buffered saline (PBS) to a concentration of 10 mg ml−1. Dilutions and aliquots were made, and all samples were kept at −80°C before thawing for use in assays and DNA extractions. Reported patient sample numbers were ordered according to TLR4-antagonistic activities for presentation purposes and relabeled independently of the assignment number at collection.
Bacterial growth.
Porphyromonas gingivalis ATCC 33277 and Fusobacterium nucleatum ATCC 25586 were grown in an anaerobic chamber at 37°C in the presence of 5% H2, 5% CO2, and 90% N2. Bacteria were grown on blood agar (Remel) and cultured for 3 to 4 days before inoculating Trypticase soy broth liquid cultures. P. gingivalis and F. nucleatum liquid medium was supplemented with 5 μg ml−1 menadione and 1 μg ml−1 hemin. For LPS extraction, P. gingivalis was grown in the presence of 10 μg ml−1 hemin to enrich for antagonistic LPS, which is designated Pg1435LPS.
LPS extraction.
Porphyromonas gingivalis and F. nucleatum bacterial cultures were grown to late logarithmic phase prior to harvesting. To isolate antagonistic LPS (Pg1435LPS), late-logarithmic growth P. gingivalis was treated using a modified Darveau-Hancock LPS extraction procedure (13), followed by a phenol-water repurification (14). The antagonistic activity of Pg1435LPS has been previously described to antagonize TLR4 activation by E. coli LPS (9, 15–17). F. nucleatum agonistic LPS (FnLPS), which bears similar TLR4 activity to E. coli LPS, was extracted using a modified trireagent protocol, as previously described (18, 19). Lipid A structure was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis, as previously reported (18).
Luciferase reporter assay.
The human embryonic kidney (HEK) 293 assays were performed as previously described (20). Briefly, cells were plated at a density of 4 × 104 cells per well and transfected 24 h later by calcium phosphate precipitation. Stimulations were performed, in triplicate wells, 20 to 24 h after transfections, followed by incubation for 4 h at 37°C and 5% CO2. For correlation studies, 1, 10, 100, and 1,000 μg ml−1 plaque samples were introduced directly to serum-free Dulbecco modified Eagle medium containing 10% fetal bovine serum as a source of soluble CD14. Synthetic lipopeptide Pam3CSK4 (Invivogen) was used as a positive control for TLR2 activation, extracted FnLPS was used as a positive control for TLR4 activation and extracted Pg1435LPS, a known TLR4 antagonist, was used to demonstrate low TLR2 and TLR4 activity. For antagonisms, 100 ng ml−1 FnLPS was mixed with one of following: healthy plaque samples, diseased plaque samples, Pg1435LPS, or PBS.
Prior to introducing plaque and/or LPS, cells were transfected with the following amounts of plasmid DNA per well: pβ-actin Renilla Luc (0.0004 μg), pNF-κB-TA-Luc (0.02 μg), pMD-2 (0.0025 μg), and pTLR4 (0.002 μg) or pTLR2 (0.002 μg) for activation studies or pTLR4SV1 (0.006 μg) for antagonism studies. The expression of TLR4 splice variant, SV1, was previously shown to enhance antagonism by Pg1435LPS (9). Empty expression vector, pDisplay was used to adjust total amount of DNA per well to 0.1 μg. After 4 h of stimulation, the cells were rinsed with PBS and lysed with passive lysis buffer (Promega, Madison, WI). The luciferase activity of each lysate was measured by using a dual luciferase assay reporter system (Promega). For correlation studies with clinical measurements, data are expressed as NF-κB activation, which represents the ratio of NF-κB-dependent firefly luciferase activation to β-actin promoter-dependent Renilla luciferase activation. Antagonism data are presented as the percent stimulation above that for F. nucleatum LPS alone.
E-selectin expression assay.
Human umbilical vein endothelial cells (Lonza, Rockville, MD) were plated at a density of 1.4 × 104 cells per well in a gelatin-coated 96-well plate in the presence of M199 growth medium (Life Technologies, Carlsbad, CA) supplemented with 4 mM l-glutamine, 90 μg ml−1 heparin, 1 mM sodium pyruvate, 30 μg ml−1 endothelial cell growth supplement (BD Biosciences, Franklin Lakes, NJ), and 20% fetal bovine serum (HyClone). As with the HEK293 assays 100 ng ml−1 F. nucleatum LPS was premixed mixed with one of following: healthy plaque samples, diseased plaque samples, Pg1435LPS, or PBS. Mixtures were then introduced to the cell monolayer in triplicate wells in the presence of stimulation medium supplemented with 10% human serum. After 4 h of incubation, the cells were washed with PBS, fixed in 0.5% glutaraldehyde for 10 min, and then blocked in PBS containing 3% goat serum. E-selectin expression was detected by a previously described enzyme-linked immunosorbent assay (ELISA) protocol (7). Antagonism data are presented as the percent stimulation above that for FnLPS activation alone.
Genomic DNA isolation.
Overnight bacterial cultures and 200 μg of plaque samples were pelleted in screw-cap microcentrifuge tubes. The following was added to each pellet: 0.4 g of 0.1-mm zirconium/silica beads (Biospec, Bartlesville, OK), 10% sodium dodecyl sulfate (Sigma-Aldrich, St. Louis, MO), 1× TE buffer (Sigma-Aldrich), and TE buffer-saturated phenol (pH 8.0; Sigma-Aldrich). Cells were disrupted in a FastPrep-24 tissue homogenizer (MP Biomedicals, Santa Ana, CA) for 50 s at a speed of 5.0 m/s and then spun at top speed to separate phases. Upper aqueous phases were subject to a series of chloroform-isoamyl alcohol and phenol phase extractions. DNA was precipitated with isopropanol and 3 M sodium acetate (Sigma-Aldrich) and then pelleted. Pellets were washed with ethanol and allowed to air dry. All DNA samples were dissolved in 1× TE buffer. Concentrations of double-stranded DNA were determined by using a Quant-iT PicoGreen assay kit (Life Technologies) and used to create standard curves for each target.
Quantitation of total bacterial load.
Absolute quantitation using quantitative PCR was performed on a LightCycler 480 (Roche Applied Science, Indianapolis, IN) targeting 16S rRNA. Portions (2 μl) of genomic DNA from plaque or controls were added to 5 μl of TaqMan Fast Advanced master mix (Life Technologies), 400 nM concentrations (each) of forward primer (5′-CGCTAGTAATCGTGGATCAGAATG-3′) and reverse primer (5′-TGTGACGGGCGGTGTGTA-3′), and 200 nM TaqMan probe (5′-FAM-CACGGTGAATACGTTCCCGGGC-TAMRA-3′) (Life Technologies). These primers and probes specifically target the 16S rRNA gene (21). Nuclease-free water was added to bring the total volume of the reaction mixture to 10 μl. Real-time PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, and then either 58 or 60°C for 1 min. The number of bacteria was determined with LightCycler 480 software using the Second Derivative Maximum method. Serial dilutions of Prevotella intermedia ATCC 25611 genomic DNA were used for internal standard curves, which were of high quality, with efficiencies of 1.8 to 2.0 and errors of <0.20.
Statistical analysis.
To examine percent antagonism or activation of plaque samples against FnLPS in both HEK293 cell and human umbilical vein endothelial cell (HUVEC) assays, one-way analysis of variance with Tukey-Kramer post hoc test was performed using GraphPad Prism v6.03 (GraphPad Software, San Diego, CA) statistical software package. All other tests are indicated in figure text. A P value below 0.05 was considered significant.
RESULTS
Characterization of healthy and diseased sites in chronic periodontitis patients.
Subgingival plaque was sampled from healthy and diseased sites in 15 chronic periodontitis patients and specific clinical measurements were taken for all sites sampled. The mean measurements of PD, CAL, and PI in diseased sites were significantly greater than matched healthy sites (Table 1). In addition, all healthy sites sampled demonstrated no bleeding on probing.
TABLE 1.
Summary of clinical characteristics of matched healthy and diseased sites in subjects
| Parameter | Mean ± SDa |
|
|---|---|---|
| Healthy sites (n = 15) | Diseased sites (n = 15) | |
| Probing depth (mm) | 2.2 ± 0.4 | 5.9 ± 1.8** |
| Clinical attachment level | 3.7 ± 0.9 | 7.5 ± 1.2** |
| Plaque index | 1.4 ± 0.5 | 2.8 ± 0.4* |
| Bleeding-on-probing status | − | + |
All values represent means and standard deviations for all measured sites in these subjects, except for the “bleeding on probing status.” Individual means were used for paired t test calculations (*, P < 0.05; **, P < 0.01).
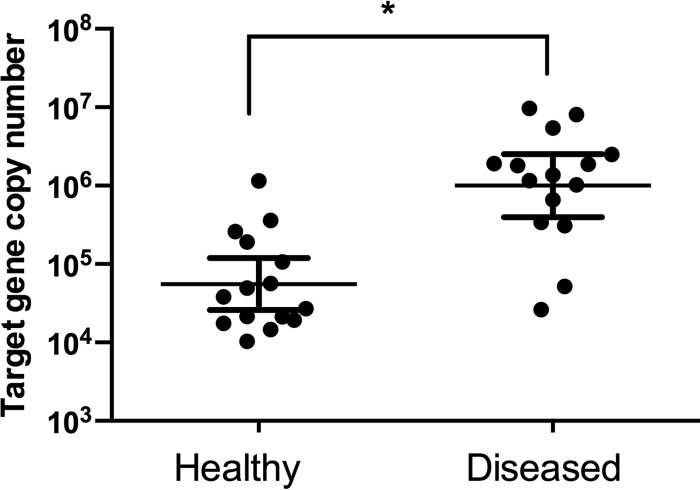
To determine the numbers of total bacteria, quantitative PCR was performed on genomic DNA preparations on equivalent dry weights of plaque samples. Bacterial load was significantly greater in diseased plaque with approximately 106 mean copy numbers, whereas healthy plaque made up >10-fold less bacteria (Fig. 1).
FIG 1.
Bacterial load of matched healthy and diseased plaque samples. The copy number of 16S rRNA genes represent the total bacterial load for each sample. Differences were tested using a Wilcoxon matched-pair signed-rank test to determine the significance (*, P < 0.05). The results shown are means ± the standard deviations (SD) of three independent assays.
Plaque activation of TLR2 and TLR4.
To quantify the ability of matched subgingival plaque samples to activate either TLR4 or TLR2 independent of corresponding clinical measurements, we measured the NF-κB activity elicited by each sample and compared it to different clinical measurements specific to that sample. Since HEK293 cells can be transfected with TLRs of choice, including TLR4 and TLR2, we can determine pathway specificity using a highly sensitive and pathway-specific reporter assay. HEK203 cells transfected with TLR4 or TLR2 were treated with 1, 10, 100, and 1,000 μg of subgingival plaque sample ml−1 and then the NF-κB activity was measured. Both TLR2 and TLR4 activities displayed a dose response, with significant decreases at 1 μg ml−1 (data not shown). The highest dose of plaque at 100 μg ml−1 was chosen for further association studies with clinical measurements since the activities were in the linear range of the assay which permitted direct comparison among the samples.
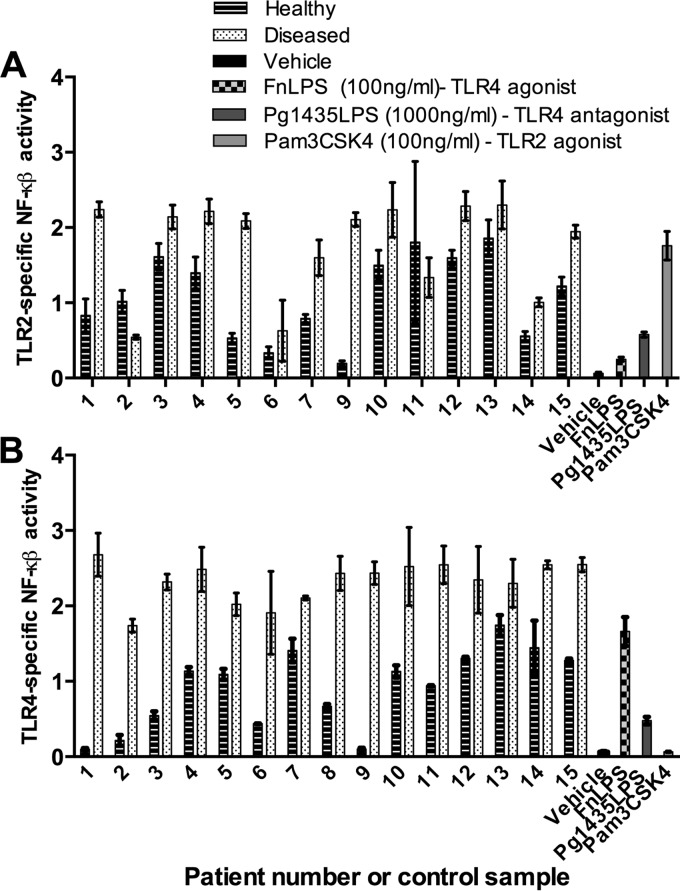
Plaque samples from diseased sites potently activated at both TLR4 and TLR2 whereas matched plaque from healthy sites activated through either TLR significantly less and with more variability at a dose of 100 μg ml−1 (Fig. 2). When we analyzed specifically the TLR4 activation of each sample, the general trend demonstrated a significant difference between healthy sites and matched diseased sites in the same patients. These measurements established a baseline of activity to further investigate possible inhibition or antagonism of TLR4 activation and also to elucidate possible relationships with clinical measurements of disease.
FIG 2.
TLR2 (A) and TLR4 (B) activation by 100 μg ml−1 subgingival plaque from matched healthy or diseased sites in chronic periodontitis patients. The TLR4- or TLR2-specific NF-κB activity was determined by calculating the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity (internal control). The results shown are means ± the standard deviations of three independent assays.
Association of clinical measurements of disease to TLR2 and TLR4 activation.
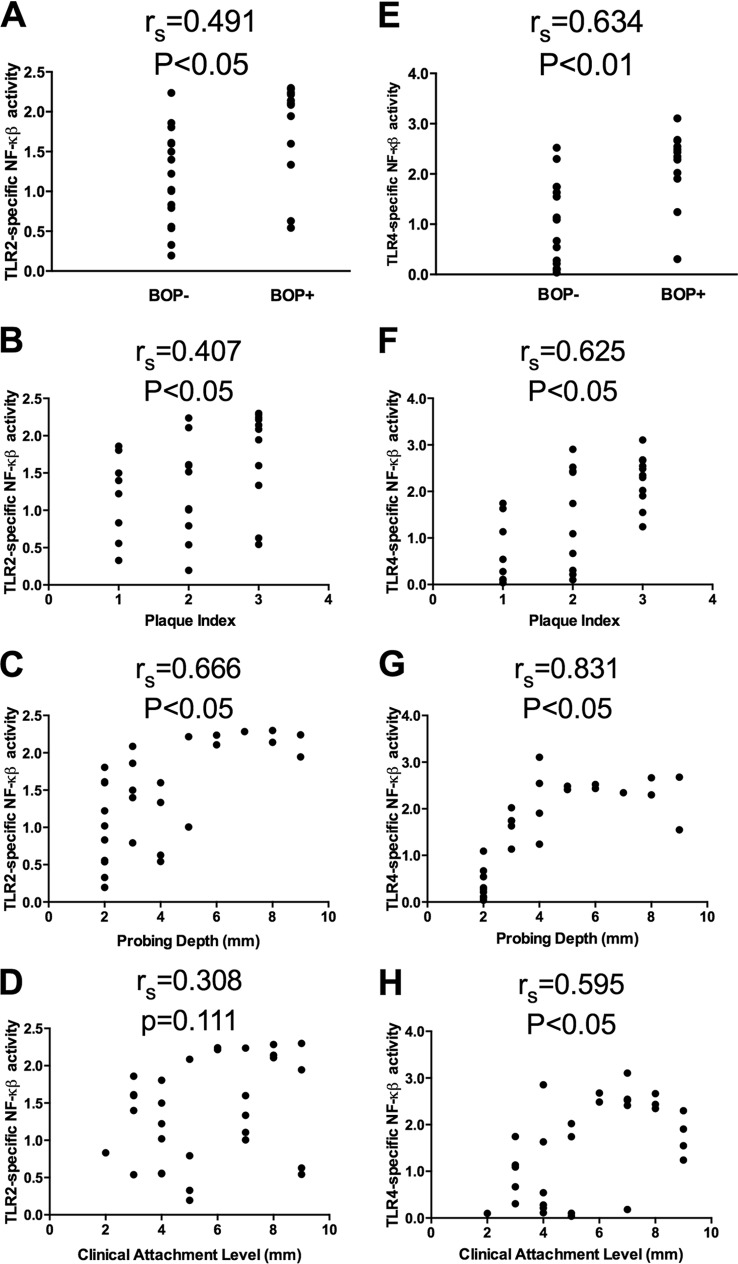
Since previous work has demonstrated a relationship of certain clinical measurements of supragingival plaque to its ability to stimulate TLR4 and TLR2 (17), we likewise determined the associations, if any, of clinical measurements to the ability of our subgingival plaque samples to activate these TLRs. These correlations determined by Spearman rank are represented as rs values (Fig. 3). The rs values were interpreted to represent high positive correlation with values of 0.7 to 1.0 and to have negligible correlation with values between 0.0 to 0.3 (22). TLR2-specific activation had a low to moderate association with the BOP status (Fig. 3A), PI (Fig. 3B), and PD (Fig. 3C). There was no significant association of TLR2 activation with CAL (Fig. 3D). In contrast, TLR4-specific activation had a moderate to high association with all of the clinical measurements examined (Fig. 3E to H), with rs values ranging from 0.595 (CAL) to 0.831 (PD) and P values of <0.05. These data demonstrate that both TLR2 and TLR4 activity increases in clinically diseased sites and that the increase in TLR4 activity demonstrated a stronger correlation to disease measurements.
FIG 3.
Relationships between TLR2- or TLR4-specific activation and clinical measurements of chronic periodontitis. TLR2 (A to D) and TLR4 (E to H) activation is represented by the NF-κB activity, and associations were determined with the corresponding clinical measurements: BOP status (A and E), plaque index (B and F), probing depth (C and G), and clinical attachment levels (D and H). The correlation was determined by calculating using the Spearman rank (rs) to determine the significance (P < 0.05).
Plaque antagonism of TLR4 activation.
Next, the ability of these matched plaque samples to antagonize TLR4 activation was determined. It was hypothesized that a mixed microbial plaque sample could potentially inhibit TLR4 activation due to either the presence of antagonistic LPS (7) or lipoteichoic acid (8) in the subgingival microbial community.
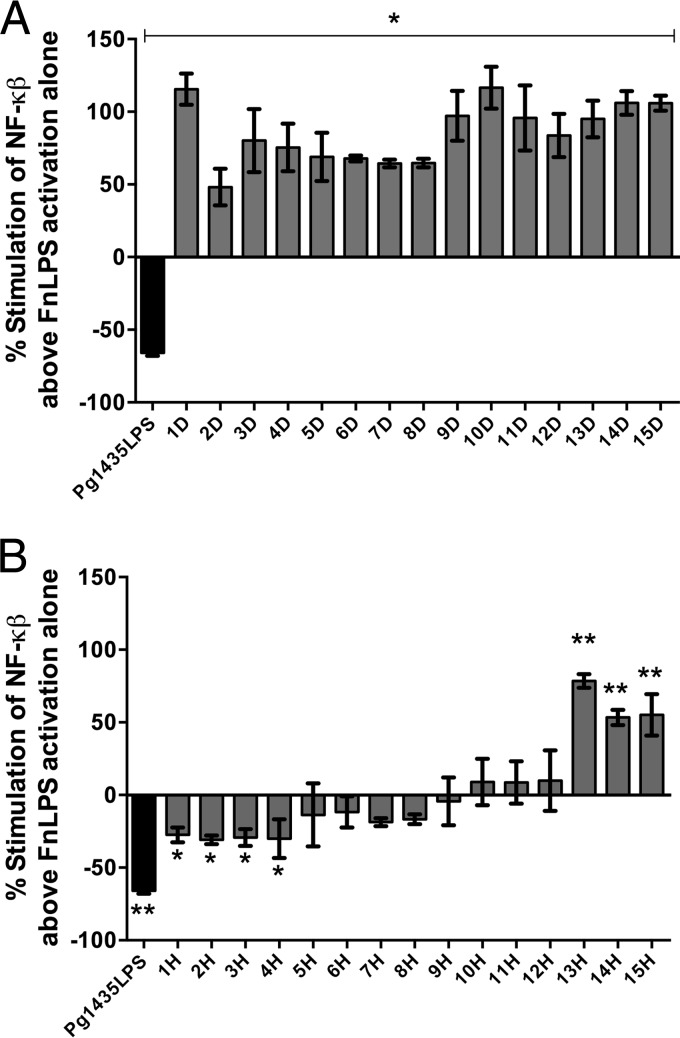
In these experiments the matched plaque samples obtained from either clinically healthy or diseased sites were combined with 100 μg ml−1 F. nucleatum LPS, in order to determine whether the plaque sample could reduce TLR4 activation in response to this TLR4 agonist. The percent stimulation above or below F. nucleatum LPS activation alone was determined for each of the patient plaque samples. P. gingivalis LPS was added as a TLR4 antagonist control. None of the subgingival samples obtained from diseased sites displayed TLR4 antagonistic activity and, in fact, the samples significantly activated TLR4 above that observed with F. nucleatum LPS alone (Fig. 4A). In contrast, plaque samples taken at healthy sites displayed a wide range of responses from significant antagonism to significant activation (Fig. 4B).
FIG 4.
TLR4 antagonistic or stimulatory potential of matched diseased (A) and healthy (B) subgingival plaque. Each patient was designated a number and “D” or “H” label corresponds to diseased or healthy plaque, respectively. The percent stimulation of NF-κB above that of F. nucleatum LPS (FnLPS) activation alone was calculated by using the formula [(y − x)/x] × 100, where x is the NF-κB activity due to TLR4 stimulation with 100 ng/ml FnLPS alone, and y is activity of FnLPS combined with either plaque at 100 μg ml−1 or Pg1435LPS control at 1 μg ml−1. The results shown are means ± the standard deviations of triplicate wells determined in one of three independent assays. *, P < 0.05; **, P < 0.01.
Plaque inhibition of E-selectin expression on HUVECs.
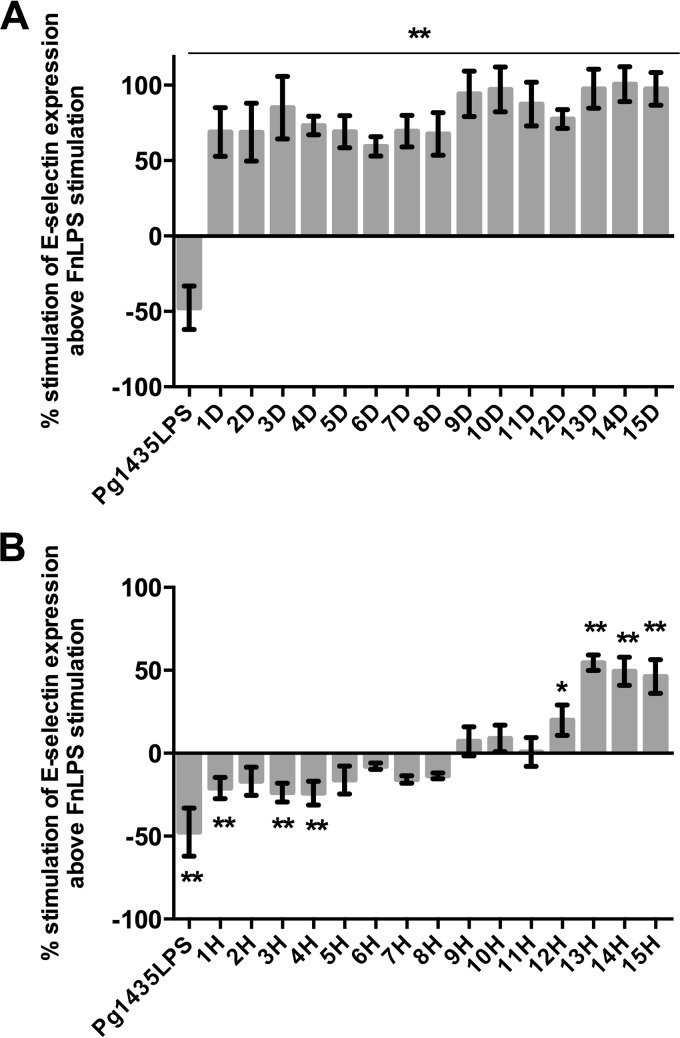
To determine whether TLR4-dependent antagonism can occur in primary human cells where TLR4/MD-2 complex is occurring at naturally expressing levels, we measured the presence of E-selectin by endothelial cells stimulated with mixtures of the plaque and F. nucleatum LPS agonist. A similar pattern of TLR4 activation and antagonism was observed for each of the plaque samples demonstrating that TLR4 antagonism in clinically healthy samples occurs at levels of TLR4 and MD-2 found in primary human cells (Fig. 5). More specifically, plaque from diseased sites all potently stimulate E-selectin expression, whereas plaque from healthy sites displayed a wide variation of E-selectin expression levels from antagonism to potent stimulation above FnLPS activation alone.
FIG 5.
Antagonism and stimulation of E-selectin expression of endothelial cells by matched diseased (A) and healthy (B) subgingival plaque. Each patient was designated a number and “D” or “H” label corresponds to diseased or healthy plaque, respectively. The percent stimulation of E-selectin expression above that of FnLPS activation alone was calculated as described for Fig. 4. The results shown are means ± the standard deviations of triplicate wells determined in one of three independent assays. *, P < 0.05; **, P < 0.01.
DISCUSSION
This study found that subgingival plaque samples from diseased sites demonstrated a significant increase in both TLR2 and TLR4 activation. The increase in both TLR2 and TLR4 activation in clinically diseased sites and the significant correlations with an increase in TLR4 activity are consistent with the demonstrated increase in microbial load and the characteristic shift to Gram-negative bacteria described for diseased sites (23–25). As the number of bacteria increase with disease, so do the number of TLR2 and TLR4-stimulatory ligands. The slightly stronger linear association of TLR4 stimulation to clinical measurements may be partly due to the stringency of ligand recognition in lipid A from only Gram-negative bacteria, whose relative abundance significantly increases in the dysbiotic community associated with periodontitis. In addition, all plaque samples from diseased sites also induced increased the TLR4-dependent expression of E-selectin in endothelial cells. This strong activation of E-selectin in disease is corroborated by a body of work demonstrating increase detection of this leukocyte extravasation molecule in inflamed tissues of chronic periodontitis patients (26). The clinical associations with clinical measurements described here agree with a previous study investigating supragingival plaque stimulation of TLR4 and TLR2 (27). However, since the composition of the subgingival microbial community examined here is in close juxtaposition to periodontal tissue and has been found to be clinically related to the presence of periodontitis, the findings of increased TLR2 and TLR4 activation in plaque sample from diseased sites directly validates the notion that an increase in the bacterial load will result in an increased inflammatory load.
In contrast to the increased TLR4 activation observed in periodontitis-affected sites, healthy sites from the same individual displayed diverse TLR4 activities, including both potent agonist and antagonist activities. It is unknown what contributes to the wide variations of TLR4 and E-selectin antagonism and stimulation in these healthy sites. It is possible that healthy plaque which demonstrate potent TLR4 and E-selectin-stimulatory behavior (Fig. 4b and 5b) may represent a microbial community or environment shifting toward disease on a molecular level but not yet manifested as such by clinical definitions. Conversely, this potent stimulation by healthy plaque, but lack of disease in the periodontal pocket, may instead represent immune homeostasis between the microbial community and a healthy and active host site. Nevertheless, this curious observation of TLR4 and E-selectin stimulation by healthy plaque warrants further characterization.
In addition to this potent stimulation, both HEK-TLR4 reporter and E-selectin ELISAs demonstrated TLR4 antagonism by other healthy plaque. Likewise, it is not clear whether one or multiple factors contribute to the TLR4 antagonism observed in subgingival plaque samples obtained from clinically healthy sites. For example, within the microbial community, P. gingivalis produces a potent TLR4 antagonistic lipid A structure that is environmentally regulated (28) and lipoteichoic acid produced by Gram-positive bacteria maybe present in sufficient quantities to contribute to the antagonism observed (8). Likewise, host components that differ in relative abundance when clinically healthy and diseased sites are compared may contribute to TLR4 antagonism. For instance, lipopolysaccharide binding protein which is expressed locally in the periodontal tissue and CD14 expression has been shown to be interrelated in periodontal tissue with significantly increased expression in clinically healthy sites (11). These key components of the TLR4 activation pathway, as well as the levels of TLR4 and MD-2, may all modulate TLR4 activation.
Importantly, the stark difference in TLR4 activation observed between clinically healthy and diseased sites emphasizes the need to further define health in addition to disease in the periodontium and the underlying mechanisms which maintain or dysregulate the homeostasis with its resident microbial community. The data suggest that TLR4 antagonism may be a normal component of healthy homeostasis, specifically antagonism may protect a healthy site from progression to disease by modulating inflammatory mediator expression. For example, the subgingival microbial community in cooperation with locally expressed host components may regulate E-selectin expression whose expression is known to be significantly increased in diseased sites function (29, 30). E-selectin, as a key mediator of neutrophil diapedesis, may be downregulated in clinically healthy sites to maintain the appropriate number of neutrophils to enter the gingival crevice to perform their immune-inflammatory surveillance. Furthermore, the role of TLR2 and the possible dampening of this pathway by dental plaque is yet to be characterized or observed. Although outside the scope of this work, deeper characterization of TLR2 activation mechanisms by dental plaque is important for understanding the dynamic between the host and the adjacent polymicrobial community.
Examination of the inflammatory activity of clinically healthy sites as performed in this study reveals the need to more fully understand periodontal inflammatory surveillance mechanisms. Examination of diseased sites is important to study with respect to understanding mechanisms underlying the relationship between a dysbiotic periodontal community and disease. However, subtle differences in the relationship between the subgingival microbial community and the host such as antagonism in healthy sites may be obscured in the presence of overwhelming proinflammatory activities. Understanding how healthy homeostasis is maintained may lead to more effective intervention strategies for the treatment of periodontitis.
ACKNOWLEDGMENTS
This study was funded by National Institute of Dental and Craniofacial Research grants awarded to the University of Washington School of Dentistry and T.T.T. (T90DE21984) and awarded to R.P.D. (R01DE012768).
REFERENCES
- 1.Armitage GC. 1996. Periodontal diseases: diagnosis. Ann Periodontol 1:37–215. doi: 10.1902/annals.1996.1.1.37. [DOI] [PubMed] [Google Scholar]
- 2.Ximénez-Fyvie LA, Haffajee AD, Socransky SS. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol 27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- 3.Darveau RP, Tanner A, Page RC. 1997. The microbial challenge in periodontitis. Periodontol 2000 14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 4.Berezow AB, Ernst RK, Coats SR, Braham PH, Karimi-Naser LM, Darveau RP. 2009. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb Pathog 47:68–77. doi: 10.1016/j.micpath.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid a phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun 79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lappin DF, Sherrabeh S, Erridge C. 2011. Stimulants of Toll-like receptors 2 and 4 are elevated in saliva of periodontitis patients compared with healthy subjects. J Clin Periodontol 38:318–325. doi: 10.1111/j.1600-051X.2011.01702.x. [DOI] [PubMed] [Google Scholar]
- 7.Coats SR, Reife RA, Bainbridge BW, Pham T-TT, Darveau RP. 2003. Porphyromonas gingivalis Lipopolysaccharide Antagonizes Escherichia coli Lipopolysaccharide at Toll-Like Receptor 4 in Human Endothelial Cells. Infect Immun 71:6799–6807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara S, Arakaki R, Rikiishi H, Takada H. 1999. Lipoteichoic acid acts as an antagonist and an agonist of lipopolysaccharide on human gingival fibroblasts and monocytes in a CD14-dependent manner. Infect Immun 67:1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol 175:4490–4498. doi: 10.4049/jimmunol.175.7.4490. [DOI] [PubMed] [Google Scholar]
- 10.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 11.Ren L, Leung WK, Darveau RP, Jin L. 2005. The expression profile of lipopolysaccharide-binding protein, membrane-bound CD14, and Toll-like receptors 2 and 4 in chronic periodontitis. J Periodontol 76:1950–1959. doi: 10.1902/jop.2005.76.11.1950. [DOI] [PubMed] [Google Scholar]
- 12.Armitage GC. 1999. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Darveau RP, Hancock RE. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol 155:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manthey CL, Vogel SN. 1994. Elimination of trace endotoxin protein from rough chemotype LPS. J Endotoxin Res 1:84–91. [Google Scholar]
- 15.Coats SR, Do CT, Karimi-Naser LM, Braham PH, Darveau RP. 2007. Antagonistic lipopolysaccharides block Escherichia coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol 9:1191–1202. doi: 10.1111/j.1462-5822.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- 16.Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, Howald WN, Darveau RP. 2006. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol 8:857–868. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 17.Herath TDK, Darveau RP, Seneviratne CJ, Wang C-Y, Wang Y, Jin L. 2013. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-κB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One 8:e58496. doi: 10.1371/journal.pone.0058496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to Porphyromonas gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol 11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from Gram-negative bacteria. Analyst 125. [DOI] [PubMed] [Google Scholar]
- 20.Jain S, Coats SR, Chang AM, Darveau RP. 2013. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect Immun 81:1277–1286. doi: 10.1128/IAI.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida A, Suzuki N, Nakano Y, Oho T, Kawada M, Koga T. 2003. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol 41:863–866. doi: 10.1128/JCM.41.2.863-866.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinkle DE, Wiersma W, Jurs SG. 2003. Applied statistics for the behavioral sciences. Houghton Mifflin, New York, NY. [Google Scholar]
- 23.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Socransky SS, Haffajee AD, Dzink JL. 1988. Relationship of subgingival microbial complexes to clinical features at the sampled sites. J Clin Periodontol 15:440–444. doi: 10.1111/j.1600-051X.1988.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 25.Marsh PD. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 26.Rezavandi K, Palmer RM, Odell EW, Scott DA, Wilson RF. 2002. Expression of ICAM-1 and E-selectin in gingival tissues of smokers and non-smokers with periodontitis. J Oral Pathol Med 31:59–64. doi: 10.1046/j.0904-2512.2001.joptest.doc.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoshioka H, Yoshimura A, Kaneko T, Golenbock DT, Hara Y. 2008. Analysis of the activity to induce Toll-like receptor (TLR)2- and TLR4-mediated stimulation of supragingival plaque. J Periodontol 79:920–928. doi: 10.1902/jop.2008.070516. [DOI] [PubMed] [Google Scholar]
- 28.Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. 2006. Hemin-dependent modulation of the lipid a structure of Porphyromonas gingivalis lipopolysaccharide. Infect Immun 74:4474–4485. doi: 10.1128/IAI.01924-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonetti MS, Imboden MA, Lang NP. 1998. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol 69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- 30.Gemmell E, Walsh LJ, Savage NW, Seymour GJ. 1994. Adhesion molecule expression in chronic inflammatory periodontal disease tissue. J Periodontal Res 29:46–53. doi: 10.1111/j.1600-0765.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]