Abstract
Salmonella enterica serovar Typhimurium is a facultative intracellular human and animal bacterial pathogen posing a major threat to public health worldwide. Salmonella pathogenicity requires complex coordination of multiple physiological and virulence pathways. DksA is a conserved Gram-negative regulator that belongs to a distinct group of transcription factors that bind directly to the RNA polymerase secondary channel, potentiating the effect of the signaling molecule ppGpp during a stringent response. Here, we established that in S. Typhimurium, dksA is induced during the logarithmic phase and DksA is essential for growth in minimal defined medium and plays an important role in motility and biofilm formation. Furthermore, we determined that DksA positively regulates the Salmonella pathogenicity island 1 and motility-chemotaxis genes and is necessary for S. Typhimurium invasion of human epithelial cells and uptake by macrophages. In contrast, DksA was found to be dispensable for S. Typhimurium host cell adhesion. Finally, using the colitis mouse model, we found that dksA is spatially induced at the midcecum during the early stage of the infection and required for gastrointestinal colonization and systemic infection in vivo. Taken together, these data indicate that the ancestral stringent response regulator DksA coordinates various physiological and virulence S. Typhimurium programs and therefore is a key virulence regulator of Salmonella.
INTRODUCTION
Salmonella enterica is a facultative intracellular human and animal bacterial pathogen responsible for global pandemics of food-borne infections that pose a major threat to public health. This highly versatile pathogen can infect a broad range of hosts and causes different clinical outcomes, ranging from asymptomatic carriage to systemic life-threatening disease (1). The single species S. enterica includes more than 2,600 serovars that share high sequence similarity and are taxonomically classified into six subspecies (2). Estimations suggest that Salmonella causes 93.8 million human infections and 155,000 deaths annually worldwide (3). The majority of nontyphoidal Salmonella (NTS) infections in humans present as gastroenteritis; however, about 5% may be invasive and manifest as bacteremia or other extraintestinal infections (4). Many of the NTS serovars are capable of colonizing the intestines of livestock with potential risk of contaminating the food chain, and therefore, salmonellosis is often associated with animal products and produce (5). One of the most common serovars worldwide is S. enterica serovar Typhimurium, ranking first in prevalence in North America and Oceania (6).
Salmonella intestinal colonization is a complex phenotype essential for establishing disease. Salmonella colonization requires synchronized function of both conserved and host-specific colonization factors such as fimbrial adhesins, invasion factors (e.g., SiiE, MisL, and ShdA) and genes encoded within Salmonella pathogenicity island 1 (SPI-1) and SPI-2 (reviewed in references 7 and 8). Both SPIs encode distinct type III secretion systems (T3SSs) and designated translocated effectors required to manipulate various host pathways during specific stages of the infection. The SPI-1-encoded type III secretion apparatus (T3SS-1) plays a pivotal role in intestinal invasion, while a second type III secretion system (T3SS-2) is central for intracellular survival and replication of Salmonella following invasion (9).
Environmental, commensal, and pathogenic bacteria have evolved to tightly regulate their metabolic, physiological, and virulence pathways using sophisticated sensing and regulatory circuits. One of the most important adaptive responses is the stringent response during nutritional deprivation, which provides a rapid adaptation to variety of growth-inhibiting stresses (10). This regulatory response is mediated by the intracellular accumulation of two small molecules called guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), together referred to as ppGpp. These secondary messengers interact with the RNA polymerase (RNAP) in concert with a 17-kDa RNAP-regulatory protein named DksA to execute a global transcriptional reprogramming in response to various nutrient limitations. This stringent response typically results in repressing transcription of tRNA, rRNA, and ribosomal proteins and the activation of amino acid biosynthesis genes (11, 12).
DksA belongs to a unique group of transcription factors that bind to the RNAP secondary channel (reviewed in reference 13), and in Escherichia coli, about 7% of all genes have been shown to be directly or indirectly regulated by DksA (14). Several studies have described a role for DksA in regulation of pathogenicity in different Gram-negative pathogens, including Vibrio cholerae (15), Pseudomonas aeruginosa (16), Shigella flexneri (17, 18), enterohemorrhagic E. coli (19), Campylobacter jejuni (20), Erwinia amylovora (21), and Haemophilus ducreyi (22). In S. Typhimurium, DksA was found to be involved in bacterial defense against oxidative (23) and nitrosative (24) stress in vitro, and this phenotype has been suggested as a possible explanation for the attenuation of a S. Typhimurium dksA mutant in the mouse typhoid model (23–25).
Here we show that DksA is required for S. Typhimurium growth in minimal medium, motility, and biofilm formation. Additionally, we establish that DksA regulates the SPI-1 and motility regulons and is essential for Salmonella host cell invasion and macrophage uptake but not for host cell adhesion. Using streptomycin-pretreated mice, we demonstrated that dksA is induced in vivo at an early stage of the infection in the midcecum and is required for intestinal colonization and systemic infection in the colitis mouse model.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains utilized in this study are listed in Table S1 in the supplemental material. Bacterial cultures were routinely maintained in Lennox Luria-Bertani (LB; BD Difco) or defined M9 or M63 minimal medium supplemented with 1% (wt/vol) glucose or glycerol and 0.135 mM histidine at 37°C. Xylose lysine deoxycholate (XLD) agar plates were used to determine bacterial loads following mouse infections. When appropriate, 100 μg/ml ampicillin, 50 μg/ml kanamycin, or 25 μg/ml chloramphenicol was added to the growth medium.
Cloning and mutant construction.
All primers used in this study are listed in Table S2 in the supplemental material. In-frame deletion of dksA in S. Typhimurium SL1344 was constructed with the λ-Red-mediated recombination system (26). For complementation, dksA was amplified by PCR using the primers clone dksA Fw and clone dksA Rv, digested with SacI and XbaI, and cloned into the low-copy-number vector pWSK29. For in vivo imaging of dksA expression, the dksA promoter region (290 bp upstream of the first methionine) was PCR amplified using the primers dksA promoter Fw and dksA promoter Rev, digested with BamHI and XhoI, and cloned into pCS26. C-terminal two-hemagglutinin (2HA)-tagged versions of SopB, SopE2, and FliC from S. Typhimurium were constructed within pWSK29 or pACYC184 as described elsewhere (27).
Motility assay.
A 10-μl portion of overnight Salmonella cultures grown in LB broth at 37°C were placed onto LB, M9, or M63 0.3% agar plates. Motility plates were incubated for 5 h (for LB plates) or 21 h (for M9 or M63 plates) at 37°C without being inverted.
Biofilm formation.
Overnight cultures grown in LB-Lennox broth (to an optical density at 600 nm [OD600] of 4.5) were diluted 1:100 into fresh LB medium without NaCl (10 g/liter peptone, 5 g/liter yeast extract) supplemented with 50 μg/ml streptomycin (and ampicillin for the pWSK29-harboring strains), and 150 μl was added to cell culture-treated 96-well microplates (Greiner Bio-one). Negative controls included an S. Typhimurium fliC fljB mutant strain and LB broth only as a blank. The plates were incubated at 28°C for 96 h. Planktonic cells were discarded, and attached cells were fixed for 2 h at 60°C. Fixed bacteria were stained with 150 μl of 0.1% crystal violet for 10 min at room temperature. The plates were washed with phosphate-buffered saline (PBS), and the dye bound to the adherent bacteria was resolubilized with 150 μl of 33% acetic acid. The optical density of each tube was measured at 560 nm.
Tissue cultures.
All cell lines were purchased from the American Type Culture Collection. Caco-2 cells were grown in Dulbecco's modified Eagle medium (DMEM)–F-12 medium supplemented with 20% heat-inactivated fetal bovine serum (FBS) and 2 mM l-glutamine. HeLa and Raw 264.7 cells were cultured in a high-glucose (4.5 g/liter) DMEM supplemented with 10% FBS, 1 mM pyruvate, and 2 mM l-glutamine. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2. Epithelial cells and macrophages were seeded at 5 × 104 and 2.5 × 105 cells/ml, respectively, in a 24-well tissue culture dish 18 h prior to bacterial infection. Host cells were infected with Salmonella cultures at multiplicity of infection (MOI) of ∼1:50 for epithelial cells and ∼1:10 for macrophages. Infection experiments were carried out using the gentamicin protection assay as previously described (27). Salmonella invasion was determined by the number of intracellular Salmonella cells at 2 h p.i. divided by the number of infecting bacteria. Adhesion was determined using cytochalasin D to inhibit actin cytoskeleton rearrangement and bacterial cell invasion in an actin-dependent manner. Cells were incubated with fresh medium containing 1 μg/ml cytochalasin D 1 h before infection. Bacteria were added and allowed to adhere for 30 min in the presence of 1 μg/ml cytochalasin D. Cells were washed four times with PBS and harvested by addition of lysis buffer (1% Triton X-100, 0.1% SDS in PBS). Salmonella adhesion was determined by the number of adherent Salmonella cells at 30 min postinfection (p.i.) divided by the number of infecting bacteria.
Western blotting.
Salmonella cultures grown in LB to the mid-logarithmic phase (OD600 ∼ 1) were OD600 normalized and centrifuged, and the pellets were resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Boiled samples were separated on 12% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories). Blots were probed with anti-2HA antibody (ab18181; Abcam) or anti-RpoD antibody (SC56768; Santa Cruz Biotechnology). Goat anti-mouse antibody conjugated to horseradish peroxidase (ab6721; Abcam) was used as a secondary antibody, followed by detection with enhanced chemiluminescence reagents (Amersham Pharmacia).
RT-PCR.
RNA was extracted from Salmonella cultures grown aerobically to the mid-logarithmic phase using the Qiagen RNAprotect bacterial reagent and the RNeasy minikit (Qiagen) according to the manufacturer's instructions, including an on-column DNase digest. Purified RNA was secondarily treated with an RNase-free DNase I followed by ethanol precipitation, and 200 ng of DNase I-treated RNA was subjected to first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Real-time PCRs and data analysis were performed as recently described (27).
In vivo mouse infections.
Mouse experiments were conducted according to the ethical requirements of the Animal Care Committee of the Sheba Medical Center (approval 933/14) and in line with national guidelines. Female C57/BL6 mice (Harlan Laboratories, Israel) were infected at an age of 7 to 8 weeks. Food and water were provided ad libitum. Streptomycin (20 mg per mouse) was given by oral gavage 24 h prior to the infection. S. Typhimurium SL1344 dksA-null mutant strains harboring pWSK129 (Kmr) or pWSK29::dksA (Ampr) were grown in LB with the appropriate antibiotic for 16 h and diluted in 0.2 ml saline. Equal numbers (∼5 × 106 CFU) of each strain were administered to the mice by oral gavage. At day 4 postinfection, mice were euthanized and tissues were collected on ice and homogenized in 0.7 ml saline using a BeadBlaster 24 homogenizer (Benchmark Scientific) for bacterial enumeration. Serial dilutions of the homogenates were plated on XLD agar plates under ampicillin and kanamycin selection, incubated overnight, and counted to calculate bacterial tissue burdens. The competitive index (CI) was calculated as [dksA pWSK129 (Kmr)/dksA pWSK29::dksA (Ampr)]output/[dksA pWSK129/dksA pWSK29::dksA]input. A CI experiment, in which mice were coinfected with Salmonella strains carrying pWSK29 and pWSK129 demonstrated a CI value of 1 (data not shown), indicating equal virulence for strains carrying pWSK29 and those carrying pWSK129.
Bioluminescence imaging of Salmonella during murine infection.
Wild-type S. Typhimurium harboring the dksA regulatory region fused to the luxABCDE operon (pCS26::pdksA) or the empty vector (pCS26) (43) as a negative control were grown in LB supplemented with kanamycin at 37°C. Female C57BL/6 mice were pretreated with streptomycin as described above and 24 h later were orally infected with 5 × 106 CFU of S. Typhimurium/pCS26::pdksA and 8 × 106 CFU of S. Typhimurium/pCS26 in 0.2 ml of saline. At 24 h p.i., mice were anesthetized, and the gastrointestinal tracts, spleens, and livers were optically imaged using a photon-counting system (Photon-Imager, Biospace Lab, France). To determine the total numbers of colonizing Salmonella (CFU), organs were homogenized in saline, diluted, and spread plated on XLD agar supplemented with kanamycin.
RESULTS
DksA is required for S. Typhimurium growth in minimal medium.
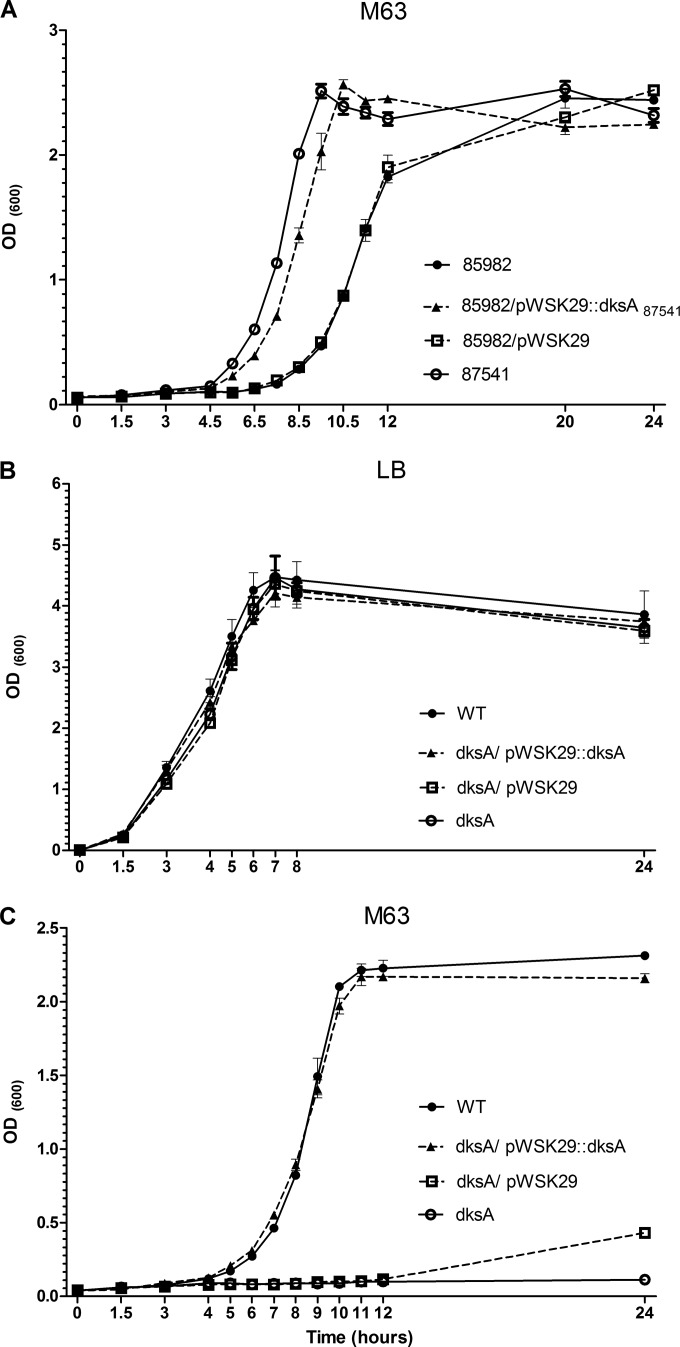
Our interest in S. Typhimurium DksA arose when we studied a clinical case of a 9-month-old male who was infected with S. Typhimurium. Whole-genome sequencing of an earlier and a later stool isolate (numbers 85982 and 87541, respectively), separated by 36 days, from this patient indicated the presence of a single nucleotide polymorphism (SNP), which has led to an amino acid substitution from asparagine (AAT) to aspartate (GAT) at position 88/151 of DksA in isolate 85982 (an additional SNP was found in isolate 87541 [harboring a WT dksA sequence], in a noncoding region, equivalent to position 3490472 in the S. Typhimurium LT2 genome). During our work to understand how such a change affects Salmonella physiology and pathogenicity, we found that while these isolates grow similarly in LB broth (see Fig. S1A in the supplemental material), in M63 minimal medium, isolate 85982 (harboring the point mutation in dksA) exhibits compromised growth compared to isolate 87541 (harboring wild-type dksA). When we cloned the dksA gene from isolate 87541 (dksA87541) and expressed it in isolate 85982, the lag in growth of the latter was largely complemented (Fig. 1A). These observations suggested that functional DksA is required for S. Typhimurium growth in minimal medium.
FIG 1.
DksA is required for S. Typhimurium growth in minimal medium but not in rich LB broth. (A) The growth of the clinical isolates 85982 (containing the amino acid substitution N88D in DksA) and 87541 (harboring wild-type DksA) and that of isolate 85982 harboring pWSK29 or pWSK29::dksA87541 were compared in M63 minimal medium at 37°C under aerobic growth conditions. (B and C) S. Typhimurium SL1344 (WT), its derivative dksA-null mutant strain, and the dksA mutant harboring the plasmid pWSK29 or the dksA gene (from SL1344) cloned into pWSK29 were grown in LB for 16 h, diluted 1:100 into fresh LB (B) or M63 (C) and grown at 37°C with aeration. OD600 was recorded at the indicated time points. Data are mean OD600 for three to four independent cultures and standard errors of the means (SEM).
Based on these results and considering the reported role of DksA in the growth of V. cholerae (15) and P. aeruginosa (16) in minimal medium, we were interested in characterizing in more detail the role of the DksA in S. Typhimurium physiology. To this end, a dksA-null mutant strain was constructed in the S. Typhimurium SL1344 reference strain background, and its growth was studied in nutrient-rich LB and in minimal M63 medium. We found that while S. Typhimurium SL1344 and its isogenic dksA-null mutant strain grew similarly in rich LB medium (Fig. 1B), the dksA-null mutant strain was unable to grow in M63 minimal medium, presenting an even more extreme phenotype than strain 85982 bearing the point mutation in dksA. Complementation of dksA in trans from a low-copy-number plasmid (pWSK29::dksA) but not the presence of the empty vector (pWSK29) restored the dksA mutant growth to levels similar to those of the wild-type strain (Fig. 1C). Similar observations were also found using M9 minimal medium (see Fig. S1B in the supplemental material). These results indicated that DksA is essential for S. Typhimurium growth in minimal medium but not in rich LB broth and showed that the amino acid asparagine at position 88 is important for this function.
DksA plays a role in S. Typhimurium motility and biofilm formation.
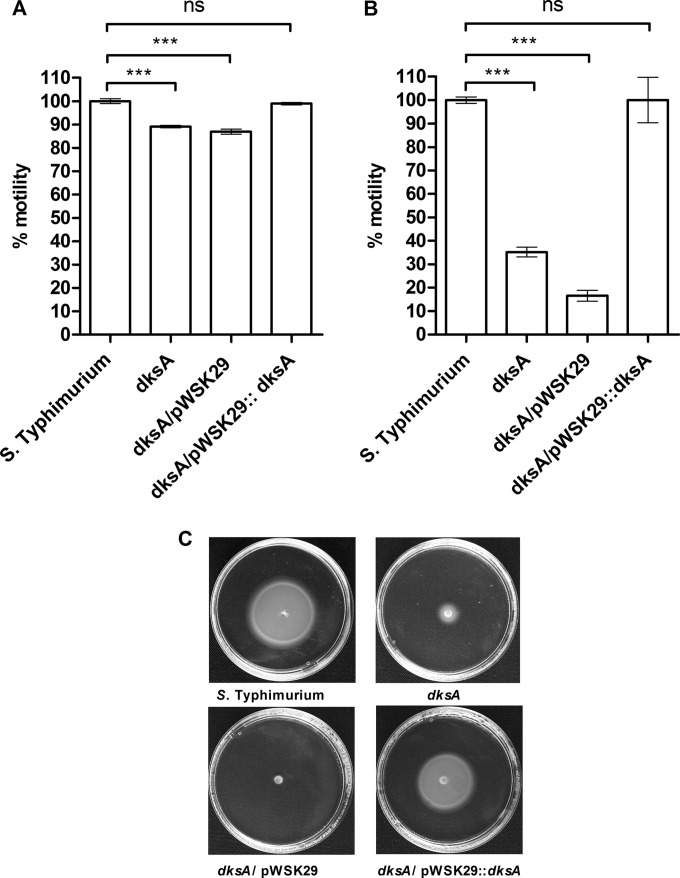
Motility and biofilm are two virulence-associated phenotypes relevant to bacterial pathogenicity in vivo (28). Since DksA was shown to be involved in these phenotypes in other pathogen (15), we were interested in characterizing the possible role of DksA in Salmonella motility and biofilm formation. In the absence of DksA, a mild but significant reduction of 11% to 13% in S. Typhimurium motility was observed on LB soft agar (0.3%) plates. Introducing dksA ectopically (pWSK29::dksA) but not the empty vector (pWSK29) complemented the impaired motility of the dksA strain to the level of the wild-type background (Fig. 2A). Similar experiments using equal inocula (of overnight LB-grown cultures), which were placed on soft agar M9 and M63 minimal medium (supplemented with 1% glucose as a carbon source) plates, demonstrated much more pronounced motility deficiency in the dksA mutant strain. Under these conditions, the S. Typhimurium dksA strain exhibited only 28% and 35% of the wild-type motility in M63 (data not shown) and M9 (Fig. 2B and C), respectively. Again, the impaired motility of the dksA strain was fully complemented by the expression of dksA in trans (pWSK29::dksA).
FIG 2.
DksA is required for S. Typhimurium motility. S. Typhimurium SL1344, its corresponding dksA mutant strain, and the dksA strain harboring the plasmid pWSK29 or pWSK29::dksA were grown in LB for 16 h. A 10-μl portion of each culture was placed in the middle of 0.3% agar LB or M9 plates. LB plates were incubated at 37°C for 5 h (A), and M9 plates were incubated for 21 h (B). Bars show the mean motility (from three independent plates) relative to that of the wild-type strain. One-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test was implemented to compare the motility of the different strains with the wild-type motility. ***, P < 0.0001; ns, not significant. (C) Representative swimming plates. The motility halo of each strain on M9 plates was imaged by a Pentax K-3 II digital SLR camera.
Furthermore, we observed that under biofilm-inducing conditions (LB without NaCl), the ability of S. Typhimurium to form biofilms was significantly decreased in the absence of DksA, to levels similar to that of the fliC fljB mutant (known to be attenuated in biofilm formation [29]), and that this impaired phenotype was complemented when the pWSK29::dksA construct was expressed in the S. Typhimurium dksA mutant strain (Fig. 3). We concluded from these experiments that DksA is required for S. Typhimurium motility and biofilm formation in vitro.
FIG 3.
DksA is required for S. Typhimurium biofilm formation. S. Typhimurium SL1344, its derivative dksA mutant strain, the dksA strain harboring the plasmid pWSK29 or pWSK29::dksA and a fliC fljB mutant (which was used as a negative control) were grown on LB medium lacking NaCl (biofilm-inducing conditions) at 28°C for 96 h. Biofilm formation was assayed by crystal violet staining. Data are means and SEM for eight biological repeats. ANOVA with Dunnett's multiple-comparison test was used to determine differences between data sets.
DksA is required for S. Typhimurium host cell entry.
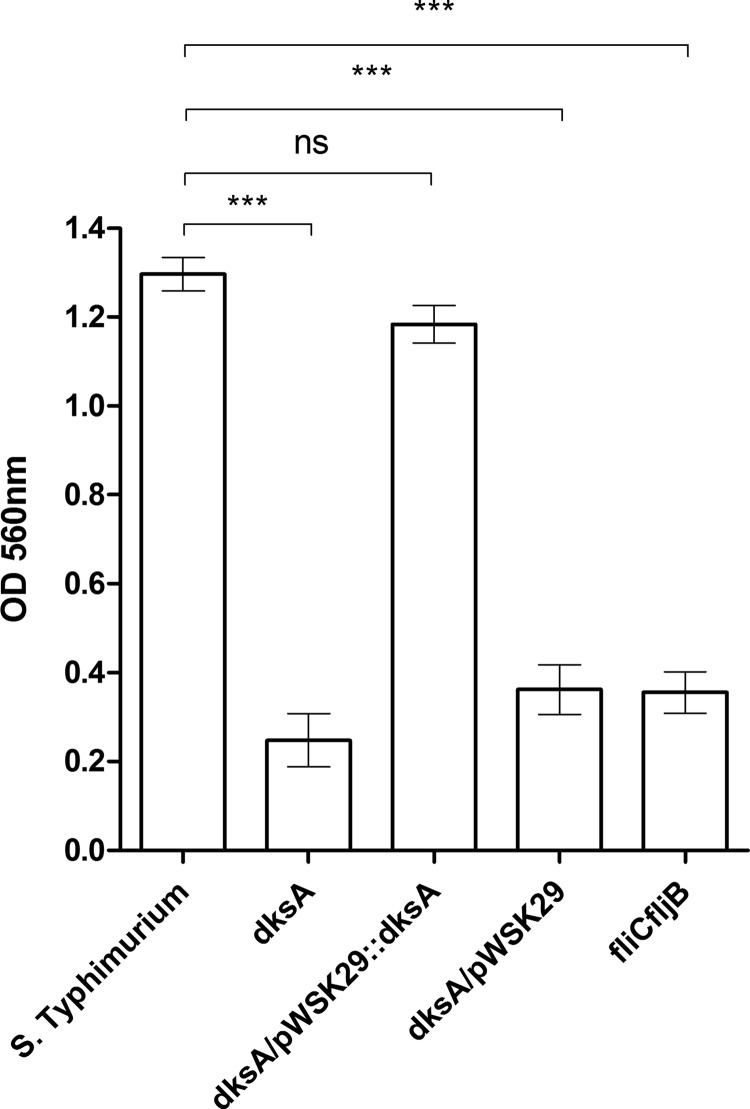
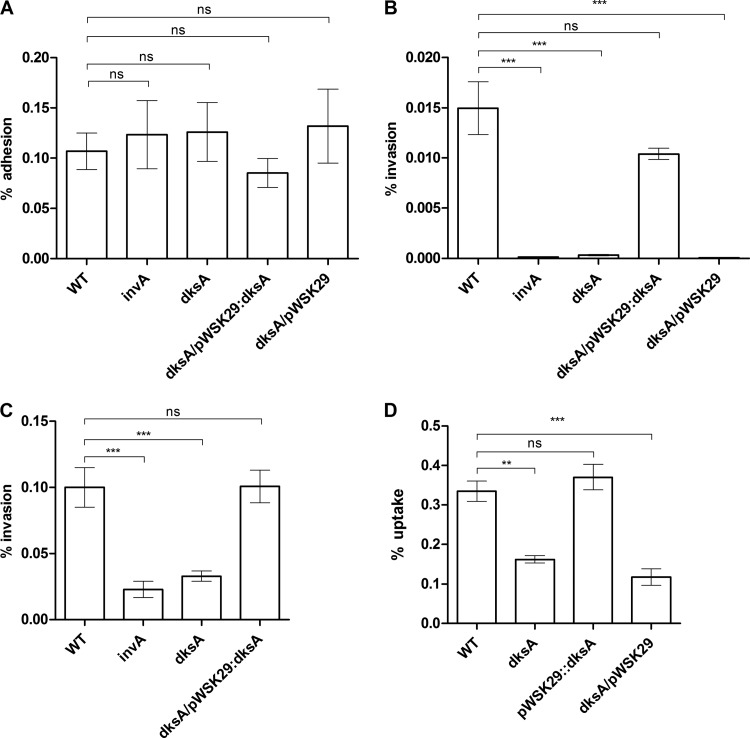
Next, we examined the potential involvement of DksA in S. Typhimurium adherence to and invasion into nonphagocytic cells. As a control for S. Typhimurium invasion, we included the invA mutant strain, known to be deficient in epithelial cell invasion (30). These experiments showed adhesion of the dksA mutant strain that was similar to that of the wild-type background in HeLa (Fig. 4A), Caco-2 (see Fig. S2A in the supplemental material) and Raw 264.7 (see Fig. S2B in the supplemental material) cell lines, suggesting that DksA is not involved in the host cell adhesion phenotype of S. Typhimurium. Nevertheless, the invasion by a dksA strain of both HeLa (Fig. 4B) and Caco-2 (Fig. 4C) epithelial cells was dramatically impaired and was comparable to the poor invasion of the invA mutant. Ectopic expression of dksA from pWSK29 complemented the reduced invasion of the dksA strain to levels similar to that of the parental strain. It is noteworthy that similar results were also obtained when a mild centrifugation (500 rpm for 5 min) was implemented immediately after cell infection (see Fig. S1C in the supplemental material), indicating that the impaired invasion phenotypes of the dksA mutant strain are not due to its reduced motility per se.
FIG 4.
DksA is required for S. Typhimurium host cell entry. S. Typhimurium SL1344 (WT), its derived invA and dksA-null mutant strains, and the dksA mutant harboring the plasmid pWSK29 or pWSK29::dksA were grown in LB at 37°C and used to infect epithelial cell lines. (A) Adhesion to HeLa cells was determined in the presence of cytochalasin D. Data are the percentage of cell-associated bacteria from the total number of CFU used to infect the cells. Invasion of HeLa cells (B) and Caco-2 cells (C) and uptake by Raw 264.7 macrophages (D) were determined using the gentamicin protection assay and calculated as the percentage of intracellular bacteria (CFU) recovered at 2 h p.i from the total number of CFU used to infect the cells. Data are means and SEM for 4 or 5 biological replicates. ANOVA with Dunnett's multiple-comparison test was used to determine differences between data sets. **, P < 0.001; ***, P < 0.0001; ns, not significant.
Furthermore, the S. Typhimurium dksA mutant presented a decrease in uptake by RAW264.7 macrophages compared to the wild-type strain, which was also complemented with the introduction of pWSK29::dksA into the dksA background (Fig. 4D). These results are in close agreement with the recently published report that showed a similar phenotype for a S. Typhimurium dksA strain in HeLa cells (31) and together establish that DksA plays a key role in S. Typhimurium invasion into nonphagocytic cells and uptake by macrophages.
DksA positively regulates motility and SPI-1 genes in S. Typhimurium.
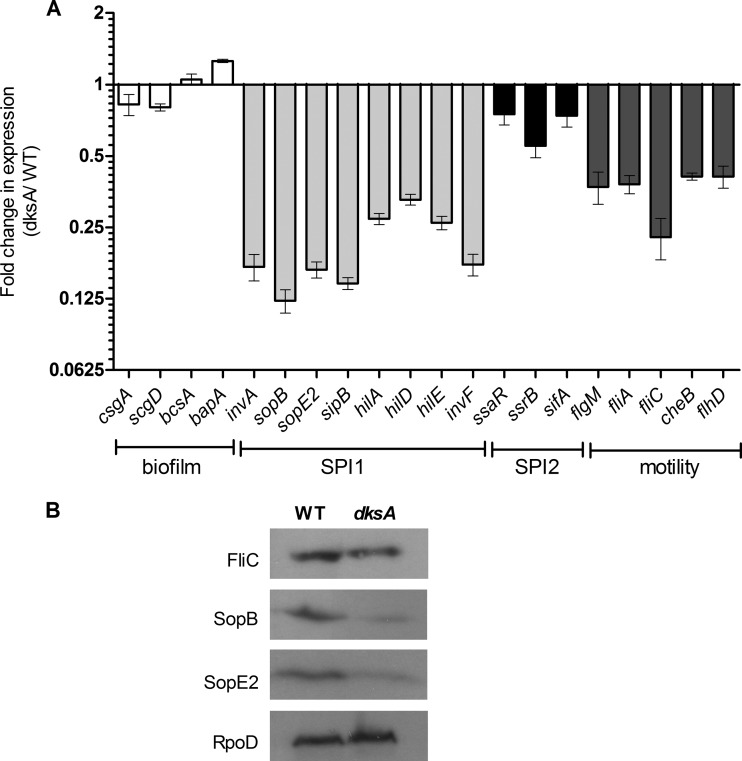
To better understand how DksA affects biofilm formation, motility, and host cell invasion, we determined by means of RT-PCR the expression of representative SPI-1, SPI-2, flagellar (motility), and biofilm-associated genes during the mid-logarithmic growth phase. While we found no significant change in the transcription of the biofilm-associated genes csgA, csgD, bcsA, and bapA and only a mild decrease in the expression of SPI-2 genes (ssaR, ssrB, and sifA), we found a 3- to 8-fold reduction in the transcription of various SPI-1 genes encoding a structural T3SS-1 component (invA), T3SS-1 effectors (sopB, sipB, and sopE2), and T3SS-1 regulatory genes (hilA, hilD, hilE, and invF) and a 2.5- to 5-fold decrease in expression of motility-flagellum genes (fliA, fliC, and flhD) (Fig. 5A). Western blotting against a 2HA-tagged version of SopB, SopE2 (T3SS-1 effectors), and FliC (flagellin subunit) confirmed the RT-PCR results and showed, on the protein level, lower expression of these proteins in the dksA mutant strain than in the wild-type background while revealing similar levels of RpoD (Fig. 5B).
FIG 5.
DksA positively regulates the transcription of SPI-1 and flagellar genes. (A) Total RNA was harvested from S. Typhimurium SL1344 and its isogenic dksA mutant strain cultures grown to mid-logarithmic phase at 37°C and was subjected to qRT-PCR. The change in the abundance of the indicated transcripts (normalized to rpoD) in the dksA mutant strain relative to wild-type S. Typhimurium is shown. Data are means and SEM from 3 to 6 independent RT-PCRs. (B) SDS-PAGE Western blot analysis of bacterial cell lysate from S. Typhimurium and dksA mutant strains expressing SopB-2HA, SopE2-2HA, and FliC-2HA grown to the mid-logarithmic phase. Protein fractions were probed using anti-HA antibody and anti-RpoD as a control.
These results indicated that DksA is a positive regulator of genes belong to the SPI-1 and the motility-chemotaxis regulons and suggested that the impaired motility, biofilm formation, and invasion phenotypes of the dksA mutant were at least in part due to the lower expression of these genes in the absence of DksA.
DksA is induced during the exponential phase and in the midcecum during early gastrointestinal infection.
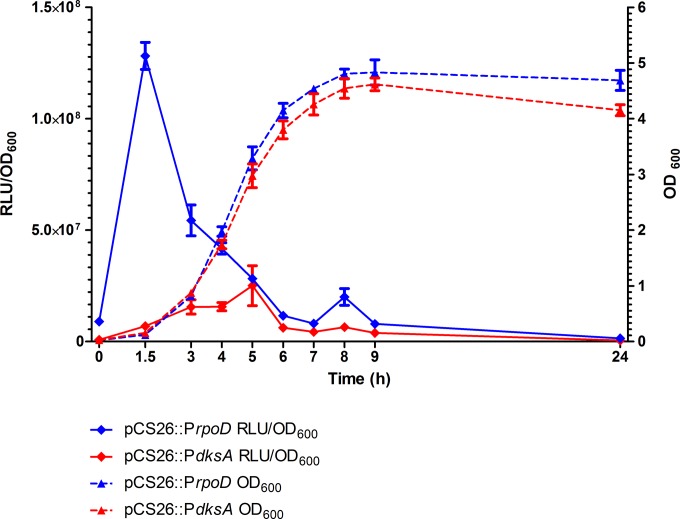
The involvement of DksA in epithelial cells invasion, motility, biofilm formation, and SPI-1 genes expression prompt us to study its expression pattern and possible role in intestinal colonization. To monitor dksA expression, the regulatory region of the dksA gene from S. Typhimurium SL1344 (PdksA) was cloned into the vector pCS26 upstream from the bioluminescence reporter operon (luxABCDE). The expression of this reporter system (PdksA::lux) was compared to that of the rpoD promoter cloned into the same vector (PrpoD::lux). Distinctly, while the expression of PrpoD::lux peaked during the early logarithmic phase (OD600 = 0.1, as expected from the vegetative sigma factor gene), the activity of PdksA::lux gradually increased during the exponential phase and reached its maximum during the mid-logarithmic to late logarithmic phase (OD600 = 3) (Fig. 6).
FIG 6.
S. Typhimurium dksA is induced at the mid-exponential to late exponential phase in vitro. S. Typhimurium SL1344 harboring the regulatory regions of dksA and rpoD cloned into pCS26 (pCS26::PdksA and pCS26::PrpoD, respectively) were grown in LB broth for 24 h. At the indicated time points, optical density (OD600) and the activity of the promoters, presented in relative luminescence units (RLU), were determined. The means and SEM for three independent cultures are shown for each time point.
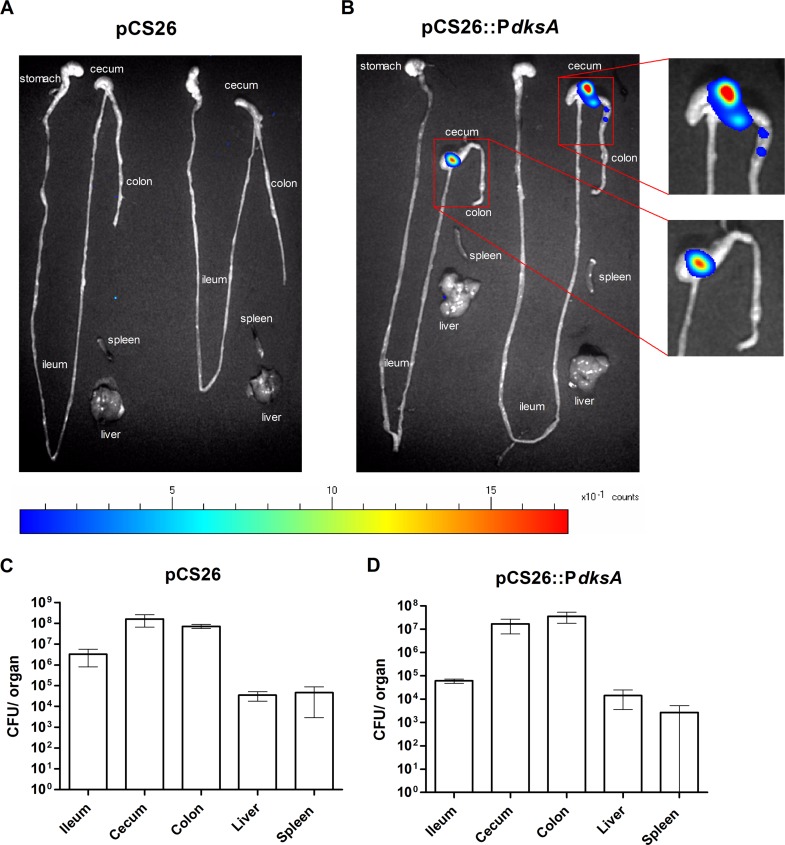
To study the expression pattern of dksA in vivo, we have next implemented the colitis mouse model (32). C57BL/6 mice that were pretreated with streptomycin were infected with S. Typhimurium harboring pCS26::PdksA (the luminescence of this inoculum was 20,000 relative luminescence units [RLU]). As a control, a second group of mice were infected with S. Typhimurium harboring the promoterless pCS26 plasmid. At day 1 postinfection, the gastrointestinal tracts and systemic sites (liver and spleen) were removed, and luminescence was detected using a photon-counting system. While high bacterial loads (107 to 108 CFU) were isolated from the colon and the cecum of all mice, we did not isolate high numbers of salmonellae from the spleen (in three of six mice, salmonellae in the spleen were undetected), indicating an early stage of the infection. Interestingly, although the colon and the cecum were colonized with similar loads, high expression of the dksA promoter was evident exclusively in the midcecum (Fig. 7). These results indicated a specific and spatial induction of dksA in the intestine at an early stage of the infection and suggested that DksA is involved in intestinal colonization of Salmonella in vivo.
FIG 7.
dksA is expressed at the midcecum during intestinal colonization in vivo. (A and B) Streptomycin-pretreated C57BL/6 mice were infected with 5 to 8 × 106 CFU of S. Typhimurium harboring pCS26 (A) or pCS26::PdksA (B). At 24 h p.i., the intestinal tracts and systemic sites (liver and spleen) were removed, and bioluminescence was imaged using a photon-counting system. Organs from two mice are shown from each infection. (C and D) To determine the total numbers of colonizing Salmonella (CFU), organs were homogenized in saline, diluted, and spread plated on XLD agar supplemented with kanamycin. Data are mean bacterial loads and SEM for three mice infected with S. Typhimurium carrying pCS26 (C) or pCS26::PdksA (D).
DksA is required for intestinal colonization and systemic infection in vivo.
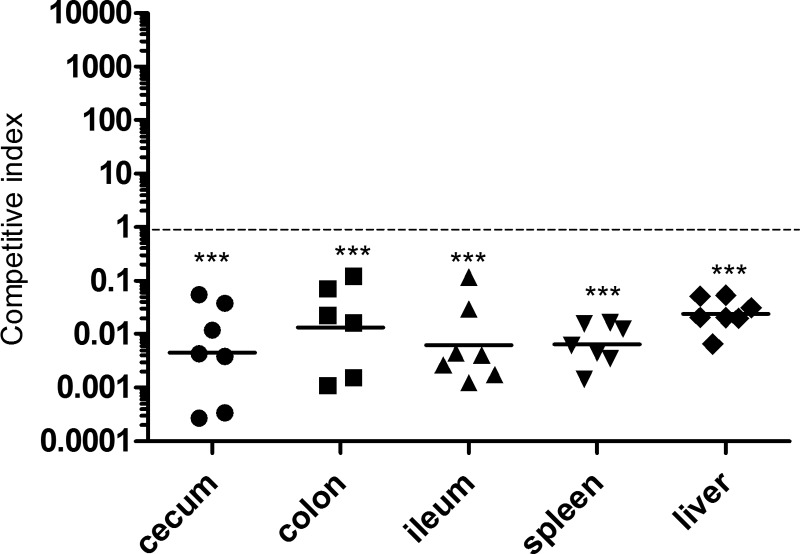
To study the possible role of DksA in Salmonella intestinal colonization, we conducted a competitive index infection experiment using the streptomycin-pretreated mouse model. After infection with S. Typhimurium, streptomycin-pretreated C57BL/6 mice develop an acute colitis which resembles a gastrointestinal disease (33). We used this model and coinfected a group of 7 mice with equal numbers of S. Typhimurium dksA mutant organisms carrying an empty vector (pWSK129, Kmr) or pWSK29::dksA (Ampr). While similar bacterial loads were recovered from mice infected with Salmonella strains carrying pWSK29 and pWSK129 (CI = 1; data not shown), the dksA mutant strain harboring the complementation construct pWSK29::dksA outcompeted the noncomplemented strain (harboring pWSK129) 42- to 222-fold in the intestinal organs (ileum, cecum, and colon) as well as at systemic sites (liver and spleen) at day 4 postinfection (Fig. 8). These results established that DksA is required not only for the systemic stage of a typhoid-like disease but also for intestinal colonization by S. Typhimurium.
FIG 8.
DksA is required for intestinal and systemic colonization by S. Typhimurium. Streptomycin-pretreated C57BL/6 mice were coinfected with equal numbers of CFU of the S. Typhimurium dksA-null mutant carrying pWSK129 (Kmr) or pWSK29::dksA (Ampr) by oral gavage. At 4 days p.i., the competitive index (CI) was determine by plating tissue homogenates on XLD plates containing kanamycin or ampicillin. Each point shows the obtained CI in one mouse, and the geometric mean is indicated by a horizontal line. A one-sample t test against a theoretical mean of 1.0 (assumes equal fitness of both strains) was used to determined statistical significance.
DISCUSSION
In E. coli, DksA acts as a transcriptional cofactor that directly binds the secondary channel of RNAP, potentiating the effect of ppGpp during a stringent response (13). In addition to its particular role in the stringent response, DksA regulates transcription of many other genes in a ppGpp-independent manner, by modulating RNAP distribution. In E. coli and H. ducreyi, for example, DksA was shown to control the expression of about 7% and 17% of their entire open reading frames, respectively, indicating a pleiotropic regulatory role of genes involved in transcription, macromolecules synthesis, protein fate, cell envelope biogenesis, energy metabolism, transport/binding, and amino acid biosynthesis (14, 22).
Here we showed that S. Typhimurium requires DksA for growth in minimal medium but not in rich LB broth. A mutation that was identified in a clinical isolate of S. Typhimurium demonstrated that a conserved asparagine in position 88 located in the middle of DksA is important for this phenotype, as an amino acid substitution to aspartate (N88D) considerably reduced the ability of this clinical isolate to grow in minimal defined medium and a null deletion of dksA abolished S. Typhimurium growth in minimal medium completely. A similar role for DksA in V. cholerae, which also needs dksA for growth in M9 medium, was reported (15). This phenotype can be explained by the positive regulation of amino acid biosynthesis operons by DksA; de novo synthesis of the amino acids is essential for prototrophic growth in minimal defined medium (34).
Another phenotype that was reported to be regulated by DksA is bacterial motility. Nonetheless, the role of DksA in bacterial motility and flagellar gene expression is somewhat disputed. E. coli strains lacking dksA were shown to express higher levels of chemotaxis and flagellum genes, resulting in overproduction of flagellin, hyperflagellated cells, and increased motility, possibly due to the inhibition of flhDC and fliA promoters by DksA (14, 35). On the other hand, Magnusson et al. reported that E. coli dksA mutants were less motile than the wild type, suggesting a positive regulation on the flagellar expression by DksA (36). Similarly, in Pseudomonas putida (37) and V. cholerae (15), deletion of dksA leads to a decrease in bacterial motility, and Legionella pneumophila requires DksA for flagellar gene activation and motility in the stationary growth phase (38). In this study, we found that S. Typhimurium DksA positively regulates the chemotaxis-motility regulon and that its absence leads to a severe motility deficiency under nutrient limitation conditions. Consistent with this phenotype, we demonstrated that in the absence of DksA, the transcription of various flagellar genes, including the regulatory genes fliA and flhD, is significantly decreased. Western blotting results were also in line with the transcription data and showed a lower level of 2HA-tagged FliC in the dksA mutant than the parental strain. Low expression of flagellar and motility genes in the dksA background (and unchanged levels of other biofilm-associated genes) can also explain the impaired biofilm formation of this strain, which was similar to the attenuated phenotype of the fliC fljB mutant.
Besides motility, an additional Salmonella regulon that was found to be downregulated in the absence of DksA is SPI-1, which is pivotal for Salmonella invasion of nonphagocytic host cells. A dksA strain showed 4- to 8-fold-lower transcription of multiple SPI-1 genes. These results were also observed on the protein level, as Western blotting showed significantly lower levels of 2HA-tagged SopB and SopE2 but similar levels of RpoD in the absence of DksA. In accordance with the decreased expression of SPI-1, we were able to demonstrate a dramatic reduction in the invasive phenotype of a dksA mutant strain into both HeLa and Caco-2 human epithelial cells. Interestingly, in contrast to H. ducreyi, where DksA was shown to be required for host cell adhesion (18), we could not assign a role to S. Typhimurium DksA in attachment to HeLa, Caco-2, and RAW264.7 cells, indicating that at least in vitro, Salmonella invasion but not adhesion is controlled by DksA.
While our manuscript was in preparation, Rice and colleagues (31) reported the results of a microarray-based transcriptomic analysis presenting increased levels of SPI-1 genes in a S. Typhimurium dksA mutant strain grown to the stationary phase, compared to the parental strain. On the other hand, the authors reported a decrease transcription of the sicA operon (sicA and sipBCDA) during the late logarithmic phase, lower expression of a sipC::lacZ fusion, undetectable levels of intracellular SipC in the dksA background, and attenuation of this strain (grown to the early stationary phase) in HeLa cell invasion. Thus, in comparison, our invasion results are in close agreement with those reported by Rice et al., but the expression results in the two studies are somewhat inconsistent. This discrepancy may possibly be explained by the different methods used or the different growth phase of the analyzed cultures (mid-log phase in our study versus stationary phase in the other study) and could suggest that the SPI-1 regulation by DksA is growth phase dependent. The induction of dksA::lux expression during the exponential phase and its decline during the stationary phase (Fig. 6) indicates a timely and coordinated expression during Salmonella growth in vitro and therefore supports this possibility.
The dramatic role of DksA in host cell invasion in vitro prompted us to study its potential contribution to Salmonella intestinal colonization. In vivo imaging of the dksA promoter activity showed a specific spatial induction at the midcecum during an early stage of infection. These results show for the first time induction of dksA expression in vivo during intestinal colonization and suggest that DksA regulates the expression of virulence factors required to establish an intestinal colonization, including motility, biofilm, and SPI-1 genes. A competitive infection study using the relevant colitis mouse model demonstrated clear attenuation of the dksA mutant in intestinal colonization as well as in systemic-site infection. These results are consistent with a previous study showing that DksA is required for S. Typhimurium colonization of the chicken alimentary tract (39) and together demonstrate an important role for DksA in gastrointestinal infection by S. Typhimurium in different animal hosts.
Previously, an attenuated virulence of a S. Typhimurium dksA mutant strain was exhibited in a murine model of acute salmonellosis (25). Additionally, Henard and colleagues have elegantly shown that DksA is critical for the resistance of S. Typhimurium to oxidative stress and reactive nitrogen species during the systemic phase in a murine model of typhoid-like disease (23, 24). Collectively, these previously published reports and the results presented here illuminate DksA as a key virulence regulator in Salmonella, which is required both at the early colonization stage and later, during systemic infection, suggesting a role in both gastrointestinal and systemic disease manifestations.
To establish a successful infection, a pathogen must tightly coordinate the expression of multiple virulence traits to ensure their accurate temporal and spatial expression. S. enterica, like many other pathogens, has acquired during evolution different virulence elements by means of horizontal gene transfer that were incorporated into its core genome. One of many examples is the acquisition of SPI-1, which also led to Salmonella speciation (40). Advantageous use of horizontally acquired factors dictates effective coordination with the existing virulence and physiological setup. One way to achieve a regulatory harmonization is to integrate virulence genes under the control of an ancestral regulator. Accumulating evidence indicates that multiple virulence factors in different Gram-negative pathogens are actually regulated by the conserved transcriptional factor DksA, found in ancestral nonpathogenic species. Thus, it is tempting to speculate that different pathogens have evolved in parallel to incorporate different virulence-associated traits under the control of the pleiotropic DksA regulator. A similar idea has been also demonstrated for PhoP, which in addition to its ancestral role in adaptation to low Mg2+ environments has evolved as a key coordinator of multiple virulence genes in Salmonella (41, 42). Consistent with this notion, DksA was shown to regulate many unique virulence factors in multiple pathogens. The V. cholerae DksA positively regulates the production of the major protease HAP, which is involved in pathogenicity and the expression of the horizontally acquired cholera toxin genes (15). In P. aeruginosa, DksA is involved in the posttranscriptional control of the extracellular virulence factor LasB elastase (16). In S. flexneri, DksA is required for Hfq regulation, an important pleotropic regulator by itself (18) and in enterohemorrhagic E. coli, expression of the locus of enterocyte effacement (LEE) pathogenicity island is also controlled by DksA (19). Hence, the conserved sequence of DksA and its regulatory role in the virulence of many Gram-negative pathogens make DksA an ideal target candidate for a broad-spectrum therapeutic.
Here, we established that DksA is required for growth in minimal medium, motility, and biofilm formation in Salmonella. Additionally, we demonstrated that DksA positively regulates the SPI-1 and motility regulons and is required for host cell invasion in vitro but not for cell adhesion. Finally, using the colitis mouse model, we exhibited that dksA is induced in the midcecum during intestinal infection and required for gastrointestinal and systemic colonization in the mouse. Taken together, these data indicate that the conserved stringent response regulator DksA plays a key role in coordination of various physiological and virulence pathways in S. Typhimurium and is required for Salmonella pathogenicity in vivo.
Supplementary Material
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01135-15.
REFERENCES
- 1.Rabsch W, Tschape H, Baumler AJ. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect 3:237–247. doi: 10.1016/S1286-4579(01)01375-2. [DOI] [PubMed] [Google Scholar]
- 2.Edwards RA, Olsen GJ, Maloy SR. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol 10:94–99. doi: 10.1016/S0966-842X(01)02293-4. [DOI] [PubMed] [Google Scholar]
- 3.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.Chen PL, Chang CM, Wu CJ, Ko NY, Lee NY, Lee HC, Shih HI, Lee CC, Wang RR, Ko WC. 2007. Extraintestinal focal infections in adults with nontyphoid Salmonella bacteraemia: predisposing factors and clinical outcome. J Intern Med 261:91–100. doi: 10.1111/j.1365-2796.2006.01748.x. [DOI] [PubMed] [Google Scholar]
- 5.Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, van der Giessen J, Kruse H. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol 139(Suppl 1):S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 7.Humphries AD, Townsend SM, Kingsley RA, Nicholson TL, Tsolis RM, Baumler AJ. 2001. Role of fimbriae as antigens and intestinal colonization factors of Salmonella serovars. FEMS Microbiol Lett 201:121–125. doi: 10.1111/j.1574-6968.2001.tb10744.x. [DOI] [PubMed] [Google Scholar]
- 8.Stevens MP, Humphrey TJ, Maskell DJ. 2009. Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci 364:2709–2723. doi: 10.1098/rstb.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrahams GL, Hensel M. 2006. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol 8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 10.Magnusson LU, Farewell A, Nystrom T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol 13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 13.Zenkin N, Yuzenkova Y. 2015. New insights into the functions of transcription factors that bind the RNA polymerase secondary channel. Biomolecules 5:1195–1209. doi: 10.3390/biom5031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol 191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK. 2012. Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J Bacteriol 194:5638–5648. doi: 10.1128/JB.00518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jude F, Kohler T, Branny P, Perron K, Mayer MP, Comte R, van Delden C. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J Bacteriol 185:3558–3566. doi: 10.1128/JB.185.12.3558-3566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogull SA, Runyen-Janecky LJ, Hong M, Payne SM. 2001. dksA is required for intercellular spread of Shigella flexneri via an RpoS-independent mechanism. Infect Immun 69:5742–5751. doi: 10.1128/IAI.69.9.5742-5751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma AK, Payne SM. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol 62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol 61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 20.Yun J, Jeon B, Barton YW, Plummer P, Zhang Q, Ryu S. 2008. Role of the DksA-like protein in the pathogenesis and diverse metabolic activity of Campylobacter jejuni. J Bacteriol 190:4512–4520. doi: 10.1128/JB.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ancona V, Lee JH, Chatnaparat T, Oh J, Hong JI, Zhao Y. 2015. The bacterial alarmone (p) ppGpp activates the type III secretion system in Erwinia amylovora. J Bacteriol 197:1433–1443. doi: 10.1128/JB.02551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holley CL, Zhang X, Fortney KR, Ellinger S, Johnson P, Baker B, Liu Y, Janowicz DM, Katz BP, Munson RS Jr, Spinola SM. 2015. DksA and (p)ppGpp have unique and overlapping contributions to Haemophilus ducreyi pathogenesis in humans. Infect Immun 83:3281–3292. doi: 10.1128/IAI.00692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henard CA, Bourret TJ, Song M, Vazquez-Torres A. 2010. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem 285:36785–36793. doi: 10.1074/jbc.M110.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henard CA, Vazquez-Torres A. 2012. DksA-dependent resistance of Salmonella enterica serovar Typhimurium against the antimicrobial activity of inducible nitric oxide synthase. Infect Immun 80:1373–1380. doi: 10.1128/IAI.06316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R III, Foster JW. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol 34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elhadad D, Desai P, Rahav G, McClelland M, Gal-Mor O. 2015. Flagellin is required for host cell invasion and normal Salmonella pathogenicity island 1 expression by Salmonella enterica serovar Paratyphi A. Infect Immun 83:3355–3368. doi: 10.1128/IAI.00468-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Q, Zhou M, Zhu L, Zhu G. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 29.Crawford RW, Reeve KE, Gunn JS. 2010. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol 192:2981–2990. doi: 10.1128/JB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galan JE, Ginocchio C, Costeas P. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol 174:4338–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice CJ, Ramachandran VK, Shearer N, Thompson A. 2015. Transcriptional and post-transcriptional modulation of SPI1 and SPI2 expression by ppGpp, RpoS and DksA in Salmonella enterica sv Typhimurium. PLoS One 10:e0127523. doi: 10.1371/journal.pone.0127523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher B, Paesold G, Barthel M, Kremer M, Jantsch J, Stallmach T, Heikenwalder M, Hardt WD. 2006. Chronic Salmonella enterica serovar Typhimurium-induced colitis and cholangitis in streptomycin-pretreated Nramp1+/+ mice. Infect Immun 74:5047–5057. doi: 10.1128/IAI.00072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol 189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterberg S, Skarfstad E, Shingler V. 2010. The sigma-factor FliA, ppGpp and DksA coordinate transcriptional control of the aer2 gene of Pseudomonas putida. Environ Microbiol 12:1439–1451. doi: 10.1111/j.1462-2920.2009.02139.x. [DOI] [PubMed] [Google Scholar]
- 38.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun 66:2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumler AJ. 1997. The record of horizontal gene transfer in Salmonella. Trends Microbiol 5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 41.Gal-Mor O, Elhadad D, Deng W, Rahav G, Finlay BB. 2011. The Salmonella enterica PhoP directly activates the horizontally acquired SPI-2 gene sseL and is functionally different from a S. bongori ortholog. PLoS One 6:e20024. doi: 10.1371/journal.pone.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato A, Groisman EA, Howard Hughes Medical Institute . 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv Exp Med Biol 631:7–21. doi: 10.1007/978-0-387-78885-2_2. [DOI] [PubMed] [Google Scholar]
- 43.Bjarnason J, Southward CM, Surette MG. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J Bacteriol 185:4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.