Abstract
Preparation of chromosome spreads is a prerequisite for the successful performance of fluorescence in situ hybridization (FISH). Preparation of high quality plant chromosome spreads is challenging due to the rigid cell wall. One of the approved methods for the preparation of plant chromosomes is a so-called drop preparation, also known as drop-spreading or air-drying technique. Here, we present a protocol for the fast preparation of mitotic chromosome spreads suitable for the FISH detection of single and high copy DNA probes. This method is an improved variant of the air-dry drop method performed under a relative humidity of 50%-55%. This protocol comprises a reduced number of washing steps making its application easy, efficient and reproducible. Obvious benefits of this approach are well-spread, undamaged and numerous metaphase chromosomes serving as a perfect prerequisite for successful FISH analysis. Using this protocol we obtained high-quality chromosome spreads and reproducible FISH results for Hordeum vulgare, H. bulbosum, H. marinum, H. murinum, H. pubiflorum and Secale cereale.
Keywords: Plant Biology, Issue 106, chromosome preparation, fluorescence in situ hybridization, plants, single-copy FISH, mitotic metaphase, relative humidity
Introduction
Fluorescence in situ hybridization (FISH) is an effective tool for the physical mapping of single and high copy sequences at the chromosomal level. Prerequisite is the preparation of high quality chromosome spreads. There is no general chromosome preparation protocol that would be equally suitable for animal and plant cells. Preparation of plant chromosomes is particularly challenging due to the rigid cell wall and various cytoplasm consistency within different species. One of the favorable methods for the preparation of plant chromosomes is a so-called drop technique also known as drop-spreading technique and air-drying technique 1,2. This method was first introduced in 1958 by Rothfels and Siminovitch for in vitro grown mammalian cells 3. Later Martin et al. 4 and Kato et al. 5 adapted this method for plants.
More recently, a method named 'SteamDrop' was developed which used water steam for the preparation of non-overlapping chromosomes 6. Although, the positive influence of high humidity was observed earlier 7, 'SteamDrop' delivers a controlled workflow of high-quality chromosome preparations 6. The steam treatment causes stretching of chromosomes probably connected to some modifications of chromosomal proteins. The quality of resulting metaphase spreads is very high, although retaining of sufficient number of complete metaphase spreads for subsequent FISH experiments demands technical expertise.
Here we present a protocol for the preparation of mitotic cereal chromosomes suitable for the FISH detection of single and high copy probes 5,8. This method is an improved variant of the air-dry dropping method described by Kato 9 performed under relative humidity of 50%-55% (Figure 1). This protocol comprises a reduced number of washing steps making its application easy, efficient and reproducible. Using this protocol we obtained high-quality chromosome spreads and FISH results for Hordeum vulgare, H. bulbosum, H. marinum, H. murinum, H. pubiflorum and Secale cereale.
Protocol
1. Chromosome Preparation
- Seed germination and fixation of root tips
- Germinate 10-20 barley seeds on two layers of moist filter paper in a Petri dish under dark conditions for 2 days at 22-24 °C. Cut off vigorous roots with the length of 1-2 cm from the seed by using a razor blade.
- Prepare ice-cold water by placing a 500 ml glass bottle containing cold tap water into crushed ice-water. Aerate the ice-cold water and immerse root tips for 20 hr to increase the frequency of metaphase cells.
- Transfer roots from water to 50 ml of ethanol: acetic acid (3:1) fixative to fix them at RT for 2 days. Store roots in a freshly prepared ethanol: acetic acid (3:1) fixative at 4 °C until use up to a year.
- Washing and enzyme treatment
- Wash the 10-20 roots with 30 ml ice-cold tap water for 5 min twice using a 50 ml glass beaker. Use binocular microscope. Transfer roots one by one into 30 ml 0.01 M citrate buffer (0.01 M citric acid + 0.01 M sodium citrate, pH 4.8) using forceps and wash by shaking the glass beaker for 5 min twice. Place roots on filter paper to remove the liquid completely and cut-off undesired non-meristematic tissue using a razor blade.
- Incubate up to 20 root tips in 1 ml enzyme mixture at 37 °C for about 50 min to soften the plant tissue (Table 1) in a watch glass. Enzyme mixture contains 0.7% cellulase R10, 0.7% cellulase, 1% pectolyase and 1% cytohelicase diluted in 0.01 M citrate buffer. Store the enzyme mixture at -20 °C and reused up to five times.
- Remove the enzyme by pipetting and wash the root tips on ice with 5 ml 0.01 M citrate buffer twice to replace the residual enzyme.
- Root maceration
- Wash root tips with 1 ml 96% ethanol twice carefully in the same watch glass. Replace ethanol with freshly prepared fixative (75% acetic acid : 25% ethanol). Use 10-15 µl fixative per root tip.
- Transfer root tips together with the fixative into a 2 ml tube and disintegrate root meristems with a dissecting needle or forceps. Tap the tube 20 times to re-suspend cells to obtain a cell suspension. Store the cell suspension at -20 °C up to two months.
- Dropping of the cell suspension
- Place 2-3 layers of water-soaked paper tissue on a hot plate at 50 °C. Immerse microscopic slides in ice-cold tap water in the fridge for 30 min. And place slides on top of the moist paper tissue.
- Pipette 7-10 µl of cell suspension and drop it from a distance of 20 cm onto the cooled slide placed on the hot plate. Pipette 10 µl of acetic acid-ethanol mixture on the same place as cell suspension on the slide and keep the slide on the hot plate for additional 2 min. Place the slide on the hot plate without the wet tissue and let it dry for 1 min.
- Quality control and storage of slides
- Check slides using a phase-contrast microscope to control the quality of the chromosome spread. Use slides either the same day or store by immersing in 96% ethanol in a Coplin jar at -20 °C.
- Pretreatment of slides before FISH; all steps are carried out at RT
- Place slides in a Coplin jar containing 50 ml of 2x SSC (20x SSC contains 3 M NaCl and 300 mM trisodium citrate) for 5 min. Using forceps, transfer slides to a Coplin jar containing 50 ml of 45% acetic acid for 3-10 min.
- Transfer slides to a Coplin jar containing 50 ml of 2x SSC for 10 min. Transfer slides to a Coplin jar containing 50 ml of 4% formaldehyde (in 2x SSC) and immerse slides for 10 min to fix chromosomes.
- Remove formaldehyde by rinsing the slides 3 times for 4 min each, in a Coplin jar containing 50 ml 2x SSC. Dehydrate slides in a Coplin jar for 2 min in series of 70%, 90% and 100% ethanol, respectively and dry slides in a vertical position.
2. Fluorescent In Situ Hybridization (FISH)
For each slide, prepare a hybridization solution of 20 µl in total using 10 µl of deionized formamide, 5 µl of 4x hybridization buffer (200 µl buffer contains 80 µl 20x SSC, 8 µl 1 M Tris-HCl pH 8.0, 1.6 µl 0.5 M EDTA, 11.2 µl 10 µg/ µl salmon sperm and 99.2 µl DNase-free water), 3 µl of the probe and 2 µl of DNase-free water.
Add 20 µl of hybridization solution per slide and cover with a 24 x 32 mm cover slip and arrest the cover slip with rubber cement. Denature slides with probes simultaneously at 80 °C for 2 min on a hot plate.
Transfer slides to a moist chamber and incubate slides at 37 °C O/N avoiding light. Remove cover slips by rinsing the slides in a Coplin jar with 2x SSC. Place slides in a Coplin jar containing 55-60 °C 2x SSC and incubate for 20 min.
Place the slides to 2x SSC in a Coplin jar for 2 min at RT. Dehydrate slides in a Coplin jar for 2 min in series of 70%, 90% and 100% ethanol, respectively.
Air-dry the slides and counterstain with 1 µg/ml 4',6-diamidino-2-phenylindoline (DAPI) in antifade mounting medium, avoid intense light.
3. Microscopic Analysis and Storage
Analyze the slides using an epifluorescence microscope. The selection of filter depends on the fluorochrome used for probe labeling. If necessary, store slides at 4 °C under dark conditions up to a year.
Representative Results
Microscopic slides with the mitotic metaphase spreads were prepared by the fast air-dry dropping chromosome preparation method described above (Suppl. Figure 1). FISH analysis was carried out using both, repetitive and single-copy sequences. Images were obtained by a epifluorescence microscope with a set of filters enabling excitation of corresponding fluorophores and captured by a high-sensitivity CCD monochrome camera. For the image acquisition we used a computer with an image acquisition software. Results of the FISH experiments on mitotic metaphase chromosomes using 5S rDNA, [CTT]10, and single-copy probes were distinct and of a high quality for Hordeum vulgare (Figre 2A, B), H. bulbosum (Figure 2C), H. marinum, H. murinum, H. pubiflorum and Secale cereale (Figure 2D). Obvious benefits of this approach are well-spread, undamaged and numerous metaphase chromosomes serving as a perfect prerequisite for successful FISH analysis. It is possible to store the cell suspension at -20 °C up to two months and to prepare the chromosome spreads on the day of the FISH experiment. Freshly prepared slides can be also stored at -20 °C in 96% ethanol, though we observed that the quality of hybridization signals on such chromosomes is reduced compared to the freshly-prepared metaphase spreads. The methods can be used to prepare high-quality chromosome spreads in cereals in an easy, efficient and reproducible way and most likely can be used in other plant species too.
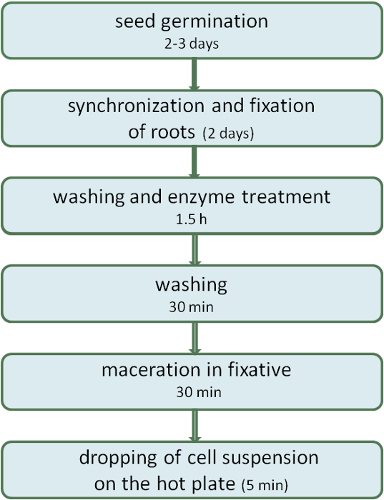 Figure 1.A scheme describing the procedure of the air-dry dropping plant chromosome preparation method.
Figure 1.A scheme describing the procedure of the air-dry dropping plant chromosome preparation method.
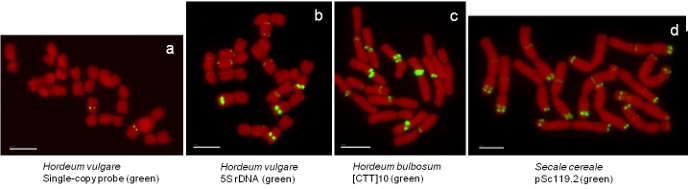 Figure 2. FISH on mitotic metaphase chromosome spreads of Hordeum vulgare, H. bulbosum and Secale cereale prepared by the air-dry dropping method. (A) H. vulgare with a single copy probe (FPct_40752) labeled with a red fluorescent dye. (B) H. vulgare with 5S rDNA probe labeled by a green fluorescent dye. (C) H. bulbosum with CTT-microsatellite labeled by a green fluorescent dye and (D) S. cereale with pSc119.2 repeat labeled by a green fluorescent dye. All chromosomes were counterstained with DAPI (in red). FISH signals are shown in yellow. Scale bar = 10 µm. Please click here to view a larger version of this figure.
Figure 2. FISH on mitotic metaphase chromosome spreads of Hordeum vulgare, H. bulbosum and Secale cereale prepared by the air-dry dropping method. (A) H. vulgare with a single copy probe (FPct_40752) labeled with a red fluorescent dye. (B) H. vulgare with 5S rDNA probe labeled by a green fluorescent dye. (C) H. bulbosum with CTT-microsatellite labeled by a green fluorescent dye and (D) S. cereale with pSc119.2 repeat labeled by a green fluorescent dye. All chromosomes were counterstained with DAPI (in red). FISH signals are shown in yellow. Scale bar = 10 µm. Please click here to view a larger version of this figure.
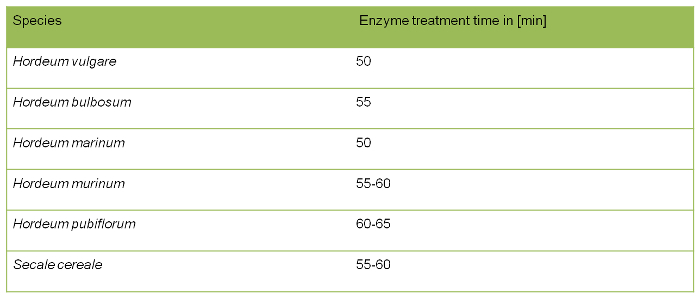
Table 1. Incubation time of enzyme treatment for different species.
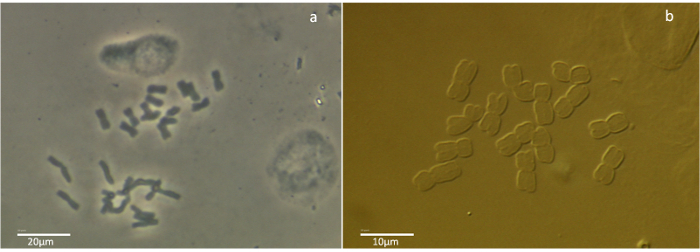 Suppl. Figure 1.Phase-contrast and differential interference contrast (DIC) images of mitotic metaphase chromosome spreads of the air-dry dropping plant chromosome preparation method on the example of Hordeum vulgare. (A) Phase-contrast imagetaken at 200X magnificationand (B) Differential interference contrast image taken at 630X magnification. Please click here to view a larger version of this figure.
Suppl. Figure 1.Phase-contrast and differential interference contrast (DIC) images of mitotic metaphase chromosome spreads of the air-dry dropping plant chromosome preparation method on the example of Hordeum vulgare. (A) Phase-contrast imagetaken at 200X magnificationand (B) Differential interference contrast image taken at 630X magnification. Please click here to view a larger version of this figure.
Discussion
The chromosome preparation experiment has been carried out using young roots of cereals belonging to the grass family (Poaceae). All analyzed species have 14 relatively long mitotic metaphase chromosomes (11-15 µm) in the diploid genome set and belong to large-genome species (5.1-7.9 Gbp).
Length of germinated roots was not more than 2 cm to obtain a maximum of meristematic tissue. Synchronization of dividing cells was achieved by a 20 hr long ice-water treatment that improved the quantity of mitotic metaphase spreads 10.
Two steps are important for the preparation of high-quality chromosome preparations: (I) the relative humidity of 50%-55% and (II) duration of the enzyme treatment. The first point was achieved by placing wet paper tissues on a hot plate in proximity of the glass slides. The relative humidity was measured with a hygrometer. The optimal humidity for the preparation of plant chromosomes was similar to the humidity reported by Kirov et al. 6. The positive effect on the chromosome quality at optimal relative humidity occurs by swelling of the cytoplasm and cell wall hydrolysis.
The duration of enzyme treatment is species dependent (Table 1). The period of enzyme treatment also depends on the time span of root fixation in ethanol/acetic acid and the size of the roots. The longer roots were stored in the fixative (up to 1 year at 4 °C), the longer it takes to digest roots to the proper grade. Insufficiently digested root material is difficult to macerate and will increase the total time of preparation as a result of long lasting maceration. Moreover, metaphase chromosomes remain embedded into cytoplasm that could hamper ensuing probe penetration during the FISH experiment. On the other hand over-digested material can influence the structure of the chromosomes themselves, and damage target DNA for the FISH analysis.
An additional factor for the improvement of the preparation is the use of the second drop of fixative (3:1, acetic acid/ethanol). High concentration of acetic acid in this mixture stimulates the digestion of cytoplasm and promotes chromosome spreading in species with large chromosomes. Cytoplasm reduction can also take place after the immobilization of the chromosomes on slides. For this purpose microscope slides carrying the chromosome spreads can be incubated in 45% acetic acid at RT for 2-10 min depending on cytoplasm level. Quality check of chromosome spreads was performed with a phase-contrast microscope without any supplementary staining (e.g., 1% aceto-carmin). Normally more than 25 slides containing high-quality chromosome spreads can be obtained from 20 roots using the method above.
Results of the FISH experiments on mitotic metaphase chromosomes using 5S rDNA, [CTT]10, and 6 kb long single-copy probe (FPct_40752) were distinct and of a high quality for all species described above (Figure 2). Obvious benefits of this approach are well-spread, undamaged and numerous metaphase chromosomes serving as a perfect prerequisite for successful FISH analysis. It is possible to store the cell suspension at -20 °C up to two months and to prepare the chromosome spreads on the day of the FISH experiment. Freshly prepared slides can be also stored at -20 °C in 96% ethanol, though we observed that the quality of hybridization signals on such chromosomes is reduced compared to the freshly-prepared metaphase spreads.
Chromosome spreads prepared by the fast air-dry dropping technique were suitable for FISH and were reproduced a number of times. Combination of this chromosome preparation method with FISH could be widely applied to explore the genome organization in plants, for instance, for karyotyping11, chromosomal mapping 12, in synthetic studies, and for the integration of physical and genetic maps13.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We gratefully thank the DFG for financial support (HO 1779/21-1) as well as Katrin Kumke and Dr. Veit Schubert (IPK, Gatersleben) for technical advice.
References
- Geber G, Schweizer D. Cytochemical heterochromatin differentiation in Sinapis alba (Cruciferae) using a simple air-drying technique for producing chromosome spreads. Pl Syst Evol. 1988;158(2-4):97–106. [Google Scholar]
- Andras SC, et al. A drop-spreading technique to produce cytoplasm-free mitotic preparations from plants with small chromosomes. Chromosome Res. 1999;7(8):641–647. doi: 10.1023/a:1009288119851. [DOI] [PubMed] [Google Scholar]
- Rothfels KH, Siminovitch L. An air-drying technique for flattening chromosomes in mammalian cells grown in vitro. Stain Technology. 1958;33(2):73–77. doi: 10.3109/10520295809111827. [DOI] [PubMed] [Google Scholar]
- Martin R, Busch W, Herrmann RG, Wanner G. Efficient preparation of plant chromosomes for high-resolution scanning electron microscopy. Chromosome Res. 1994;2(5):411–415. doi: 10.1007/BF01552801. [DOI] [PubMed] [Google Scholar]
- Kato A, Albert PS, Vega JM, Birchler JA. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech. Histochem. 2006;81(2-3):71–78. doi: 10.1080/10520290600643677. [DOI] [PubMed] [Google Scholar]
- Kirov I, Divashuk M, Van Laere K, Soloviev A, Khrustaleva L. An easy 'SteamDrop' method for high quality plant chromosome preparation. Mol. Cytogenet. 2014;7:21. doi: 10.1186/1755-8166-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck JL, Zinsmeister AR, Meyer KJ, Jalal SM. Dynamics of chromosome spreading. Am J Med Genet. 1996;61(4):387–393. doi: 10.1002/(SICI)1096-8628(19960202)61:4<387::AID-AJMG15>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Ma L, et al. Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 2010;18(7):841–850. doi: 10.1007/s10577-010-9166-3. [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U S A. 2004;101(37):13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WH, Houben A, Schlegel R. Highly effective cell synchronization in plant-roots by hydroxyurea and amiprophos-methyl or colchicine. Genome. 1993;36(2):387–390. doi: 10.1139/g93-053. [DOI] [PubMed] [Google Scholar]
- Kim JS, et al. Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome. 2002;45(2):402–412. doi: 10.1139/g01-141. [DOI] [PubMed] [Google Scholar]
- Lapitan NLV, Brown SE, Kennard W, Stephens JL, Knudson DL. FISH physical mapping with barley BAC clones. Plant J. 1997;11(1):149–156. [Google Scholar]
- Aliyeva-Schnorr L, et al. Cytogenetic mapping with centromeric BAC contigs shows that this recombination-poor region comprises more than half of barley chromosome 3H. Plant J. 2015;84:385–394. doi: 10.1111/tpj.13006. [DOI] [PubMed] [Google Scholar]


