Abstract
Organotypic cultures allow the reconstitution of a 3D environment critical for cell-cell contact and cell-matrix interactions which mimics the function and physiology of their in vivo tissue counterparts. This is exemplified by organotypic skin cultures which faithfully recapitulates the epidermal differentiation and stratification program. Primary human epidermal keratinocytes are genetically manipulable through retroviruses where genes can be easily overexpressed or knocked down. These genetically modified keratinocytes can then be used to regenerate human epidermis in organotypic skin cultures providing a powerful model to study genetic pathways impacting epidermal growth, differentiation, and disease progression. The protocols presented here describe methods to prepare devitalized human dermis as well as to genetically manipulate primary human keratinocytes in order to generate organotypic skin cultures. Regenerated human skin can be used in downstream applications such as gene expression profiling, immunostaining, and chromatin immunoprecipitations followed by high throughput sequencing. Thus, generation of these genetically modified organotypic skin cultures will allow the determination of genes that are critical for maintaining skin homeostasis.
Keywords: Developmental Biology, Issue 106, organotypic, skin, genetics, RNA interference, retroviral overexpression, keratinocytes, fibroblasts, dermis, epidermis, transduction, skin constructs, stratified epithelia, epithelium, dermatology, SNAI2, epithelial to mesenchymal transition
Introduction
The human epidermis is a stratified epithelium that connects to the underlying dermis through an extracellular matrix known as the basement membrane zone.The epidermis not only serves as an impermeable barrier to prevent the loss of moisture but also as a first line of defense to protect the body from foreign and toxic substances1. The basal layer, which is the deepest layer of the epidermis, contains the epidermal stem and progenitor cells that give rise to the differentiated progeny that form the rest of the epidermis2. As epidermal progenitor cells differentiate they migrate upwards to form the first layer of differentiated cells known as the spinous layer3. In the spinous layer, cells turn on the expression of keratins 1 and 10, which then provide the strength to withstand physical stress for the differentiated layers of the epidermis. As the spinous layer cells further differentiate, they move upwards in the epidermis to form the granular layer which is characterized by the formation of keratohyalin and lamellar granules as well as structural proteins that are assembled below the plasma membrane. As the cells proceed in the differentiation process, the proteins beneath the plasma membrane are cross linked to each other while the lamellar granules are extruded from the cells to form a lipid rich barrier called the stratum corneum4.
Diseases that involve alterations in epidermal growth and differentiation impact ~20% of the population5. Thus, understanding the mechanisms of this process is of great importance. Since manifestation of many of these diseases is contingent upon cell-cell or cell-matrix contact, organotypic cultures where the human epidermis is reconstituted in a 3D environment have been created6-10. These methods typically involve the use of primary or transformed keratinocytes seeded on extracellular matrix such as devitalized human dermis, Matrigel, or collagen.
To understand gene regulatory mechanisms that are important in epidermal growth and differentiation, keratinocytes can be genetically manipulated through retroviral vectors to knockdown or overexpress genes in 2D culture and then reconstituted in 3D. These methods have been used extensively to characterize genes involved in epidermal stem and progenitor cell self-renewal and differentiation as well as progression to neoplasia11-21. Here, an in-depth protocol on how to alter gene expression in epidermal organotypic cultures through the use of retroviruses is provided.
Protocol
The human skin protocol was performed in accordance with the guidelines of the University of California, San Diego’s Research Ethics Committee. Human skin can be obtained from discarded surgical samples or bought from skin banks (skin bank is listed in the Material/Equipment Table). The location where the skin is derived from or age of the donor is not critical to the experiment as long as the basement membrane zone proteins (collagen/laminin) in the dermis are not degraded.
1. Preparation of Devitalized Human Dermis
Upon receiving human skin, place the tissue in the tissue culture hood to thaw (~3-5 min). Depending on the size of the skin, place thawed tissue in a sterile disposable 50 ml conical tube or 125 ml bottle. The typical size of skin from the skin bank is 0.75 square feet.
Fill the container 75% full with 4x penicillin and streptomycin (pen/strep) in 1x PBS. Shake vigorously for 5 min and dispose of the liquid. Repeat this step 3 more times.
After the last wash, transfer the tissue to a new tube/bottle and add fresh 4x pen/strep/PBS to fill 75% of the container. Incubate this mixture in a tissue culture incubator at 37 °C for 2 weeks to allow the separation of the dermis from the epidermis.
Under sterile conditions, use forceps to peel away the epidermis from the dermis. The dermis is completely white while the epidermis will have coloring depending on the race of the donor (Figure 1A). Discard the epidermis according to the institution’s protocol for dealing with human tissue.
Place the dermis in a tube or bottle and wash it in 4x pen/strep diluted in 1x PBS with vigorous shaking (5 min washes for 4 times). After the last wash, transfer the dermis to a new tube/bottle in 4x pen/strep diluted in 1x PBS and store at 4 °C until ready to use. Dermis can be stored at 4 °C for up to a year.
2. Genetically Modifying Primary Human Keratinocytes
NOTE: Use amphotropic phoenix cells grown in complete DMEM media [DMEM +10% fetal bovine serum (FBS)+pen/strep] to produce viruses to infect primary human keratinocytes. Viral packaging genes that encode proteins such as gag-pol and envelope are stably integrated into the phoenix cell genome which allows for virus production when transfected with a retroviral vector. Phoenix cells can be transfected with high efficiency allowing for high viral titer production. Virus produced from this packaging line can also infect a large variety of mammalian cells including human.
Day 1: The day before transfection, seed phoenix cells in a 6-well plate at a density of 800,000 cells per well in complete DMEM media.
- Day 2: The day of transfection, use 3 µg of retroviral vectors per well. Aliquot 3 µg of retroviral vector into a 1.5 ml tube. Use a mix of 100 µl DMEM plus 6 µl of transfection reagent for every 3 µg of vector. Mix 6 µl of transfection reagent with 100 µl DMEM in a 1.5 ml tube. (The retroviral vectors that are used are listed in the equipment/materials table).
- Incubate for 5 min and add the 106 µl mixture into the pre-aliquoted tube containing the 3 µg of retroviral vector. Mix and incubate at RT for 30 min. Add this entire mixture dropwise to each well of the phoenix cells.
Day 3: The day after transfection, remove the media from the phoenix cells and add 2 ml of fresh complete DMEM media. On the same day, seed primary human keratinocytes into 6-well plates at a density of 75,000 cells per well. Maintain the keratinocytes in a keratinocyte serum free medium (KCSFM) with antibiotics (pen/strep). NOTE: Primary human keratinocytes were purchased. Cells can be purchased from a variety of vendors (listed in the Material/Equipment table).
- Day 4: Harvest the media from the transfected phoenix cells, which now contains viral particles. Pass the media through a 0.45 µm filter using a syringe (to remove any contaminating phoenix cells) and place 2 ml onto the keratinocytes (plated at 75,000 cells/well the previous day: see step 2.3). Add hexadimethrine bromide (5 µg/ml) to the media to help mediate the infection process. Viral titers are ~1 x 107 TU/ml.
- Spin the 6-well plates in a centrifuge at 200 x g for 1 hr at RT. After the spin, remove the media containing virus and wash the cells once with 1x PBS before adding KCSFM.
Day 5: Infect the same batch of keratinocytes again as in step 2.4. NOTE: Select the keratinocytes using a drug if there is a selectable marker on the retroviral vector and expand for use in downstream analysis or applications such as organotypic cultures.
3. Setting Up Organotypic Skin Cultures
- Build the organotypic cassette from common materials found in the laboratory or the cassettes can be custom made through a metal fabrication shop (Figure 2A-F). To make these cassettes from a 3.5 cm tissue culture plate, use a cautery to remove a 1.0 cm x 1.0 cm square from the center of the lid of a 3.5 cm dish (Figure 2A-B). Do this under sterile conditions.
- Attach square pegs to the bottom of the cassette (lid of 3.5 cm dish) using clear nail polish (Figure 2C). Allow 5 min for the nail polish to dry and flip the cassette over so that it is resting on the square pegs (Figure 2D). Place the cassette into a 6 cm dish. NOTE: Square pegs can be purchased from a variety of arts and crafts stores.
Prepare keratinocyte growth medium (KGM) comprised of 66% DMEM, 22% Ham’s F12, 100 units/ml penicillin, 100 µg/ml streptomycin, 10% FBS, 2.67 µg/ml adenine, 40 ng/ml hydrocortisone, 10 ng/ml cholera toxin, 4.94 µg/ml insulin, 5 µg/ml transferrin, 1.36 ng/ml Triiodo-L-thyronine, 10 ng/ml EGF, and 10 µg/ml ciprofloxacin hydrochloride. Filter the media through a 0.22 µm filter and store at 4 °C until ready to use.
Take the dermis out of the 4x pen/strep + PBS solution and wash 2 times in KGM and then incubate in KGM at 37 °C for two days prior to use.
Cut the dermis using a scalpel to 1.5 cm x 1.5 cm sized pieces and then place onto the organotypic cassette. Place the dermis with the top of the dermis facing up. The top of the dermis will be the side that comes into contact with the keratinocytes (Figure 1B).
Place the pre-cut dermis onto the square hole of the organotypic cassette with the top side of the dermis facing up (Figure 3A).
Flip the entire organotypic cassette containing the dermis over using forceps (Figure 3B). Thaw the extracellular matrix solution on ice. Use a needle and syringe to draw out 100 µl of extracellular matrix solution per cassette. Add 5 drops to the bottom of the dermis (glossy side) and shake slightly to ensure even distribution throughout the dermis (Figure 3B).
Allow 5 min for the commercial extra cellular matrix solution to completely solidify (Figure 3C). After solidification, use forceps to flip the cassette back over. Add 4 ml of KGM to the 6 cm dish.
- Place 0.5-1 million genetically modified keratinocytes onto the organotypic cassettes containing the dermis (Figure 3D).
- Trypsinize keratinocytes by placing 4 ml of 0.05% Trypsin onto 10 cm plates for 5 min and quench with 4 ml of complete DMEM media.
- Count the keratinocytes using a hemocytometer and then spin down at 200 x g for 5 min. Resuspend 0.5-1 million cells (depending on the number of cells available) in 90 µl of KGM and place all the cells onto the dermis. NOTE: It is important to place the same number of cells on dermis within the same experiment to ensure that any differences seen in thickness of the regenerated epidermis is not due to differences in starting cell number. Placing less than 0.5 million cells on dermis can lead to poor stratification and differentiation.
Change KGM media on the organotypic cultures every other day. Harvest tissue 1-14 days after placement of keratinocytes on the dermis. Full stratification and differentiation of the epidermis usually occurs after day 5.
Representative Results
The first step in generating organotypic human skin is to remove the epidermis from the dermis. The two week incubation of the skin at 37 °C in 4x pen/strep/PBS should allow the separation of the dermis from the epidermis (Figure 1A). If separating the epidermis and dermis is difficult then place the tissue at 37 °C in 4x pen/strep/PBS for another week and then try peeling again using forceps.
One of the keys to regenerating human epidermis on devitalized dermis is to ensure that the keratinocytes are placed on the correct side of the dermis. The dermis has a top and a bottom. The top of the dermis is the side that comes into contact with the keratinocytes in vivo as well as in organotypic cultures. The top of the dermis can be differentiated from the bottom of the dermis due to the glossiness of the bottom (Figure 1B). If keratinocytes are placed directly onto the bottom side (glossy) of the dermis, the epidermis will not differentiate or stratify properly. Thus, it is crucial to have the top of the dermis (non-glossy) facing up in the organotypic cassette. The dermis can be placed directly onto an organotypic cassette (Figure 2A-F). Cassettes made from tissue culture plates should be UV sterilized while metal fabricated cassettes can be autoclaved prior to the dermis being placed on. The genetically modified keratinocytes can then be placed onto the dermis. The keratinocytes are initially submerged in liquid since they were placed onto the dermis resuspended in KGM. After 24 hr the KGM will have diffused through the dermis which allows the keratinocytes to be exposed to the air-liquid interface which causes stratification and differentiation into epidermis7 (Figure 3).
RNA, protein, DNA/chromatin, as well as tissue sections can be harvested from the organotypic skin cultures which can all be used for a variety of downstream applications. RNA can be used to determine gene expression differences between control and knockdown/overexpression tissue using RT-QPCR, microarrays, or RNA-Seq. DNA/chromatin can be used for DNA or ChIP-Seq applications. The regenerated skin can be mounted in OCT and frozen sections can be used for immuno-staining or hematoxylin and eosin staining to assess tissue morphology, protein expression/localization in control and experimental groups.
An example of this is shown in Figure 4 with the hematoxylin and eosin staining of a typical organotypic skin culture. Note that all four distinct layers of the epidermis are formed with layer specific expression of differentiation related proteins (Figure 4)11,12,14,15. This system can be used to determine the impacts of overexpression or knockdown of different genes on the epidermis. Figure 5 shows regenerated epidermal tissue that expresses control, LACZ as well as tissue overexpressed for SNAI2. Overexpression of SNAI2 led to an inhibition of epidermal differentiation as noted by the lack of keratin 1 (K1) staining as well as loss of expression of differentiation induced structural genes such as TGM1 and SPRR1A (Figure 5A-C)13,22. The epidermal thickness shown in Figure 4 and Figure 5 may vary within the same sample due to more cells accumulating in the middle of the dermis versus the periphery when the cells were initially placed on the dermis. Thus, the middle sections tend to have thicker epidermis whereas the periphery tends to have thinner. For direct comparison between different samples the same regions of a section should be compared. However, the staining for the markers should stay consistent.
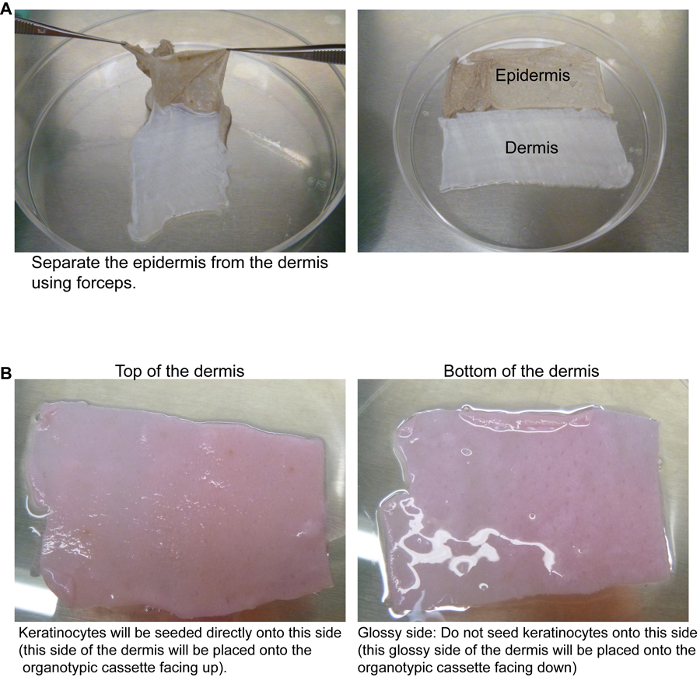 Figure 1. Preparation of the dermis from human skin. (A) After incubation of the human skin at 37 °C for 2 weeks, the dermis can be separated from the epidermis. Forceps are used to peel the epidermis (brown colored) away from the dermis (white). (B) The top of the dermis can be distinguished from the bottom by the glossiness of the tissue. The top of the dermis (the side that will naturally face the epidermis in vivo or where the keratinocytes will be seeded) is non-glossy. The bottom of the dermis is glossy. The tissue is pink due to incubation in KGM. Please click here to view a larger version of this figure.
Figure 1. Preparation of the dermis from human skin. (A) After incubation of the human skin at 37 °C for 2 weeks, the dermis can be separated from the epidermis. Forceps are used to peel the epidermis (brown colored) away from the dermis (white). (B) The top of the dermis can be distinguished from the bottom by the glossiness of the tissue. The top of the dermis (the side that will naturally face the epidermis in vivo or where the keratinocytes will be seeded) is non-glossy. The bottom of the dermis is glossy. The tissue is pink due to incubation in KGM. Please click here to view a larger version of this figure.
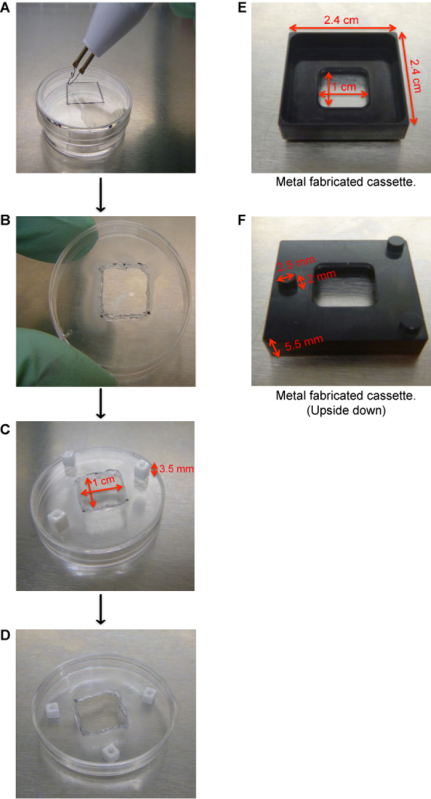 Figure 2. Generation of organotypic cassettes. (A) Use a cautery to remove a 1.0 cm x 1.0 cm square from the center of the lid of a 3.5 cm dish. (B) Remove the square from the center of the 3.5 cm dish. (C) Use clear nail polish to adhere square pegs. Allow 5 min for the nail polish to dry. (D) Flip the lid over so that it is resting on the square pegs. The cassette is now ready to be used. (E) Image of a metal fabricated organotypic cassette with its dimensions. (F) Image of a metal fabricated cassette flipped upside down to show the support pegs. Please click here to view a larger version of this figure.
Figure 2. Generation of organotypic cassettes. (A) Use a cautery to remove a 1.0 cm x 1.0 cm square from the center of the lid of a 3.5 cm dish. (B) Remove the square from the center of the 3.5 cm dish. (C) Use clear nail polish to adhere square pegs. Allow 5 min for the nail polish to dry. (D) Flip the lid over so that it is resting on the square pegs. The cassette is now ready to be used. (E) Image of a metal fabricated organotypic cassette with its dimensions. (F) Image of a metal fabricated cassette flipped upside down to show the support pegs. Please click here to view a larger version of this figure.
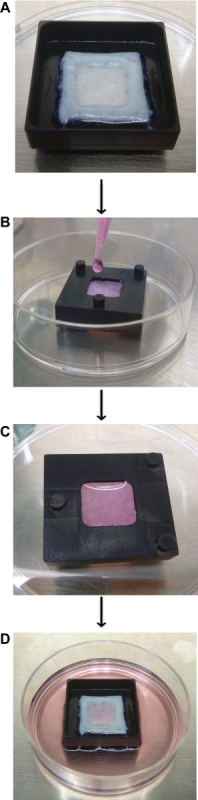 Figure 3. Generation of organotypic skin cultures. (A) Place the dermis with the top of the dermis (non-glossy side) facing up onto the organotypic cassette. (B) Flip the cassette over and add Matrigel to the glossy side of the dermis. (C) Allow 5 min for the Matrigel to solidify and then flip the cassette back over. (D) Add genetically modified keratinocytes to the dermis (0.5-1 million cells are resuspended in 90 µl of KGM). Harvest the tissue 1-14 days after seeding of the keratinocytes. Please click here to view a larger version of this figure.
Figure 3. Generation of organotypic skin cultures. (A) Place the dermis with the top of the dermis (non-glossy side) facing up onto the organotypic cassette. (B) Flip the cassette over and add Matrigel to the glossy side of the dermis. (C) Allow 5 min for the Matrigel to solidify and then flip the cassette back over. (D) Add genetically modified keratinocytes to the dermis (0.5-1 million cells are resuspended in 90 µl of KGM). Harvest the tissue 1-14 days after seeding of the keratinocytes. Please click here to view a larger version of this figure.
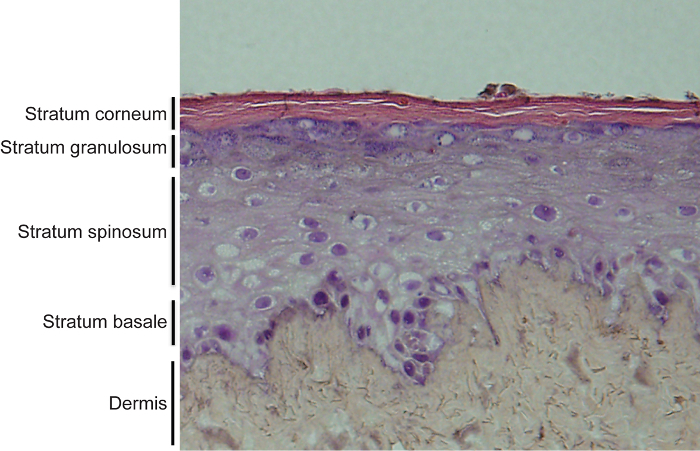 Figure 4. Hematoxylin and eosin staining of organotypic skin cultures. Regenerated human skin contains both the epidermis and dermis. The epidermis is fully stratified and differentiated with 4 distinct layers. These layers include the undifferentiated basal layer and the 3 differentiated layers including stratum spinosum, granulosum and corneum. Please click here to view a larger version of this figure.
Figure 4. Hematoxylin and eosin staining of organotypic skin cultures. Regenerated human skin contains both the epidermis and dermis. The epidermis is fully stratified and differentiated with 4 distinct layers. These layers include the undifferentiated basal layer and the 3 differentiated layers including stratum spinosum, granulosum and corneum. Please click here to view a larger version of this figure.
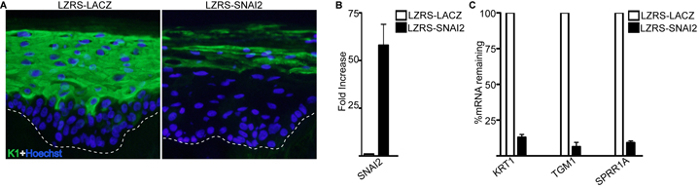 Figure 5. Immunofluorescence staining and gene expression analysis of genetically modified human epidermis. (A) Overexpression of SNAI2 inhibits epidermal differentiation. Staining for differentiation protein keratin 1 (K1) is shown in green and nuclei is marked in blue (Hoechst). (B) RT-QPCR analysis on the mRNA levels of SNAI2 in control LACZ and SNAI2 overexpressing tissue. Error bars = SD, n = 3. (C) RT-QPCR analysis on the mRNA levels of differentiation transcripts KRT1, TGM1, and SPRR1A in control LACZ and SNAI2 overexpressing tissue. Error bars = SD, n = 3. Please click here to view a larger version of this figure.
Figure 5. Immunofluorescence staining and gene expression analysis of genetically modified human epidermis. (A) Overexpression of SNAI2 inhibits epidermal differentiation. Staining for differentiation protein keratin 1 (K1) is shown in green and nuclei is marked in blue (Hoechst). (B) RT-QPCR analysis on the mRNA levels of SNAI2 in control LACZ and SNAI2 overexpressing tissue. Error bars = SD, n = 3. (C) RT-QPCR analysis on the mRNA levels of differentiation transcripts KRT1, TGM1, and SPRR1A in control LACZ and SNAI2 overexpressing tissue. Error bars = SD, n = 3. Please click here to view a larger version of this figure.
Discussion
Genetic manipulation in human skin organotypic cultures offer many advantages to commonly studied 2D cultured cells as well as mouse models. 2D cultures lack the three dimensional cell-cell and cell-extracellular matrix interactions found in intact tissues and organs. Recent studies have also found tremendous differences between 2D and 3D cultured skin cancer cells with the 3D cultured cells showing much more gene expression similarities to primary human skin tumors16. Human organotypic skin cultures also have several advantages over mouse models. Mouse models are time consuming and expensive to generate as well as several notable differences between human and mouse skin. These include different tissue turnover rates as well as the thickness of the epidermis between murine and human epidermis8. With new technologies emerging such as CRISPR-CAS, specific genes can be modified/mutated to model human skin diseases using organotypic skin cultures23. Furthermore, genes can be delivered or knocked down in these cells through a wide variety of methods besides the retroviral gene transfer described here. As long as there is robust delivery through high transduction efficiency or drug selection, the method can be employed. These delivery methods include nucleofection of siRNAs, lentiviral, adenoassociated virus, and adeno virus11,12,24-26.
The organotypic skin cultures can be modified by adding different cell types to the system. For example, normal or genetically altered human fibroblasts can be added to the devitalized dermis to study mesenchymal and epithelial crosstalk. Immune cells can also be added to the dermis to study the inflammatory response to perturbations in the skin. One limitation of the system is that the desquamation process that occurs in vivo does not occur in organotypic cultures. This prevents the use of these cultures for long term and most analysis is done between 1-14 days after seeding of the epidermal cells. For longer term assays, the organotypic cultures can be grafted onto the backs of immune compromised mice17,27.
The skin organotypic culture presented here is unique in that it utilizes intact devitalized human dermis which is the natural substrate of the epidermis in vivo whereas most other systems use collagen or Matrigel as the substrate28. Setup of organotypic skin cultures using substrates such as Matrigel results in thinner and less robust stratification of the epidermis in our experience. The dermis contains basement membrane zone proteins including laminin and collagen which allows the epidermal cells to attach, proliferate, differentiate, and stratify29. Use of dermis allows for generation of epidermis that mimics their in vivo counterpart. Thus, organotypic skin cultures can be extremely valuable in toxicological and pharmacological studies in cases where animal studies are not permitted (cosmetics industry)30.
Using devitalized dermis also comes with its own drawbacks since the thickness of the dermis between different donors or even within the same donor can vary. Epidermal cells grown on thick dermis tend to not proliferate and generate as thick of epidermis as cells grown on thinner dermis. Thus, it is critical to choose similar thickness dermis when setting up the experiments comparing multiple samples. Another critical parameter is to seed the same number of epidermal cells when comparing different groups so that any differences seen is not due to the differences in initial number of cells seeded.
In conclusion, the organotypic skin culture can serve as an advanced and efficient in vitro model for validating molecular pathways in a more physiological context, which ultimately will accelerate the understanding of gene regulatory mechanisms that are important in epidermal growth and differentiation.
Disclosures
We have nothing to disclose.
Acknowledgments
This work was supportedby the American Cancer Society Research Scholars Grant(RSG-12-148-01-DDC) and the CIRM Basic Biology Award (RB4-05779).
References
- Tadeu AM, Horsley V. Epithelial stem cells in adult skin. Current topics in developmental biology. 2014;107:107–131. doi: 10.1016/B978-0-12-416022-4.00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL. Remembering one's identity: the epigenetic basis of stem cell fate decisions. FASEB J. 2011;25:2123–2128. doi: 10.1096/fj.11-182774. [DOI] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA. Transglutaminase function in epidermis. J Invest Dermatol. 2005;124:481–492. doi: 10.1111/j.0022-202X.2005.23627.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. Genetic pathways in disorders of epidermal differentiation. Trends Genet. 2013;29:31–40. doi: 10.1016/j.tig.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Epidermal differentiation: the bare essentials. J Cell Biol. 1990;111:2807–2814. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O, Thomas J. Growth of cultured human epidermal cells into multiple epithelia suitable for grafting. Proc Natl Acad Sci U S A. 1979;76:5665–5668. doi: 10.1073/pnas.76.11.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA. Modelling cancer in human skin tissue. Nat Rev Cancer. 2006;6:270–280. doi: 10.1038/nrc1838. [DOI] [PubMed] [Google Scholar]
- Parenteau NL, Bilbo P, Nolte CJ, Mason VS, Rosenberg M. The organotypic culture of human skin keratinocytes and fibroblasts to achieve form and function. Cytotechnology. 1992;9:163–171. doi: 10.1007/BF02521744. [DOI] [PubMed] [Google Scholar]
- Oh JW, Hsi TC, Guerrero-Juarez CF, Ramos R, Plikus MV. Organotypic skin culture. J Invest Dermatol. 2013;133:e14. doi: 10.1038/jid.2013.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, et al. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Zhang K, Sen GL. SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells. 2014;32:3209–3218. doi: 10.1002/stem.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med. 2010;16:1450–1455. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Sen GL. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell. 2012;11:127–135. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson KL, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med. 2013;19:626–630. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Sen GL. Transcriptional profiling of SNAI2 regulated genes in primary human keratinocytes. Genomics data. 2015;4:43–46. doi: 10.1016/j.gdata.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebis C, et al. Efficient in vitro transduction of epithelial cells and keratinocytes with improved adenoviral gene transfer for the application in skin tissue engineering. Transpl immunol. 2002;9:323–329. doi: 10.1016/s0966-3274(02)00034-5. [DOI] [PubMed] [Google Scholar]
- Melo SP, et al. Somatic correction of junctional epidermolysis bullosa by a highly recombinogenic AAV variant. Mol Ther. 2014;22:725–733. doi: 10.1038/mt.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D, Matsushita N, Toki F, Higashiyama S. Efficient expansion of human keratinocyte stem/progenitor cells carrying a transgene with lentiviral vector. Stem cell res ther. 2013;4:127. doi: 10.1186/scrt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci NC, Cuono CB, Langdon RC, McGuire J. In vitro reconstitution of skin: fibroblasts facilitate keratinocyte growth and differentiation on acellular reticular dermis. J Invest Dermatol. 1991;97:843–848. doi: 10.1111/1523-1747.ep12491522. [DOI] [PubMed] [Google Scholar]
- Mathes SH, Ruffner H, Graf-Hausner U. The use of skin models in drug development. Adv Drug Deliv Rev. 2014;69-70:81–102. doi: 10.1016/j.addr.2013.12.006. [DOI] [PubMed] [Google Scholar]


