Abstract
The two principal cell types of importance for normal vessel wall physiology are smooth muscle cells and endothelial cells. Much progress has been made over the past 20 years in the discovery and function of transcription factors that coordinate proper differentiation of these cells and the maintenance of vascular homeostasis. More recently, the converging fields of bioinformatics, genomics, and next generation sequencing have accelerated discoveries in a number of classes of noncoding sequences, including transcription factor binding sites (TFBS), microRNA genes, and long noncoding RNA genes, each of which mediates vascular cell differentiation through a variety of mechanisms. Alterations in the nucleotide sequence of key TFBS or deviations in transcription of noncoding RNA genes likely have adverse effects on normal vascular cell phenotype and function. Here, the subject of noncoding sequences that influence smooth muscle cell or endothelial cell phenotype will be summarized as will future directions to further advance our understanding of the increasingly complex molecular circuitry governing normal vascular cell differentiation and how such information might be harnessed to combat vascular diseases.
Keywords: microRNA, Long noncoding RNA, Transcription factor binding site, Smooth muscle cell, Endothelial cell, Differentiation
Introduction
The establishment and maintenance of a fully differentiated program of gene expression in endothelial cells (ECs) and subjacent smooth muscle cells (SMCs) are indispensable for vascular homeostasis. The unique gene expression signature of ECs and SMCs is attained through DNA-binding transcription factors, associated cofactors, and the TFBS to which such transcriptional complexes bind. In addition to protein-coding transcription factors, a growing number of noncoding RNA genes have been discovered that modify normal and stress-induced conditions of gene expression in ECs and SMCs. These molecular rheostats provide an important layer of regulatory control over a cell’s transcriptome or proteome, often times by adjusting the steady-state expression of key transcription factors. Variations in nucleotide sequence or altered expression or activity of transcription factors and/or noncoding RNA genes have unfavorable effects on normal EC and SMC differentiation and, hence, vascular homeostasis. For example, acute and chronic diseases of the vessel wall (e.g., restenosis and atherosclerosis) may be manifest by reduced expression of a critical SMC cofactor called myocardin leading to SMC phenotypic modulation, wherein the normal contractile program of gene expression is subverted to a less differentiated status and either adopts a synthetic state typified by excessive cell proliferation, migration, and matrix hyper-secretion or transdifferentiates into other cell types that can perturb normal vascular function [1–3]. Although there have been challenges to the notion of SMC phenotypic modulation [4, 5], strong evidence for such a phenomenon is emerging from studies using rigorous confocal microscopy and in vivo lineage tracing [2, 3, 6–8].
Do ECs similarly exhibit phenotypic modulation? It is well documented that pulsatile blood flow and attending laminar shear stress (~15 dynes/cm2) promote a quiescent EC differentiated phenotype characterized by a gene expression program that enables normal vasoactivity, selective permeability, anti-thrombogenicity, and anti-inflammation. However, changes in laminar blood flow, such as the disturbed blood flow ECs encounter at critical branch points and curvatures of the vasculature, promote a less differentiated, pro-inflammatory EC phenotypic state, presumably through mechanosensory signals that destabilize normal EC gene expression [9]. In both cases of SMC and EC phenotypic modulation, there are antecedent changes in the molecular determinants governing normal patterns of gene expression such as altered levels or activity of DNA-binding transcription factors or associated cofactors (e.g., myocardin). Elucidating the transcriptional circuitry of SMC and EC gene expression and illuminating the basis for deviations in the homeostatic settings of such phenotypic states hold promise for the development of new diagnostic and treatment modalities for many vascular diseases.
Thanks in large part to efforts of the ENCODE (Encyclopedia of DNA Elements) Project Consortium [10–13], we have a broad understanding of functional sequences in the human genome as well as variations in nucleotide sequences that may disrupt normal cellular activity. Accordingly, the concept of “junk DNA” [14] has been challenged with the reality of “pervasive transcription” [15] as well as the prodigious number of functional regulatory elements throughout the human genome [16, 17]. Indeed, advances in genome science have yielded a bewildering number of noncoding sequences, including millions of regulatory elements and tens of thousands of noncoding RNA genes defined broadly as either short noncoding RNA (processed transcript length of less than 200 base pairs) or long noncoding RNA (processed transcript length of more than 200 base pairs). This review will highlight the biology and interplay of several noncoding sequences among three major classes (TFBS, microRNAs, and long noncoding RNAs) that control SMC and EC phenotype. Also discussed will be challenges that must be overcome to gain further insight into the function of vascular cell-associated noncoding sequences and existing opportunities for diagnostic or therapeutic intervention.
Transcription factor binding sites in vascular cells
The human genome is punctuated with millions of transcription factor binding sites (TFBS) that act as molecular zip codes for some 1400 transcription factors [18]. The combinatorial permutations for gene regulation at this level are staggering. Nevertheless, tools in computational biology and genomics have greatly advanced the historical and pain-staking reductionist approach of defining functional TFBS in vascular cells. Considered below are the major TFBS and associated transcription factors involved in the establishment and maintenance of vascular SMC and EC differentiation.
Smooth muscle cell TFBS
SMCs are derived from numerous anatomical sites during embryonic development and exhibit variations in both signaling inputs as well as functional outputs [19]. Despite such diversity in origin and function, there exists a gene expression signature that distinguishes differentiated SMCs from the remaining 250 cell types making up the mammalian body plan. The majority (25/42) of these genes harbor one or more functional CArG box, the consensus sequence of which is CC[A/T]6GG (Table 1). The CArG box is the core sequence of the serum response element, initially described in the 5′ promoter region of the Fos proto-oncogene [20]. Subsequent work revealed conserved CArG boxes in the regulatory region of several contractile genes in sarcomeric muscle [21]. The CArG box binds the widely expressed serum response factor (SRF) [22]. Alterations in SRF expression or activity have been associated with a number of diseases across many organ systems, including the cardiovascular system [23].
Table 1.
SMC transcriptome and functional TFBS (number)
| Gene symbol | Alias | CArG boxes | RBPJ sites | MEF2 sites | SMAD sites |
|---|---|---|---|---|---|
| MYH11 | SM-MHC | Yes (5) | Yes (1) | Yes (1) | No |
| MYLKv7 | Telokin | Yes (1) | No | No | No |
| SMTNB | Smoothelin B | No | No | No | No |
| HDAC8 | HDAC8 | No | No | No | No |
| KCNMB1 | Maxi-K-β1 subunit | Yes (2) | No | No | No |
| ACTG2 | SM γ-actin | Yes (5) | No | No | No |
| NOTCH3 | Notch3 | No | No | No | No |
| ITGA8 | α8 - integrin | No | No | No | No |
| PTK2v | FRNK | No | No | No | No |
| GLMN | Glomulin | No | No | No | No |
| LPP | Lipoma preferred partner | Yes (3) | No | No | No |
| CNN1 | SM-Calponin | Yes (5) | No | No | No |
| LMOD1 | SM-Leiomodin | Yes (2) | No | No | No |
| MIRN143/145 | miR-143/145 | Yes (1) | Yes (3) | No | Yes (1) |
| FHL2 | 4.5 LIM domain 2 | Yes (1) | No | No | No |
|
TGFB1I1 SPEG |
HIC-5 APEG1 |
Yes (1) No |
No No |
No No |
No No |
| SMTNA | Smoothelin-A | Yes (2) | No | No | No |
| ACTA2 | SM α-actin | Yes (3) | Yes (1) | No | Yes (1) |
| ITGA1 | α1-integrin | Yes (1) | No | No | No |
| EFNB2 | Ephrin B2 | No | No | No | No |
| TAGLN | SM22α | Yes (2) | Yes (1) | No | Yes (1) |
| HEY2 | CHF-1 | No | Yes (2) | No | No |
| CSRP1 | CRP1 | Yes (1) | No | No | No |
| MYOCD | Myocardin | No | No | Yes (1) | No |
| TPM2 | β-tropomyosin | Yes (1) | No | No | No |
| MRF2A | MRF2α | No | No | No | No |
| BARX2 | Barx-2b | Yes (1) | No | No | No |
| MEOX2 | Gax | No | No | No | No |
| ELN | Elastin | No | No | No | Yes (2) |
| DES | Desmin | Yes (1) | No | Yes (1) | No |
| DMD | Dystrophin | Yes (1) | No | Yes (1) | No |
| HRC | Histidine-rich calcium BP | No | No | Yes (1) | No |
| PGM5 | Aciculin | No | No | No | No |
| SRF | SRF | Yes (2) | No | No | No |
| MYLKv6 | smMLCK | Yes (2) | No | No | No |
| CALD1 | Heavy caldesmon | Yes (1) | No | No | No |
| VCL | meta-vinculin | Yes (1) | No | No | No |
| ACTN1 | a-actinin | Yes (2) | No | No | No |
| AEBP1 | ACLP | No | No | No | No |
| TPM1 | α-tropomyosin | Yes (1) | No | No | No |
| ARHGAP42 | GRAF3 | No | No | No | No |
There are more than 1200 permutations of the CArG box [24], and previous computational analyses have revealed thousands of conserved CArG boxes in the human genome [25, 26]. Validating the function of SRF-binding CArG boxes has been an important research objective. Historically, transgenic reporter mouse studies were done to assess the functional importance of CArG boxes in such SMC-restricted genes as Tagln [27], Acta2 [28], Telokin [29], Myh11 [30], Csrp1 [31], Kcnmb1 [32], Cnn1 [33], and Lmod1 [34]. These genetic studies offered strong support for the in vivo functionality of CArG boxes and in some cases resulted in the development of novel mouse strains that could direct transgene expression (e.g., Cre recombinase) in a SMC-restrictive manner [35, 36].
More recently, genome-wide studies have been carried out to demonstrate global SRF-binding to CArG elements, albeit studies have been limited to only a few cell types (mostly immortalized cancer cell lines) analyzed under specific cell culture conditions. Thus, ChIP-seq experiments have established SRF-binding to thousands of CArG boxes, including those in proximity to non-contractile genes [37–39]. Many of these CArG boxes were computationally predicted based on the plasticity of this TFBS in what has come to be known as the CArGome [25, 26]; however, there are a number of ChIP-seq-derived SRF binding sites that do not conform to any of the >1200 permutations of the CArG box suggesting we still have much to learn about the binding rules for SRF to this class of TFBS [37, 40].
An important outgrowth of the CArGome has been the computational identification of CArG sequence variants, such as single nucleotide polymorphisms (SNPs). These CArG-SNPs may have consequences for target gene expression in disease states, including vascular disorders. For example, there is a CArG-SNP in the first intron of KLF6 (rs10795076) that severely reduces SRF binding [26]. KLF6 is known to stimulate the pro-angiogenic factor, ALK1, in vascular cells following vascular injury [41]. Therefore, it would be of interest to know whether patients with poor angiogenic responses following myocardial infarction have reduced KLF6 due to the aforementioned CArG-SNP. To date, there are no annotated CArG-SNPs surrounding SMC contractile genes. Rare CArG-SNPs around SMC contractile genes probably do exist but their identification will require extensive sequencing across thousands of families. This “clan genomics” line of inquiry represents a powerful approach to personalized genomics because while the existence of private CArG-SNPs likely is rare, they would probably have a large effect on a phenotype [42]. Finally, it is possible that SNPs create functional CArG boxes in sequences that otherwise would not support SRF binding.
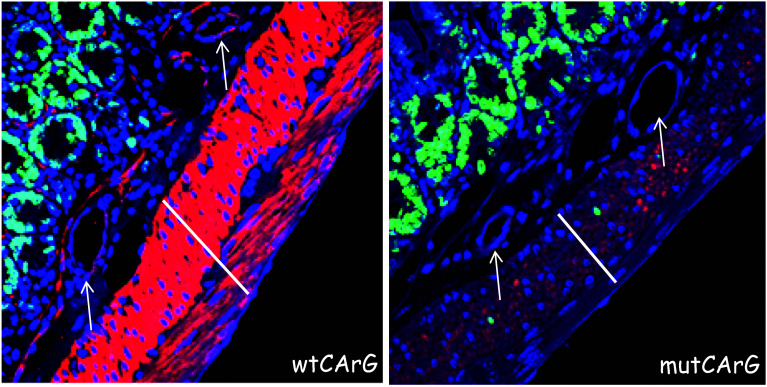
Several challenges and opportunities exist for the next generation of studies on the CArGome. First, we need to define CArG box function under various SMC phenotypic states using ChIP-seq coupled to RNA-seq following SRF knockdown. Second, the function of CArG boxes in pericytes, which have some attributes of SMC, is virtually unchartered territory as we are naïve to the gene expression profile of these cells. Third, there is a need to identify the SRF cofactor (among more than 60) facilitating CArG-dependent target gene expression under various conditions, including those related to perturbations in the SMC differentiated phenotype. Elegant ChIP-seq experiments from the Treisman lab revealed an interaction between SRF and the myocardin-related transcription factors in the serum-induced response of murine fibroblasts [43]. These and other comprehensive genomic studies will provide new and perhaps unexpected findings that will require more reductionist approaches to address such matters as linking SRF-bound CArG boxes to their respective target gene and defining whether a CArG box is functionally important for target gene expression in live animals. The advent of chromatin conformation capture assays [44] and precision-guided genome editing with clustered regularly interspaced short palindromic repeats (CRISPR) technology [45] offer innovative and powerful approaches to study the functionality of CArG boxes in their native genomic milieu. For example, CRISPR-mediated genome editing of a consensus intronic CArG box in the SMC-restricted Cnn1 gene nearly abolished this gene’s mRNA and protein expression in SMCs of the vessel wall [46]. Although other SMC differentiated markers were unaffected by the near abrogation of CNN1 expression, a moderate increase in SMC DNA synthesis was noted suggesting an important, yet poorly understood, role for CNN1 in the maintenance of a SMC quiescent state. The marked reduction of CNN1 upon mutation of an intronic CArG box is also seen in visceral SMCs and subjacent vascular SMCs (Fig. 1). These results, representing the first ever application of precision-guided genome editing of a regulatory element in an animal model, offer definitive genetic proof for the importance of a CArG box in the control of an SRF target gene [46]. CRISPR will likely be the method of choice over traditional approaches (e.g., lacZ reporter, Cre-lox excision) to the functional study of enhancers and TFBS in living animals.
Fig. 1.
CRISPR editing of a TFBS. Immunofluorescence staining for CNN1 protein (red) in vascular SMCs (white arrows) and visceral SMCs (white lines) of small intestine in mice carrying a wildtype CArG box (left) or a mutant CArG box (right). Note the presence of proliferating visceral SMCs at right (green nuclei)
CRISPR technology will also inform us as to the in vivo function of CArG boxes shown previously to be dominant over other, neighboring CArG boxes adjacent to such SMC-restricted genes as Acta2 [28], Tagln [27], and Actg2 [47]. Moreover, CRISPR will be instrumental in modeling CArG-SNPs such as the intronic CArG-SNP in the KLF6 gene in animal models [26]. The latter approach represents an exciting new chapter in genetics because traditional genome wide association studies have defined hundreds of non-coding regulatory variants, but rarely define the functional importance of the sequence variant.
The manner in which CArG-SRF directs SMC gene expression was solved with the seminal discovery of myocardin (MYOCD) in Eric Olson’s laboratory [48]. MYOCD is one of nature’s most powerful coactivators of gene expression [48]. Notably, whereas the conversion of fibroblasts to specific differentiated cell types (e.g., cardiomyocytes) required a cocktail of several gene products [49], conversion of similar cells to SMCs necessitates the ectopic expression of only MYOCD [50–52]. MYOCD can also program human embryonic stem cells to adopt a functional SMC state [53]. Thus, the CArG-SRF-MYOCD triad is the primary transcriptional switch directing the SMC differentiated state. Of note, not all genes expressed in differentiated SMCs contain functional CArG boxes (bold genes in Table 1), and MYOCD is not sufficient to activate some of these [53, 54], although at least one SMC-restricted gene (ITGA8) appears to be activated by MYOCD in a CArG-SRF-independent manner [55]. Understanding how MYOCD performs such SRF-independent actions of transcription is an open area for future investigation. One of the challenges in studying MYOCD is the lack of reliable antibodies to assess endogenous protein expression under various experimental conditions such as vascular injury responses. CRISPR may be of value in this context for epitope tag targeting of the endogenous Myocd locus.
Although the CArG-SRF-MYOCD triad is the dominant driver of SMC differentiation [56], parallel signaling pathways exist that also converge upon the genome to facilitate SMC differentiation associated gene expression. For example, the NOTCH intracellular domain (NICD) is a cofactor that interacts with the DNA binding transcription factor, Recombination signal Binding Protein for immunoglobulin kappa J region (RBPJ), to control the SMC contractile gene program [57, 58]. The core consensus TFBS for RBPJ is TGG/AGAA and recent ChIP-seq experiments revealed the presence of both NICD-dependent and NICD-independent RBPJ binding sites [59]. Key NICD-RBPJ target genes in SMC include ACTA2 [60], MYH11 [61], and the microRNA143/145 gene [62]. The latter genes also contain CArG boxes (Table 1) so there may be synergistic pathways of SMC target gene activation at these gene loci. The SMTNB isoform in the Smoothelin locus is highly specific to vascular SMCs and lacks functional CArG boxes [63], but is activated by NOTCH signaling [61]. Formal genetic proof for a functional RBPJ binding site near the SMTNB locus awaits further study. Recently, the NOTCH ligand, JAG1, was shown to mediate SMC differentiation in the developing mouse embryo by repressing a chondrogenic program of cell differentiation within progenitor cells of the sclerotome; inactivating JAG1 resulted in reduced SMC contractile gene expression and the emergence of several chondrocytic markers [64]. MYOCD can repress the skeletal muscle program of gene expression and promote a SMC contractile phenotype in progenitors of the same sclerotomal compartment of the somite [65]. Together, these studies underscore the existence of parallel transcriptional circuits for the establishment of differentiated SMCs derived from a specific developmental niche.
In addition to CArG and RBPJ sites, several SMC genes contain functional SMAD-binding elements (consensus site is GTCTAGAC) that are the convergence points of TGFβ1 signaling [66] or MEF2 binding sites (consensus element is CTA[A/T]4TAG) which regulate genes in all three muscle lineages [67] (Table 1). Gene inactivation studies have implicated SMAD4 and MEF2C in the proper differentiation of SMCs [68, 69]. Although the CArG and MEF2 TFBS have related sequences, and the associated DNA-binding transcription factors harbor similar functional domains [70], SRF and MEF2 do not exhibit reciprocal DNA-binding activities across CArG and MEF2 elements, nor do they physically interact. Collectively, there are, in addition to CArG-SRF-MYOCD, additional TFBS and associated complexes that can direct at least some aspects of the SMC differentiated state. Important future experiments include elucidating all RBPJ, SMAD, and MEF2 binding sites and associated cofactors during SMC differentiation through ChIP-seq. One important way to filter potentially functional TFBS is through comparative genomics against other species. This has been particularly powerful in defining functional CArG boxes across different animal species [25, 26]. There will also likely be SNPs within MEF2, SMAD, and RBPJ binding sites that could alter SMC gene expression. In this context, a refined comparative genomics tool called phylogenetic module complexity analysis was used to elucidate the function of a TFBS variant in the PPARG gene that appears to contribute to Type II diabetes [71]. This and other emerging technologies will be important to ascertain the clinical relevance of TFBS variants associated with vascular SMC-related disorders.
Endothelial cell TFBS
Like SMCs, ECs have diverse embryonic origins and functionality [72]. However, whereas conversion of cells to a SMC-like phenotype can occur through transient ectopic expression of just one factor (MYOCD), several transcription factors and signaling pathways appear necessary for conversion of cells to an EC-like phenotype, possibly because of the common developmental origin EC progenitors share with those of hematopoietic precursors [73]. Moreover, in vitro specification of some ECs occurs only after protracted periods of time under defined culture conditions. For example, induced pluripotency stem cell factors (e.g., KLF4 and OCT4) can convert human fibroblasts into ECs when cells are primed with various EC growth factors such as VEGF or basic FGF over many weeks [74]. Another report found that an abbreviated treatment protocol resulted in the induction of a protein known as SETSIP, which translocates into the nucleus and binds to and activates the CDH5 promoter within 1 week of conversion of cells to an EC phenotype [75]. Further, several members of the E26 transformation specific (ETS) family of DNA-binding transcription factors (e.g., ETV2, ERG1, and FLI1) could convert amniotic cells into ECs without traversing a pluripotency state if TGFβ signaling was suppressed [76]. More recently, Morita et al. [77] screened 18 transcription factors in a human fibroblast cell line (HFL-1) and discovered ectopic ETV2, in coordination with endogenous FOXC2, was sufficient for EC conversion; addition of VEGF and basic FGF to the culture medium augmented expansion of functional ECs. These latter findings will need to be replicated but from this discussion, it is clear that specification of the EC lineage is complex and not quite as straightforward as that of the MYOCD-directed program of SMC gene expression. Nevertheless, the same approaches used to define TFBS governing SMC differentiation have been employed to define the function of TFBS in ECs and this line of work will be summarized next.
Much of what we have learned about transcriptional control of EC-restricted gene expression originates from transgenic mouse studies using the lacZ reporter gene under control of promoter/enhancer sequences of candidate EC-specific genes. Hence, EC-restricted promoter/enhancer activity has been demonstrated in mice for sequences adjacent to Tek [78], VWF [79], Kdr [80], Cdh5 [81], Tie1 [82], Acvrl1 [83], PROCR [84], ROBO4 [85], FLT1 [86], and Dll4 [87]. As in the SMC field, these transgenic reporter mouse studies paved the way for the development of new and improved mice that can inducibly express transgenes in ECs [88]. Experimental characterization of sequences around EC regulatory regions revealed functional TFBS such as GC-rich binding sites for SP1 [83, 85] and EGR1/3 [86], a site for HIF2A [80], and a composite GATA/TAL1 element [84]. Recently, a functional CArG box was suggested to direct myocardin-related transcription factor binding to and activation of the CDH5 gene in human ECs [89]. The latter result, which will require validation in mice, is consistent with the established role of SRF in maintaining normal EC homeostasis in vivo [90].
Several reports have defined functional ETS elements that control EC-restricted gene expression [83–87]. The ETS family of transcription factors, with more than 25 members, is, like SRF, widely expressed and controls disparate programs of gene expression. However, a subset of ETS factors (ETS1, ETS2, FLI1, ERG, ETV6, ELK3, and ETV2) is of particular importance for EC differentiation and embryonic vascular formation [91]. These family members recognize a core DNA consensus sequence of GGAA/T, a regulatory element that is nearly ten times more frequent in the human genome than the CArG or MEF2 elements. How such a prevalent TFBS confers EC-specific gene expression is complex, but likely relates to a combination of binding partners, their post-translational modification, and flanking sequence content [91]. The latter is relevant to this discussion as an elegant computational study discovered a disproportionately large number of EC-restricted genes harboring a composite ETS-FOXC2 binding site [92]. More than 1500 composite ETS-FOXC2 sites exist in the reference human genome [92]. Recently, a composite ETS-FOXC2 element was characterized in the ECE1 gene and shown to be critical for enhancer activity in transgenic mice [93]. The ECE1 gene encodes for endothelin-converting enzyme, a protein that catalyzes the formation of one of nature’s most powerful vasoconstrictors [94]. Interestingly, the ECE1 enhancer drives arterial EC-specific activity and is co-dependent on a conserved SOX17 TFBS [93]. This highlights the subtle differences that can exist between genes similarly regulated by a common TFBS as a means to direct appropriate levels of expression of a target gene in sub-types of a specific cell lineage, such as arterial EC. Other ETS-containing EC-restricted promoters that exhibit arterial EC-specific activity include Acvrl1 [83] and Dll4 [87]. Whether a specific TFBS exists that directs venular EC-specific promoter/enhancer activity is presently unknown.
Just as CArG-SRF regulates a large number of SMC-restricted genes, ETS factors bound to ETS elements are dominant in controlling EC-specific genes (26/39 in Table 2). Conversely, not all EC-restricted genes harbor ETS or composite ETS-FOXC2 elements implying there exists parallel transcriptional circuits for the differentiation of committed cells to the EC lineage [92] (Table 2). For example, laminar shear stress (LSS) is an essential biomechanical signaling mediator for EC gene expression that antagonizes diseases of the vessel wall such as atherosclerosis [95]. One of the EC-restricted genes lacking functional ETS sites and induced with LSS is KLF2 [96]. KLF2 promoter analyses in vitro support a proximal MEF2 binding site as the LSS-responsive element [97]. KLF2 binding sites (GCCGGG) are similar to those of SP1 and EGR family members; the latter can also be induced by LSS in ECs to direct specific patterns of gene expression [98]. The existence of multiple GC-binding factors complicates the distinction between those factors binding one GC-rich TFBS over another. For example, early in vitro experiments provided evidence for a role of SP1 in the transcriptional regulation of NOS3 [99]. Subsequent work, however, revealed NOS3 to be a direct transcriptional target of LSS-induced KLF2 [100]. This emphasizes the need for deeper analyses of all TFBS in ECs and their binding partners in the regulation of EC gene expression under normal and pathological states. ChIP-seq coupled to RNA-seq following knockdown of a specific ETS factor and determining whether these or other transcription factors synergistically activate EC gene expression will be informative. Further, it will be important to pinpoint regulatory SNPs that fall within EC-restricted genes and ascertain whether such regulatory variants are associated with alterations in EC phenotype. Finally, several EC-restricted genes do not contain any of the major EC-associated TFBS (bold genes in Table 2) and have not been rigorously analyzed in an in vivo setting. Thus, it will be imperative to delineate control elements governing expression of these less characterized EC-restricted genes and determine functional regulatory variants therein.
Table 2.
EC transcriptome and functional TFBS (number)
| Gene symbol | Alias | ETS-FOXC2 site | ETS site | KLF2 site | MEF2 site | CArG box |
|---|---|---|---|---|---|---|
| KDR | VEGFR-2 | Yes (3) | Yes (3) | Yes (1) | No | No |
| CDH5 | VE-Cadherin | Yes (1) | Yes (2) | No | No | Yes (1) |
| DLL4 | Delta-like 4 | Yes (1) | Yes (1) | No | Yes (1) | No |
| TEK | TIE2 | No | Yes (2) | No | No | No |
| TAL1 | SCL | Yes (1) | Yes (1) | No | No | No |
| NFATC1 | NFAT2 | No | No | No | No | No |
| TIE1 | TIE1 | No | Yes (1) | No | No | No |
| VWF | Von Willebrand factor | No | Yes (1) | No | No | No |
| TAL1 | SCL | Yes (1) | Yes (1) | No | No | No |
| PECAM1 | CD31 | No | No | No | No | No |
| PDGFRB | PDGFR-1 | Yes (1) | Yes (1) | No | No | No |
| NOS3 | eNOS | No | Yes (1) | Yes (1) | No | No |
| NFATC4 | NFAT3 | No | No | No | No | No |
| MMRN1 | Emilin-4 | Yes (1) | Yes (1) | No | No | No |
| MEF2C | MEF2C | Yes (1) | Yes (1) | No | No | No |
| LMO2 | RHMO2 | No | Yes (3) | No | No | No |
| KLF2 | LKLF | No | No | No | Yes (1) | No |
| FLT1 | VEGFR-1 | No | Yes (1) | No | No | No |
| ESM1 | Endocan | Yes (2) | Yes (2) | No | No | No |
| FLI1 | EWSR2 | No | Yes (3) | No | No | No |
| EPHB4 | Ephrin Receptor B4 | No | No | No | No | No |
| ENG | Endoglin | Yes (1) | Yes (3) | No | No | No |
| EDN1 | Endothelin - 1 | No | No | No | No | No |
| S1PR1 | EDG1 | No | No | Yes (1) | No | No |
| S1PR3 | EDG3 | No | No | No | No | No |
| CD34 | CD34 | Yes (1) | Yes (2) | No | No | No |
| ANGPT1 | Angiopoietin-1 | No | Yes (3) | No | No | No |
| ANGPT2 | Angiopoietin-2 | No | Yes (3) | No | No | No |
| ROBO4 | MRB | No | Yes (1) | No | No | No |
| EGFL7 | VE-Statin | No | Yes (2) | Yes (1) | No | No |
| THBS1 | Thrombospondin - 1 | No | No | No | No | No |
| NRP1 | Neuropilin-1 | Yes (1) | Yes (1) | No | No | No |
| THBD | Thrombomodulin | No | Yes (3) | No | No | No |
| PODXL | PCLP - 1 | No | No | No | No | No |
| CLDN5 | Claudin - 5 | No | No | No | No | No |
| ECE1 | Endothelin converting enzyme | Yes (1) | No | No | No | No |
| PROCR | EPCR | No | Yes (2) | No | No | No |
| ACVRL1 | ALK1 | No | No | No | No | No |
| NOTCH4 | NOTCH4 | No | Yes (4) | No | No | No |
Summary
Vascular cell identity and function is conferred by distinct TFBS and associated protein complexes that drive unique signatures of gene expression in SMCs and ECs. Traditional means of analyzing vascular cell TFBS such as CArG boxes and ETS elements have been rather arduous and artificial. The revolutionary CRISPR system of genome editing will soon be commonplace in most laboratories to test the in vivo functionality of vascular cell TFBS in their native genomic milieu as well as clinically significant regulatory variants that may disturb normal SMC/EC-restricted gene expression. Ultimately, it may be instructive to screen patients for the presence of TFBS-SNPs as a means of sub-classifying disease states or responsiveness to drug therapies. Where specific vascular TFBS-SNPs contribute to a disease process, genome editing with CRISPR in patient-derived induced pluripotent stem cells may hold therapeutic promise.
Short noncoding RNA genes in vascular cells
Several classes of short noncoding RNA genes exist in mammalian genomes, including small interfering RNAs, small nuclear RNAs, small nucleolar RNAs, and microRNAs. As very little published work exists with respect to most short noncoding RNA classes in vascular cells, this review will focus on the microRNA class only. MicroRNA (miR) genes reside within introns or exons of protein coding or long noncoding RNA genes or between two gene loci, either as single or clustered, co-transcribed miRs. The biogenesis of miR genes is increasingly understood and involves step-wise trimming from a primary transcript to fully mature 20–23 nucleotide guide and passenger strand RNAs through the sequential action of Drosha/DGCR8 and Dicer [101]. The importance of these miR processing proteins for normal SMC [102–104] and EC [105, 106] phenotype has been firmly established. Mature miR transcripts almost always exert their action on target genes through either mRNA destabilization (increasingly understood) or translational repression (less understood) via Watson-Crick base pairing between the 5′ seed sequence of the miR plus a variable number of 3′ nucleotides and the target mRNA sequence found predominantly in the 3′ untranslated region of coding genes. The bias for functional miR binding sites in the 3′ untranslated region appears to relate to the interference of miR activity by active translation [107]. The net effect of most miR gene products is attenuated vascular cell gene/protein expression, though notable exceptions exist. Here, we provide an up-to-date review of several SMC- and EC-associated miR genes that are of significance in regulating the phenotype of each cell type.
Smooth muscle cell miR genes
The initial foray into SMC miR gene discovery was done in the laboratory of Chunxiang Zhang who used microarray to define differentially expressed miR genes in normal versus balloon-injured rat carotid artery [108]. This screen revealed that some miR genes are down-regulated (e.g., miR-145) and others up-regulated (e.g., miR-21) following arterial injury. Inhibiting expression of miR-21 with antisense oligonucleotides blunted the neointimal response in vivo and reduced proliferation of cultured SMCs through elevations in programmed cell death or the de-repression of PTEN [108]. In contrast, another group showed miR-21 to promote human pulmonary artery SMC differentiation via BMP4 or TGFβ1 signaling pathways [109]. These disparate findings are commonplace in biology and underscore the context-dependent effects that occur across different species and types of SMC, particularly those grown in vitro. The miR-221/222 gene cluster was then shown to be induced by growth factors and to mediate SMC proliferation in vitro and in vivo, presumably through the targeting of key negative growth regulators [110, 111]. Another miR gene that promotes SMC proliferation is miR-146a, and reducing this miR gene’s expression could attenuate the neointimal response to injury [112]. The miR-130a gene stimulated vascular SMC proliferation in the context of hypertension, and the growth-arrest homeobox gene product, MEOX1, was shown to be a target of miR-130a [113]. In addition to proliferation, miR genes have been demonstrated to regulate SMC inflammatory phenotypes [114, 115] or help mediate transitions to another cell type [116]. It is relevant to point out that many of the aforementioned miR genes have yet to be inactivated in vascular SMCs, which is necessary to gain definitive proof for their importance in controlling vascular SMC phenotypes. Nevertheless, there is much interest in using miRs as biomarkers of vascular disease and/or developing therapeutic strategies to blunt their action [117]. One of the challenges in blocking the action of a miR is the unanticipated consequence related to the broad spectrum of mRNA targets that any one miR can bind and negatively regulate.
There are a number of miR genes that counteract the growth promoting effects of miR-21, miR-221/222, and miR-146 in SMCs as well as the miR-mediated SMC pro-inflammatory and transdifferentiating phenotypes. For example, MYOCD induces miR-1, a weakly expressed miR gene in SMCs that appears to inhibit SMC proliferation possibly through its repression of the Pim1 target mRNA [118]. There is also some evidence to support miR-1 directing the differentiation of ES cells into a SMC-like phenotype by targeting KLF4 [119]. On the other hand, miR-1 knockout mice exhibit aberrant SMC differentiation marker expression in the developing heart because of sustained expression of a miR-1 target mRNA, Myocd [120, 121]. These results provide insight into how the expression of several CArG-SRF-MYOCD-dependent SMC markers in the heart is attenuated during embryonic development and point to the in vivo interplay that exists between TFBS, miR genes, and the targets of mature miR transcripts.
There are two virtually identical miR-1 genes located on separate chromosomes and each is co-transcribed with highly homologous miR-133a genes. In humans, the miR1-1/miR-133a-2 and miR-1-2/miR133a-1 gene pairs are contained within introns of longer “host genes” (http://genome.ucsc.edu/). Though each miR-133a and miR-1 pair of paralogs is highly similar in sequence, miR-1 and miR-133a exhibit low sequence homology indicating non-overlapping mRNA targets. The induction of both miR-1 genes by MYOCD occurs through conserved SRF-binding CArG boxes located in enhancers that drive expression primarily in cardiac and skeletal muscle cells [122].
As for miR-133a, there are conflicting reports on its role in SMC phenotypic control. An early report from the Olson lab [123] showed that suppression of miR-133a resulted in aberrant expression of SMC markers in the developing heart, much like the aforementioned results seen with miR-1 inactivation. On the other hand, levels of miR-133a have been demonstrated to correlate with the differentiation of adult vascular SMCs, and miR-133a appears to target the SP1 transcription factor [124], a known repressor of the gold standard marker for SMC lineages, Myh11 [125]. Accordingly, the down-regulation of SP1 upon miR-133a overexpression had the effect of increasing Myh11; however, several other CArG-SRF-dependent SMC genes such as Cnn1 and Acta2 were reduced [124]. This highlights rare uncoupling of the expression of markers for the SMC differentiated state. Gain-of-function experiments showed that miR-133a suppressed SMC proliferation and migration, two key processes in the development of vascular lesions [124]. Further studies showed that neointimal formation could be attenuated with miR-133a and exacerbated with anti-miR-133a [124]. While miR-133a expression was noted in vascular SMCs and a clear effect on phenotype was seen upon manipulating its expression, levels of miR-1 were not detected. This raises the challenge of elucidating the differential stability of co-transcribed miRs and the variation in stability of a mature miR transcript under different biological conditions.
Another MYOCD-induced miR defined in the context of SMC phenotypic modulation is miR-29a [126]. This miR transcript reduces SMC migration, an effect proposed as a mechanism for lower neointimal formation seen in the injured arterial wall of mice transduced with adenovirus carrying MYOCD. Evidence was provided to support the Pdgfrb mRNA as a probable target of miR-29a suggesting the observed sessile SMC phenotype stems from miR-29a targeting this growth factor receptor [126]. Inhibition of neointimal formation with MYOCD overexpression provides support for harnessing this cofactor as a therapeutic intervention for the treatment of vascular disorders, and recent experimental data support this concept [127].
One of the most studied SMC miR genes is the bicistronic cluster comprising miR-143 and miR-145. This gene cluster, as with an increasing number of other miR genes, lies within a long noncoding host gene (http://genome.ucsc.edu/, see below). Both miR-143 and miR-145 are co-transcribed in an SRF-dependent manner through a conserved upstream CArG box [128, 129] as well as binding sites for SMAD3/4 [130] and RBPJ [62] (Table 1). The miR-143/145 gene cluster is highly specific for SMCs [129, 131] and promotes SMC contractile gene expression through the repression of several transcription factors that antagonize the SMC contractile gene program [128, 132]. An important aspect of the SMC contractile phenotype is the expression of numerous ion channels and the establishment of proper currents across the SMC membrane. Evidence shows that miR-145 controls key ion channel genes in SMCs [104, 133]. Levels of miR-143/145 are reduced under conditions of SMC phenotypic modulation as seen in acute vascular injury or atherosclerosis [128, 132] and this likely contributes to the attending decrease in SMC contractile markers. On the other hand, levels of miR-145 are elevated in both experimental and clinical pulmonary arterial hypertension. In the experimental setting, disease progression can be attenuated when levels of miR-145 are reduced either genetically or pharmacologically [134]. The disparate direction of miR-145 expression under conditions of SMC phenotypic modulation highlight important differences in the pathogenesis of arterial disease and suggest unique pathways of activation or transcript stability worthy of future study.
It is interesting to note that miR-145 may directly bind and activate Myocd mRNA through a site in the 3′ untranslated region [128]. Similarly, miR-34a appears to bind and activate one of its target mRNAs, SIRT1, and this deacetylase goes on to bind and activate SMC promoters in a CArG-dependent fashion [135]. The miR-34a transcript is associated with SMC differentiation of both mouse and human embryonic stem cells and appears to function upstream of the SRF-MYOCD molecular switch [135]. MicroRNA-mediated activation of target mRNAs is unusual given the near universal repressive mode of action of miRs on target mRNA sequences. The precise mechanism by which miR-145 and miR-34a promote activation of their respective target mRNAs is unknown; possibilities include mRNA stabilization through displacement of mRNA destabilization factors or enhanced translation. There are likely other miR genes with direct activation properties similar to those of miR-145 and miR-34a, particularly given the recent surge in miR gene identification [136].
Ectopic expression of miR-145 reduces neointimal burden in vivo [132, 137, 138] and partially rescues the reduction in SMC contractile genes seen with loss of Dicer in vitro [104]. Several independent knockouts of miR-143/145 have been carried out with varying results [129, 131, 137]. In general, loss in miR-143/145 compromised expression of SMC contractile genes and caused a mild hypotensive state. On the other hand, there were observed differences in lesion formation; two studies reported mild increases in neointimal formation [131, 137] whereas another study showed the lack of a neointima upon acute vascular injury [129]. Variations in the phenotype of miR-143/145 null mice likely relates to genetic background, the mode of vascular injury, and perhaps subtle differences in the loss of genetic information around the locus following Cre-mediated excision. The effect of miR-34a on the neointimal response to injury is unknown; however, there may be no effect since the influence of miR-34a on SMC differentiation is only seen in the context of embryonic stem cell differentiation to the SMC lineage [135]. Given the effects of miR-145, particularly, on SMC phenotype and vascular lesion formation, there may someday be a direct test of its efficacy in limiting lesion formation [138]. Challenges exist, however, in fully elucidating the targets of miR-145 (or any miR brought to the clinic) in order to gauge potential off targets and attending toxicity issues. Other miR genes that effect changes in the SMC differentiated state include miR10a [139], miR-1 [140], and miR-26a [141]. Table 3 summarizes many miR genes implicated in SMC differentiation and phenotypic changes associated with vascular disease.
Table 3.
SMC miR function and targets
| miR | Function | Target mRNA | Species | References |
|---|---|---|---|---|
| 1 | ↓ SMC proliferation | PIM1 | Hsa | [118] |
| 1 | ↑ SMC differentiation in ES cells | Klf4 | Mmu | [119] |
| 10a | ↑ SMC differentiation in ES cells | Hdac4 | Mmu | [139] |
| 124 | ↓ SMC proliferation/↑ SMC differentiation | Nfatc1 | Hsa | [235] |
| 125b | ↑ SMC inflammation | Suv39h1 | Mmu | [236] |
| 130a | ↑ SMC proliferation | Meox1 | Rno | [113] |
| 132 | ↓ SMC proliferation/↓ neointima | Lrrfip1 | Rno | [237] |
| 133a | ↓ SMC proliferation/↓ SMC migration | Sp1 | Rno | [124] |
| 138 | ↓ SMC apoptosis | Mst1 | Rno | [238] |
| 143/145 | ↓ SMC proliferation/↑ SMC differentiation | Kfl4/Elk1/Camk2d | Mmu | [128] |
| 143/145 | ↑ SMC differentiation/↓ neointima | Kfl5 | Rno | [132] |
| 143/145 | Cytoskeletal dynamics | Klf4/Klf5/Add3 | Mmu | [129] |
| 146a | ↑ SMC proliferation/↑ neointima | Kfl4 | Rno | [112] |
| 195 | ↓ SMC proliferation/↓ SMC migration | CDC42/CCND1 | Hsa | [239] |
| 200 | ↑ SMC inflammation in diabetes | Zeb1 | Mmu | [114] |
| 204 | ↓ SMC proliferation/↑ SMC apoptosis | SHP2 | Hsa | [240] |
| 205 | ↓ SMC calcification | Runx2 | Hsa | [241] |
| 206 | ↓ SMC proliferation/↑ SMC differentiation | Notch3 | Hsa | [242] |
| 21 | ↑ SMC differentiation | PDCD4 | Hsa | [109] |
| 21 | ↓ SMC differentiation/↑ neointima | Pten | Rno | [108] |
| 210 | ↓ SMC apoptosis | E2F3 | Hsa | [243] |
| 221/222 | ↑ SMC proliferation/↓ SMC differentiation | KIT/CDKN1B | Hsa | [110] |
| 221/222 | ↑ SMC proliferation/↑ neointima | Cdkn1b/Cdkn1c | Rno | [111] |
| 22 | ↑ SMC differentiation | Mecp2 | Mmu | [244] |
| 224 | ? | TCF21 | Hsa | [168] |
| 24 | ↓ SMC inflammation/↓ SMC migration | CHI3L1 | Hsa | [115] |
| 26a | ↑ SMC proliferation/↓ SMC differentiation | SMAD1 | Hsa | [141] |
| 29a | ↓ SMC migration | Pdgfrb | Mmu | [126] |
| 30b/30c | ↓ SMC calcification | RUNX2 | Hsa | [116] |
| 31 | ↑ SMC proliferation | Lats2 | Rno | [245] |
| 31 | ↓ SMC differentiation | CREG | Hsa | [246] |
| 322 | ↓ SMC proliferation/↓ SMC migration | Ccnd1/Ppp3 cc | Rno | [247] |
| 34a | ↑ SMC differentiation | Sirt1 | Mmu | [135] |
| 365 | ↓ SMC proliferation | Ccnd1 | Rno | [248] |
| 638 | ↓ SMC proliferation/↓ SMC migration | NR4A3 | Hsa | [249] |
| 663 | ↓ SMC proliferation/↑ SMC differentiation | JUNB | Hsa | [250] |
| 96 | ↓ SMC differentiation | TRB3 | Hsa | [251] |
| Let7a | ↓ SMC proliferation/↓ SMC migration | Myc/Kras | Rno | [252] |
Endothelial cell miR genes
The initial reporting of EC-associated miR genes was from the laboratory of Giuseppe Rainaldi who used a microarray to define 15 abundantly expressed miR genes in human umbilical vein ECs, including miR-221/222 which appeared to be necessary for normal EC migration and tube formation through the repressive action on KIT [142]. Since laminar shear stress (LSS) is central to a normal EC phenotype, there has been much work devoted to the definition of miR genes responsive to changes in blood flow [143, 144]. For example, the miR-101 and miR-19a genes are induced by LSS and promote a less proliferative phenotype in ECs by repressing expression of MTOR and CCND1, respectively [145, 146]. The SMC enriched miR-143/145 cluster is induced by LSS in a KLF2-dependent manner and is transported via exosomes from ECs to SMCs to confer an atheroprotective SMC phenotype [147]. AMP activated protein kinase alpha 2 subunit phosphorylates p53 which appears to up-regulate miR143/145 in ECs via post-transcriptional processing [148]. Elevated EC miR-143/145 targets angiotensin-converting enzyme, thus, maintaining normal vascular homeostasis [131, 148]. Remarkably, TGFβ stimulates the transfer of miR-143/145 from SMCs to ECs through “tunneling nanotubes” and this has the effect of reducing EC proliferation and tubulogenesis thereby implicating the miR-143/145 cluster in the control of angiogenesis [149]. MicroRNA-10a, which is also active in SMCs (Table 3), is induced by LSS and exerts an anti-inflammatory phenotype in ECs by targeting regulators of the IκBα gene. Accordingly, miR-10a levels are low in atherosusceptible regions of the vasculature where disturbed flow patterns exist [150]. Recently, miR-10a was shown to be transferred from quiescent ECs to monocytes where the NF-kB pathway of inflammation could be suppressed [151]. Thus, the movement of mature miRs (and other RNA products) from one vascular cell type to another cell type represents a juxtacrine/endocrine signaling mechanism of fine-tuning local or remote programs of gene expression that may function to maintain or perturb vascular homeostasis.
There are some miR genes whose expression is reduced with LSS leading to EC phenotypic modulation. For example, LSS-mediated attenuation of miR-663 is linked to increased monocyte adhesion to ECs, and blocking miR-663 with a locked nucleic acid antagonist reversed this phenotype [152]. The EC enriched miR-92a gene can also be reduced with LSS leading to elevations in KLF2, a transcription factor critical for mediating an EC differentiated state [153]. Another miR that is suppressed in ECs by LSS is miR-34a [154]. Here, low miR-34a promotes a normal EC differentiated phenotype (opposite to what it does in SMC precursor cells, above), presumably through reduced degradation of its target, SIRT1, thereby inhibiting adhesion molecule expression (VCAM-1) and subsequent monocyte binding and attending inflammatory events on the surface of ECs [154]. Taken together, these few examples highlight the important role LSS has on miR gene expression in ECs and the regulation of EC differentiation or phenotypic modulation.
The most abundant miR gene in ECs is miR-126 [155, 156]. This miR transcript is co-expressed with the EC-restricted gene EGFL7 (Table 2) through two ETS elements [157] and effects several biological processes through context-dependent transcript targeting. An early study showed that miR-126 repressed the vascular cell adhesion molecule, thus maintaining an anti-inflammatory phenotype in mature ECs [155]. Gene knockout experiments demonstrated miR-126 to play an important role during developmental angiogenesis via negative regulation of SPRED1, an anti-angiogenic factor [156, 158]. Of note, the vascular defect seen with miR-126 inactivation is very similar to the defective sprouting angiogenesis phenotype observed with initial knockout studies of Egfl7 [159]. It was subsequently discovered, however, that the Egfl7 null phenotype was a consequence of inadvertent disruption of the miR-126 gene [160]. This highlights the importance of carefully thought out gene inactivation strategies and revisiting previously defined phenotypes from the conditional knocking out of protein coding genes where Cre-mediated deletion of exons and introns may also have disrupted non-coding genes or key TFBS.
Important phenotypic responses of ECs during development and disease processes include their proliferation and migration, which lead to the augmentation of vascular channels from pre-existing vessels, a process known as angiogenesis. Not surprisingly, the majority of miR genes studied in ECs exerts some effect on the establishment of this critical biological process (Table 4). Accordingly, the miR-17-92 cluster is expressed abundantly in ECs, and miR-92a targets pro-angiogenic genes, including Itga5 [161]. Over-expressing miR-92a inhibits angiogenic responses both in cultured ECs and an animal model of neovascularization [161]. Interestingly, miR-34a attenuates an angiogenic response through the induction of EC senescence, which is linked to miR-34a-mediated repression of SIRT1 [162]. A major driver for the angiogenic response is hypoxia and some miR genes are induced by this powerful stimulus. For example, miR-424 (ortholog is miR-322 in mice) is induced by hypoxia in human ECs and stabilizes HIF1Α through the targeting of CUL2, a component of the ubiquitin ligase system [163]. Transactivation of miR-424 was also shown to occur through the ETS factor, SPI1 (aka PU.1), which in turn was activated by RUNX1 [163]. Since both SPI1 and RUNX1 are transcription factors found in hemogenic endothelium, the phenotypic modulation of differentiated ECs in this context may involve their transition through this specialized type of EC during the angiogenic response. There is a major effort towards developing therapeutic interventions targeting specific miR genes for measured angiogenic responses under ischemic conditions, most notably peripheral artery disease associated with type II diabetes [164]. A challenge in the development of miR-based therapy for the promotion of angiogenesis is limiting the response to the desired tissue and avoiding the activation of dormant tumors.
Table 4.
EC miR function and targets
| miR | Function | Target mRNA | Species | References |
|---|---|---|---|---|
| 10 | ↑ Angiogenesis | FLT1 | Hsa | [253] |
| 10a | ↓ EC inflammation | MAP3K7/BTRC | Hsa | [150] |
| 100 | ↓ Angiogenesis | Mtor | Mmu | [254] |
| 101 | ↑ With LSS/↓ EC proliferation | MTOR | Hsa | [145] |
| 107 | ↑ With hypoxia/↓ EPC differentiation | Arnt | Rno | [255] |
| 126 | ↓ EC inflammation | VCAM | Hsa | [155] |
| 126 | ↑ Angiogenesis | Spred1 | Mmu | [156] |
| 126-5p | ↑ EC proliferation | Dlk1 | Mmu | [256] |
| 130a | ↑ Angiogenesis | MEOX1/HOXA5 | Hsa | [257] |
| 132 | ↑ Angiogenesis | RASA1 | Hsa | [258] |
| 146 | ↓ EC inflammation | ELAV1 | Hsa | [259] |
| 147b | ↓ EC permeability | ADAM15 | Hsa | [260] |
| 149 | ↓ Angiogenesis | GPC1/FGFR1 | Hsa | [261] |
| 15a | ↓ Angiogenesis | FGF2/VEGF | Mmu/Hsa | [262] |
| 152 | ↓ EC proliferation/↓ EC migration | ADAM17 | Hsa | [263] |
| 155 | ↓ Vasodilation | NOS3 | Hsa | [264] |
| 155 | ↓ EC inflammation | BACH1 | Hsa | [265] |
| 17-3p | ↓ Angiogenesis | KDR | Hsa | [266] |
| 181a | ↑ EC differentiation | PROX1 | Hsa | [267] |
| 181b | ↑ EC inflammation | KPNA4 | Hsa | [268] |
| 19a | ↑ With LSS/↓ EC proliferation | CCND1 | Hsa | [146] |
| 200b | ↓ Angiogenesis | ETS1 | Hsa | [269] |
| 200c | ↓ EC proliferation/↑ EC apoptosis | ZEB1 | Hsa | [270] |
| 200c | ↑ EC differentiation | ZEB1 | Hsa | [271] |
| 21 | ↑ EC differentiation from iPSC | Pten | Mmu | [167] |
| 21 | ↑ With LSS/↓ EC apoptosis/↑ EC NO | PTEN | Hsa | [272] |
| 210 | ↑ With hypoxia/↑ EC migration | EFNA3 | Hsa | [273] |
| 214 | ↓ Angiogenesis | QKI | Hsa | [274] |
| 217 | ↓ Angiogenesis/↑ EC senescence | SIRT1 | Hsa | [275] |
| 218 | ↓ Vasculogenesis | ROBO1/2/GLCE | Hsa | [276] |
| 221 | ↑ Angiogenesis | ZEB2 | Hsa | [277] |
| 221 | ↑ Angiogenesis | Cdkn1b/Pik3r1 | Dre | [278] |
| 223 | ↓ Angiogenesis/↓ EC proliferation | ITGB1 | Hsa | [165] |
| 23/27 | ↑ Angiogenesis | Spry2/Sema6a | Mmu | [279] |
| 29a | ↑ Angiogenesis | PTEN | Hsa | [280] |
| 30b | ↑ Angiogenesis | Dll4 | Dre | [281] |
| 34a | ↑ EC inflammation | SIRT1 | Hsa | [154] |
| 424 | ↑ With hypoxia/↑ angiogenesis | CUL2 | Hsa | [163] |
| 492 | ↓ EC migration | RETN | Hsa | [282] |
| 503 | ↓ EC proliferation/↓ EC migration | Ccne1/Cdc25a | Mmu | [283] |
| 6086/6087 | ↓ EC differentiation | CDH5/ENG | Hsa | [284] |
| 663 | ↓ With LSS/↑ EC inflammation | ? | Hsa | [152] |
| 92a | ↓ Angiogenesis | Itga5 | Mmu | [161] |
| 92a | ↓ With LSS/↑ EC differentiation | KLF2 | Hsa | [153] |
Several additional miR genes play a role in the maintenance of the normal EC phenotype. The miR-223 gene, which is abundant in freshly dispersed ECs but then is transcriptionally down-regulated upon further growth in vitro, was shown to promote EC quiescence through the targeting of integrin beta 1, thus minimizing the action of growth factor signaling [165]. In human embryonic stem cells induced to an EC lineage, several miR genes (miR-99b, miR-181a/b) were induced and promoted the expression of EC restricted genes (CDH5 and PECAM1) as well as increases in nitric oxide production and the potentiation of revascularization in a hind limb ischemia model [166]. Conversely, knockdown of Dicer blocked EC gene expression and normal morphology [166], consistent with pioneering Dicer knockdown studies in cultured adult ECs [105, 106]. Finally, the miR-21 gene was shown to facilitate EC differentiation and angiogenic potential in an induced pluripotent cell model of EC differentiation [167]. Table 4 summarizes many miR genes expressed in ECs, their associated function, and validated targets.
Summary
A problematic task in studying the biological significance and potential therapeutic efficacy of miR genes in EC/SMC biology continues to be accurate target gene identification. This is further complicated by the recent surge in novel, human miRs showing Dicer-dependency and bonafide target RNA binding [136]. Thus, reductionist studies will be needed to assess new human-specific miR genes in vascular cells and their modulated expression during vascular cell differentiation and phenotypic modulation. The fact that more than 6500 miRs now exist in the human genome (a likely underestimate of the true total) [136] compels investigators to be more mindful of the increasingly complex interplay between multiple miR genes, including those modulated by LSS and hypoxia, over individual target transcripts as well as the ever-present need to elucidate the mechanisms underlying miR-mediated target transcript destabilization or translational arrest. As with TFBS, elucidating nucleotide variants in either miR genes or their target sequences, and the functional importance therein, represent potentially fruitful areas of future research. For example, a cardiovascular disease-associated SNP (rs12190287 at 6q23.2) located in the 3′ untranslated region of the TCF21 gene creates a favorable seed binding sequence for miR-224 facilitating repression of the target mRNA and perturbed expression of this important transcription factor in human coronary artery SMC [168].
Long noncoding RNA genes in vascular cells
The emergence of high-throughput transcriptomics heralded the concept of pervasive transcription and the expansive class of long noncoding RNA (lncRNA) genes [169–171]. Indeed, the number of lncRNA genes in the human genome is soaring with recent totals surpassing all known protein-coding and miR genes [172]. LncRNA genes are transcribed by RNA polymerase II, exist as single or multi-exonic transcripts, undergo 5′ capping and splicing, and can either be polyadenylated or non-polyadenylated [173]. In general, compared to protein-coding genes, lncRNA genes are poorly conserved and weakly expressed. Nevertheless, lncRNA genes are regulated by external stimuli such as hypoxia [174] and inflammatory agonists [175] and exert important functions even at low copy number [176]. In fact, recent knockout studies have clearly demonstrated a biologically relevant role for many lncRNAs [177]. In this context, whereas miR genes almost always function to down-regulate mRNA and/or protein expression, lncRNA genes exhibit unpredictable and increasingly diverse functions related to the induction or suppression of transcription, translation, and signaling as well as the modification of chromosome structure and function and the formation of nuclear microdomains [178, 179]. It is possible that the varied functions associated with lncRNAs, while still in the embryonic stages of our understanding, may someday rival those of protein coding genes.
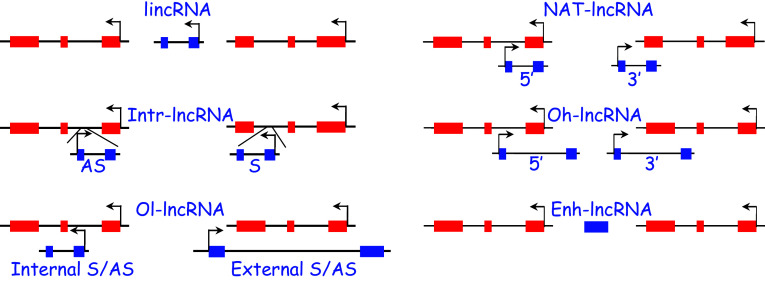
Several sub-classes of lncRNAs exist based on their genomic position relative to other gene loci. These include promoter-containing long intervening noncoding RNAs (lincRNAs), which do not overlap with other genes and are often found in what was once referred to as gene deserts [180]; natural antisense transcript lncRNAs (NAT-lncRNAs) whose one or more exons overlap with exons of an opposing transcribed gene [181]; intronic lncRNAs (Intr-lncRNAs) [182]; and the less numerous 5′ or 3′ overhanging lncRNAs (oh-lncRNAs) and internally or externally overlapping lncRNAs (ol-lncRNAs) (Fig. 2). Other subclasses of lncRNA genes include enhancer RNAs, which play an ill-defined role in directly mediating gene transcription [183], some pseudogenes, whose RNA functions are only beginning to be realized [184], and circular RNAs [185].
Fig. 2.
LncRNA classification based on orientation. In each example, exons of the lncRNA gene are blue while those of neighboring genes are red. Bent arrows indicate the direction of transcription. See text for further details. S sense, AS antisense
One method of functionally defining lncRNA genes is based on their directionality of transcription. For example, a large number of lncRNAs are transcribed in the antisense (or opposite strand, os) orientation to the nearest transcription unit; these may be NAT-lncRNAs (e.g., MEF2C-AS1 and GHRL-OS) or non-NAT-lncRNAs (e.g., DIAPH2-AS1 and DNM3-OS). Expression of antisense-sense gene pairs may be concordant or discordant. Concordantly expressed gene pairs may share similar regulatory regions (such as promoters). Discordant expression of antisense lncRNA-sense RNA gene pairs may reflect the negative regulation of the former on the latter. For example, the NAT-lncRNA, APOA1-AS, recruits the demethylase, LSD1, over the APOA1 gene and represses its expression both in vitro and in vivo [186]. Thus, a first approximation of the function of an antisense lncRNA may be through an examination of its expression with respect to the overlapping mRNA. It is important to stress that while such correlative data have often been used to advance the function of an antisense lncRNA, deeper analyses are required to formally elucidate the function of any lncRNA (see “Summary” below).
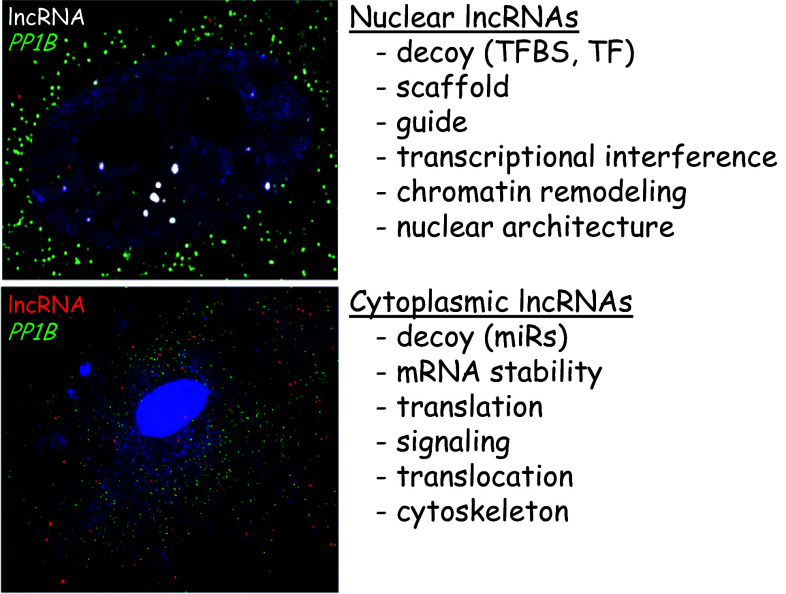
Another method of functionally categorizing lncRNA genes relates to their localization within a cell as demonstrated by RNA-fluorescence in situ hybridization (RNA-FISH, Fig. 3). Accordingly, nuclear localized lncRNAs function primarily in the regulation of gene transcription either in cis (local) or trans (distal) by acting as guides or scaffolds that localize chromatin remodeling factors such as the polycomb repressor complex to specific gene loci. Nuclear lncRNAs may also function as decoys for key transcription factors or TFBS or through the very act of being transcribed [179]. Cytoplasmic lncRNAs can mediate changes in the level of gene/protein expression by acting as decoys (or competing endogenous RNAs) for microRNAs [187] as well as direct effectors of mRNA stability or translation [188, 189]. Cytoplasmic lncRNA genes have also been shown to regulate nuclear translocation of transcription factors and mediate critical signaling pathways via RNA–protein interactions [190–192]. While lncRNAs can suppress or stimulate transcription, translation and signaling, there are virtually no known features of lncRNAs that would be predictive of their function in a cell. One notable exception is a recent report that defined a sequence element (AGCCC plus A/T at -8 and G/C at -3) shown to be both necessary and sufficient for nuclear localization of a lncRNA [193]. This result suggests there may be consensus nucleotide sequences (much like protein domains) that portend lncRNA function. Given the low sequence conservation of lncRNAs across species, the latter is likely to be the exception rather than the rule. Consequently, although it may now be convenient (even necessary) to sort lncRNAs based on genomic and cellular locale as well as their transcriptional directionality with respect to a neighboring gene locus, these categorizations will not be predictive of lncRNA function. It is possible that families of lncRNAs will emerge based on related structural characteristics that may serve as non-sequence specific interaction domains for particular RNA-binding proteins. In the end, the functions of lncRNAs may have more to do with the complex structures they assume in association with other macromolecules than their precise nucleotide sequence content.
Fig. 3.
LncRNA classification based on spatial localization. Single molecule RNA-FISH of a nuclear lncRNA (white dots in the DAPI-stained nucleus, upper panel) versus a cytoplasmic lncRNA (red dots around DAPI-stained nucleus, lower panel). Both panels depict RNA-FISH in human coronary artery SMCs. The green dots represent RNA-FISH of a housekeeping gene which is used to demarcate cell borders for quantitative purposes
In general, lncRNA genes have short open reading frames (<100 amino acids) and, by definition, do not encode for proteins, a concept that has support from ribosomal profiling studies [194]. However, recent studies have demonstrated the presence of small, conserved open reading frames in annotated lncRNAs that encode for previously unrecognized peptides [195, 196]. Thus, an important undertaking will be to carefully assess whether a mRNA is in fact masquerading as a lncRNA. The idea that human-specific lncRNAs encode for novel peptides is an intriguing concept that may elucidate subtle differences in vascular physiology between humans and other mammals. However, there are no known lncRNAs encoding for a human-specific SMC- or EC-associated peptide.
Whereas much is known about miR genes in vascular cells, there are only a handful of vascular lncRNA genes described at this time, though this will change dramatically in the coming years as more RNA-seq data accumulate. Here, we provide an up-to-date summary of experimentally defined lncRNAs in vascular cells and their putative function in regulating vascular cell phenotype.
Smooth muscle cell lncRNA genes
The first report of a lncRNA in vascular SMCs occurred more than 20 years ago with the cloning of H19 [197]. H19 is a conserved, imprinted NAT-lncRNA that serves as a host gene for miR-675 (http://genome.ucsc.edu/). H19 is expressed in SMCs of developing arteries, but then levels are reduced in adult vessels until atherosclerotic lesions are manifest or following acute vascular injury [198, 199]. Although the precise function of H19 in SMCs is unknown, recent data suggest a role as a sponge RNA for the let-7 family of miRs [200]. In this context, overexpression of let-7g in oxidized LDL-treated SMCs attenuated autophagy, apoptosis, and reactive oxygen species through the targeted repression of LOX1 [201]. Reduced expression of H19 in vascular lesions may therefore limit the local effects of oxidized LDL in intimal SMCs by permitting elevated expression of let-7g, though it remains to be determined whether let-7g levels are increased in intimal cells of SMC origin. Of note, H19 overlaps in the antisense orientation with a tumor suppressor (HOTS); no studies have yet examined the interplay between these transcripts in vascular SMCs [202]. Another well-known lncRNA expressed in SMCs is CDKN2B-AS1 (aka ANRIL), which resides on chromosome 9p21, a hotspot for variations linked to cardiovascular disease and cancer [203]. This nuclear antisense ol-lncRNA mitigates neighboring cell cycle gene expression through the recruitment of the polycomb repressor complex [204]. Human SMCs derived from patients carrying SNPs in the ANRIL locus exhibit elevated cell proliferation in vitro [205], a finding supported by genetic deletion of orthologous sequences in the mouse [206]. It will be important to determine whether correction of variants in ANRIL, using CRISPR, rescues the SMC growth phenotype. SNP variants in ANRIL highlight the critical importance of defining functional variations in other noncoding sequences in ECs and SMCs.
A conserved antisense transcript was discovered and found to have at least 3 untranslated exons overlapping exons at the 3′ end of the NOS3 gene [207]. This antisense transcript may, therefore, be categorized as a NAT-lncRNA. Expression of the sense NOS3/NAT-lncRNA gene pair is discordant in SMCs with higher levels of the antisense lncRNA over NOS3. Similarly, in situ hybridization reveals little to no overlap in co-expression of these two transcripts. Knockdown of the NAT-lncRNA results in increases in NOS3 providing a post-transcriptional mechanism for low-level expression of NOS3 in SMCs [207]. The NOS3 NAT-lncRNA, known as ATG9B, is a rare example of a transcript annotated as both a protein-coding gene (involved in the regulation of autophagy) and a NAT-lncRNA gene (http://genome.ucsc.edu/). There are likely to be additional examples of such bifunctional RNA transcripts given the extensive number of sense-antisense gene pairs in the human genome.
A few lncRNAs have been defined in SMCs using next generation sequencing. The first reported use of RNA-seq for the discovery of SMC lncRNAs was in rat aortic SMCs stimulated with angiotensin II. This study revealed numerous lncRNA genes including one that is a host gene for the miR-221/222 cluster [208]. This lncRNA, provisionally named here as MIR221HG, appears to be conserved in the human genome. Knockdown of MIR221HG resulted in reduced miR221/222 expression and attenuated SMC growth responses in rat SMCs [208]. An increasing number of lncRNAs are found to be host genes for miRs (e.g., Fig. 4). Whether these host lncRNA genes have functions independent of the microRNA genes they harbor is unclear at this time, but should be a fruitful avenue for future investigation.
Fig. 4.
UCSC Genome Browser screenshot of a host gene for the miR-143/145 cluster
In an effort to define unannotated lncRNA genes in human SMCs, RNA-seq was done in human coronary artery SMCs and an antisense lncRNA, called SENCR, was found to overhang the 5′ end of the EC-restricted FLI1 gene [209]. SENCR exons do not overlap those of FLI1 and so SENCR is not a NAT-lncRNA and does not appear to regulate FLI1 expression. The largely cytoplasmic localization of SENCR supports an extra-nuclear function. To gain insight into the function of SENCR, RNA-seq experiments were done following its knockdown in human coronary artery SMCs. This approach showed that MYOCD and its contractile target mRNAs were reduced with attenuated SENCR suggesting some loss in SMC differentiated properties. On the other hand, numerous genes involved in cellular migration were elevated upon SENCR knockdown. Accordingly, SMCs were shown to be hyper-motile with knockdown of SENCR, a phenotype that could be completely rescued by simultaneous knockdown of motility genes that were up-regulated upon knockdown of SENCR. Thus, SENCR appears to play some role in the maintenance of the sessile, SMC contractile phenotype. There is a 61 amino acid open reading frame in SENCR that, if translated, would encode a novel small peptide. However, no evidence exists to support an encoded peptide within the SENCR transcript. Moreover, there are no known RNA-binding proteins associated with SENCR. The latter represents an important gap in understanding the function of this new vascular lncRNA.
A microarray revealed hundreds of lncRNAs differentially expressed between control and varicose saphenous veins, and several were validated by qRT-PCR. Many represent antisense lncRNAs and expression analysis revealed concordant expression with neighboring mRNAs [210]. In a follow-up report, reduced expression of the snoRNA-containing GAS5 lncRNA in varicose veins correlated with increases in SMC proliferation and migration, two contributing factors to this pathology; overexpression of GAS5 reduced SMC proliferation [211]. Importantly, RNA pulldown experiments showed an interaction between GAS5 and a calcium-dependent RNA binding protein called ANXA2. The enhanced proliferation and migration of SMCs with attenuated GAS5 could be rescued upon simultaneous knockdown of ANXA2 suggesting that GAS5 functions in some manner to regulate the expression and/or activity of ANXA2 [211]. Curiously, a single-exon, NAT-lncRNA (GAS5-AS1) exists at the 3′ end of GAS5 though no investigation of this lncRNA was reported. Another NAT-lncRNA (HAS2-AS1), however, was shown to directly mediate the transcription of the overlapping HAS2 gene in human SMCs by recruiting the p65 subunit of NFκB to the adjacent HAS2 promoter [212]. HAS2-mediated increases in the glycosaminoglycan, hyaluronan, are found in atherosclerotic lesions, suggesting that therapeutic targeting of HAS2-AS1 could have a desirable impact in models of atherosclerosis. Another atherosclerosis-related lncRNA is lincRNA-p21, which was reduced in the ApoE null model of atherogenesis as well as human atherosclerotic lesions [213]. Gain-of-function studies revealed that lincRNA-p21 represses SMC proliferation while loss-of-function in lincRNA-p21 exaggerated the neointimal response to acute injury. RNA pulldown experiments showed an interaction between lincRNA-p21 and the E3 ubiquitin-protein ligase, which had the effect of de-repressing p53-dependent target genes, including several apoptosis-related genes [213]. Thus, targeting SMCs with lincRNA-p21 could be of clinical value in chronic or acute vascular diseases. Another recent study showed that the NAT-lncRNA, HIF1A-AS1, was increased in the serum of patients with aortic aneurysms; targeted reduction of this lncRNA in SMCs reduced apoptotic related genes such as caspase 3 and caspase 8 while increasing BCL2 [214]. Neither the mechanism of HIF1A-AS1 in regulating SMC apoptosis nor its effects on the overlapping HIF1A mRNA was assessed. Table 5 lists the lncRNA genes that have been identified and studied in vascular SMCs.
Table 5.
SMC lncRNAs
| LncRNA | Type/localization | Putative function in SMC | References |
|---|---|---|---|
| H19 | NAT-lncRNA/cytoplasmic | ? | [197] |
| CDKN2B-AS1 | Antisense external ol-lncRNA/nuclear | ↑ SMC proliferation? | [205] |
| ATG9B | NAT-lncRNA/? | ↓ NOS3 | [207] |
| MIR221HG | LincRNA/? | ↑ SMC proliferation | [208] |
| SENCR | 5′ Antisense oh-lncRNA/cytoplasmic | ↑ SMC differentiation/↓ migration | [209] |
| HIF1A-AS1 | NAT-lncRNA/? | ↑ SMC apoptosis | [214] |
| LincRNA-p21 | LincRNA/nuclear | ↓ SMC proliferation | [213] |
| HAS2-AS1 | NAT-lncRNA/? | Matrix regulation | [212] |
| GAS5 | LincRNA/cytoplasm | ↓ SMC proliferation/↓ migration | [211] |
Endothelial cell lncRNA genes
The first lncRNA to be described in ECs was an antisense transcript with several exons overlapping exons of the EC-restricted NOS3 gene [207]. Unlike SMCs, expression of the overlapping transcript to this NAT-lncRNA is low in ECs whereas NOS3 mRNA is high. Interestingly, overexpression of the overlapping NAT-lncRNA sequences in ECs attenuated levels of NOS3 protein with little effect on NOS3 mRNA suggesting a block in translation of the processed NOS3 transcript [207]. Subsequent studies showed that hypoxia augmented expression of the NOS3 antisense lncRNA (now annotated as a noncoding isoform of ATG9B) in both cultured ECs and rat aortic tissue [215]. Hypoxia-induced decreases in NOS3 were blocked when the NAT-lncRNA, ATG9B, was down-regulated prior to the hypoxic stimulus in cultured ECs [215]. These results support a post-transcriptional mechanism for the known decrease in NOS3 under hypoxic conditions.
As ECs are the primary effector cells for angiogenesis, it should be no surprise that lncRNA genes, like miR genes, influence the angiogenic response whether developmental or pathological. Accordingly, the first angiogenesis-related lncRNA discovered in ECs was a NAT-lncRNA that overlaps the 3′ untranslated region of TIE1 [216]. This lncRNA, called TIE1-AS, is conserved in mouse, human and zebrafish and its expression and spatial localization are concordant with that of the overlapping TIE1 mRNA in developing zebrafish. Interestingly, bioinformatic and hybridization assays support a direct association between TIE1-AS and TIE1 mRNA. Consistent with this interaction, forced expression of TIE1-AS could reduce levels of TIE1 mRNA, and specific defects in EC junctional complexes and vascular patterning were seen in zebrafish injected with TIE1-AS. Interestingly, expression of TIE1-AS was reported to be elevated in human hemangiomas suggesting this lncRNA could be a viable target of therapy for vascular malformations [216].
More recently, RNA-seq analysis of human umbilical vein ECs revealed high level expression of MALAT1 (aka NEAT2), a conserved nuclear lincRNA that could also be induced by hypoxia [217]. Knockdown of MALAT1 in ECs evoked a pro-migratory phenotype in vitro and reduced capillary density and blood flow in a hind limb ischemia model. The latter phenotype was suggested to occur through reduced proliferation of stalk cells [217]. While gain-of-function studies were not conducted, an independent study revealed increases in EC viability with overexpression of MALAT1 [218]. In this context, elevated levels of MALAT1 were observed upon treatment of ECs with high levels of glucose, a stimulus that increases inflammatory mediators associated with faulty angiogenic responses [218, 219]. Consistent with this observation, MALAT1 RNA was increased in the defective retinal vasculature of diabetic or obese mice [218]. Both the diabetic retinopathy and the inflammatory phenotype of ECs could be reversed with targeted suppression of MALAT1 [218, 219]. Perturbations in retinal angiogenesis were also noted in MALAT1 knockout mice suggesting that a critical level of this lncRNA is needed for normal microvascular patterning [217]. Given the widely reported effects of MALAT1 in regulating migration of various cancer cells and metastasis, it will be important to differentiate between effects of MALAT1 suppression on cancer cells versus local tumor vasculature. It will also be important to assess the role of SENCR in EC phenotypes, since levels of this lncRNA are higher in ECs than SMCs [209].
A second conserved nuclear lncRNA implicated in pathological angiogenesis is MIAT. Like MALAT1, MIAT is induced in ECs treated with high glucose [220]. Knockdown of MIAT attenuated high glucose-mediated EC proliferation, migration, and tubulogenesis in vitro and improved diabetes-induced microvascular pathology in vivo. Importantly, multiple miR-150-5p binding sites were found in the VEGF transcript as well as MIAT suggesting this lncRNA could function as a sponge RNA and regulator of VEGF levels. Indeed, a series of studies provided direct support for this concept, thus offering new insight into pathological angiogenesis and a new potential target for the treatment of aberrant angiogenesis [220]. The results of the MIAT study also support the idea of RNA sponging of miRs within the nucleus. Another EC sponge RNA was recently reported to regulate autophagy in human ECs. This lncRNA (TGFB2-ot1) is a rare example of a sense lncRNA embedded within the 3′ untranslated region of a protein-coding gene. Consequently, loss-of-function studies are impossible to interpret since targeted knockdown of the lncRNA also reduces TGFB2 [221]. Nevertheless, gain-of-function studies showed that TGFB2-ot1 sponges miR-4459, which can target the autophagy-related 13 (ATG13) mRNA thereby increasing MTOR, a key regulator of autophagy [221]. There will, no doubt, be more examples of vascular lncRNAs that function as competing endogenous RNAs for miRs. Several databases are available to assist investigators in this context including StarBase (http://starbase.sysu.edu.cn/mirLncRNA.php) [222] and LNCipedia (http://www.lncipedia.org/) [172].
Two very recent reports have defined lncRNAs that effect changes in EC phenotype associated with angiogenesis. First, Kurian et al. [223] studied an annotated NAT-lncRNA located at the 3′ end of the AGAP2 mRNA. The AGAP2-AS1 lncRNA is elevated in human embryonic stem cells induced to differentiate to the EC lineage. Morpholino knockdown of AGAP2-AS1 in zebrafish caused defective vascular branching leading to abnormal vascular patterning in the embryo. This phenotype could be rescued by introducing the human ortholog of AGAP2-AS1 into mutant fish. In human ECs, knockdown of AGAP2-AS1 resulted in reduced EC proliferation, tubulogenesis, and acetylated LDL uptake [223]. Though expression of EC-restricted markers was not reported, results suggest AGAP2-AS1 functions downstream of EC lineage commitment. It will be important to ascertain the effects of loss or gain of AGAP2-AS1 on AGAP2 mRNA to determine whether effects of the lncRNA relate to changes in expression of the overlapping protein-coding gene. Second, Li et al. [224] discovered a multi-exonic NAT-lncRNA overlapping the EC-restricted DLL4 gene. The DLL4-AS and DLL4 gene pairs are concordantly expressed at baseline and under stimulatory conditions. Overexpression of two isoforms of DLL4-AS can increase DLL4 protein levels whereas knockdown of DLL4-AS decreases DLL4 mRNA. The precise mechanism of DLL4-AS regulation of DLL4 is unknown at this time. Intriguingly, there appears to be a shared promoter between the DLL4 sense-antisense gene pair [224]. Formal testing of the function of this bi-directional promoter could be done with CRISPR-mediated editing and subsequent measuring of each transcript. Functional studies show that, like SENCR in SMCs and MALAT1 in ECs, knockdown of DLL4-AS in ECs results in a hyper-motile state. Further, increases in EC proliferation and spouting were observed though the latter was considered to be non-productive [224]. These results indicate an important layer of regulation of DLL4 expression and hence EC phenotype. Table 6 lists the known lncRNA genes and their putative functions in ECs.
Table 6.
EC lncRNAs
| LncRNA | Type/localization | Putative function in EC | References |
|---|---|---|---|
| ATG9B | NAT-lncRNA/nuclear+cytoplasmic | ↓ NOS3 following hypoxia | [215] |
| TIE1-AS | NAT-lncRNA/cytoplasmic | Regulation of EC contact junctions | [216] |
| NRON | Intr-lncRNA/cytoplasmic | ↓ EC proliferation/↓ migration | [285] |
| SENCR | antisense oh-lncRNA/cytoplasmic | ? | [209] |
| MALAT1 | LincRNA/nuclear | Proper angiogenesis | [217] |
| TGFB2-ot1 | sense ol-lncRNA/? | ceRNA regulating autophagy | [221] |
| MIAT | LincRNA/nuclear | ceRNA regulating EC function | [220] |
| ASncmtRNA-2 | Stem loop/nucleus | Replicative senescence | [286] |
| AGAP2-AS1 | NAT-lncRNA/cytoplasm | Proper angiogenesis | [223] |
| HIF1A-AS1 | NAT-lncRNA/? | ↑ EC apoptosis/↓ EC proliferation | [287] |
| DLL4-AS | NAT-lncRNA/cytoplasm | ↑ EC proliferation/↑ EC migration | [224] |
Summary
Our understanding of lncRNA expression and function in vascular cells is rather primitive at this time. As more labs perform RNA-seq experiments under various experimental conditions in ECs and SMCs, a treasure trove of annotated lncRNAs will emerge for further study. Given the increasing number of non-polyadenylated lncRNA genes [225], and the fact that many deep sequencing experiments utilize polyA+ RNA, a vast number of non-polyadenylated lncRNA genes will need to be characterized. Depending on the manner in which the RNA-seq data are analyzed, final datasets will include new and already annotated transcripts with the latter designated as pseudogene, antisense RNA, opposite strand RNA, eRNA, and overlapping transcripts, to name a few [226]. Regarding the formal naming of lncRNA genes, there are established guidelines based on function or genomic position that investigators should follow [227]. Many lncRNA genes will likely have functions related to phenotypic modulation or differentiation of vascular cells. Each putative lncRNA, whether novel or already identified and annotated in public databases, will require in depth analyses. Accordingly, we propose a logical order of operations for the systematic study of new and annotated transcripts of unknown expression and function in vascular cells (Fig. 5).
Fig. 5.
Order of operations for the study of a lncRNA
The first step in analyzing new lncRNA genes is to carefully examine the transcript on the UCSC Genome Browser and define such features as transcript abundance, conservation, and chromatin marks by exploiting the vast data from the ENCODE Consortium [228]. The presence of miR-binding sequences should be examined as an increasing number of lncRNAs function as sponge or competing endogenous RNAs [220]. Further bioinformatics might include the definition of open reading frames (e.g., NCBI, ORF Finder, http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and evaluating each ORF for the presence of known protein domains (e.g., Pfam databases, http://pfam.xfam.org). Much of this information can also be gleaned quickly by simply turning on tracks within the UCSC Genome Browser. An important question in this context is whether a transcript engages the ribosomal machinery and generates a translated peptide. This will require in vitro transcription/translation and/or epitope-tagging of the transcript followed by Western blotting. Whether or not a cytoplasmic lncRNA is translated, its association with ribosomes could have a regulatory role in fine-tuning translation through effects on the ribosomal machinery itself or mRNA stability. We have found several un-annotated transcripts (mostly single exon) of high abundance and conservation to be probable paralogs of known protein-coding genes (unpublished). This indicates there will be ongoing revision of the human genome as previously annotated lncRNAs emerge as mRNAs. Whether encoding a peptide or not, it seems that a noncoding function of any transcript can no longer be ignored [215, 229]. Finally, it will be important to determine whether any relevant SNPs exist in a putative lncRNA using LincSNP (http://210.46.85.180:8080/LincSNP/search.jsp) [230] or lncRNASNP (http://bioinfo.life.hust.edu.cn/lncRNASNP/) [231].
The second step in analyzing a lncRNA is to validate expression of the transcript in independent isolates of cells under conditions of the original RNA-seq experiment. Further confirmation of expression should include cell line and tissue profiling by qRT-PCR and Northern blotting, the latter representing a convenient method of ascertaining transcript length and alternative splice/polyadenylation variants. More often than not, rapid amplification of cDNA ends (RACE) will extend existing exons and reveal new exons with attending splice variants that could exhibit differential functions. The third step in lncRNA analysis should be to define the spatial localization of the transcript under baseline and stimulatory states. RNA fractionation followed by qRT-PCR is a reasonable first approximation; however, single molecule RNA-FISH offers much more quantitative data. The incorporation of a second set of fluorescent probes in RNA-FISH and scoring coincident signals following knockdown of the lncRNA represents a powerful approach to the spatial localization of a lncRNA [209]. Understanding where a new transcript resides will assist greatly in defining how to proceed in the functional analysis of a lncRNA (Fig. 3). Of course, such assays only represent a snapshot of lncRNA localization and cannot predict dynamic nucleocytoplasmic shuttling. Thus, real-time imaging of lncRNAs will be an important development to further elucidate lncRNA biology in vascular cells.
The fourth step in analyzing a lncRNA gene relates to unveiling its function. Thus, loss-of-function assays should be done through appropriate means including dicer substrate siRNA or GapmeR knockdown approaches. Given the ease in which genes can now be inactivated using CRISPR, inserting a strong polyadenylation signal in the first exon of a lncRNA could be quite informative, particularly for conserved lncRNAs that can be inactivated in model organisms. CRISPR-mediated deletions are more difficult and potentially confounding with possible excision of other sequences independent of the lncRNA sequence. Thus, where possible, rescue experiments should be undertaken to address this possibility as well as distal off-targeting effects. Whether the goal is to delete an exon or entire transcript or insert a polyadenylation signal for premature transcriptional arrest, great care must be taken in fully elucidating the 5′ end of the lncRNA (using RACE) and strategizing in such a manner that other genes or regulatory elements are not disturbed [232]. Gain-of-function studies require complete definition of the full-length transcript by 5′ and 3′ RACE. Full-length lncRNAs may then be integrated in a suitable viral system (e.g., lentivirus) for delivery to cells or tissues. Whether the aim is to knockdown or overexpress the lncRNA, unambiguous phenotyping of the cells or animal will be of paramount importance towards understanding lncRNA function. One common approach is to knockdown the lncRNA and then conduct RNA-seq to assess the resultant transcriptome [209, 217, 223]. However phenotyping is done, the last step in the process of studying a lncRNA is elucidating its mechanism of action (Fig. 5). Several biochemical assays have been developed to understand the physical interaction between a lncRNA and a protein, a lncRNA and another RNA, or a lncRNA and DNA. These assays (e.g., CLIP-seq, PAR-CLIP, ChIRP, etc.) generally involve the crosslinking of a biotinylated lncRNA or tiled oligonucleotides that are antisense to an endogenous lncRNA followed by streptavidin bead capture of co-precipitating lncRNA and associated macromolecules. It is important to recognize that there likely will be issues of oligonucleotide accessibility to the lncRNA. Thus, it may be prudent to first demonstrate enrichment of the lncRNA following crosslinking and pulldown before proceeding with RNA binding protein or RNA/DNA sequence identification. Ultimately, interacting proteins, RNAs, or DNA can be identified by gel electrophoresis and mass spectrometry, RNA-seq, and qRT-PCR, respectively [233]. Obtaining such information will be critical in carrying forward additional experiments that would define interaction domains and 3-D structures with the goal of developing small molecule inhibitors that disrupt or stabilize lncRNA interactions with other macromolecules in the cell as well as constructing more sophisticated interactome maps under normal and pathological conditions. Finally, as additional assays emerge to detect macromolecules that bind lncRNA transcripts, a challenge will be interpreting the function and mechanism of action of the far more numerous ultra-low abundant lncRNA transcripts in vascular cells.
Perspective
The human genome represents a veritable digital library of information that includes a massive number of functional noncoding sequences, including TFBS that direct correct spatio-temporal and context-specific expression of protein coding mRNAs as well as the continually growing number of miR and lncRNA genes. The interplay among these three classes of noncoding sequences within and between ECs and SMCs is largely speculative at this time and will require integrated studies to build more complete interactome maps (Fig. 6). Methodological reductionism will continue to be of importance in understanding the basics of noncoding sequences (e.g., regulatory processes underlying miR stability and transcriptional control of miR/lncRNA expression), but the isolated study of any one mRNA/protein, TFBS, miR, or lncRNA can no longer proceed without addressing such issues as the existence of lncRNAs that act as TFBS decoys or whether a lncRNA is stabilized by a mRNA or some other lncRNA that sponges a miR. Accordingly, the next generation of scientists will have a rich resource of data and increasingly sophisticated tools to generate new hypotheses underlying vascular cell differentiation and phenotypic change in disease. The ever-present challenge will be supplementing our two-dimensional, reductionist approach to biology with higher dimensional thinking so as to make better sense of what clearly is a biological symphony of interactive processes necessary for vessel wall homeostasis. For example, one might imagine data derived from deep transcript profiling of individual vascular cells under various physiological, pathophysiological, or pharmacological conditions and the biophysical study of individual and interactive coding/noncoding genes being used to identify and integrate dominant mRNA-miR-lncRNA associations in vascular cells over time and developing models that will predict specific biological outcomes. Coupling these data with an individual’s genomic sequence and the ~3 million sequence variants that confer individual genetic identity, should facilitate improved tailoring of an interventional approach (preventive or otherwise) that maintains vascular cell homeostasis or reverts a modulated, disease-dependent phenotypic state to one favoring health [234]. Achieving this and other goals will require a concerted effort by basic vascular biology labs that do cell, molecular, and animal modeling of disease (particularly CRISPR mice), clinical labs that furnish appropriately coded patient data, and computational labs that distill enormous wet (e.g., ChIP-seq) and dry (e.g., TFBS predictions) lab data to manageable datasets for interrogation and integration with the aforementioned information. Clearly, 21st century vascular biology has ushered in unprecedented challenges that demand cooperation along the lines of the human genome project in order to fully advance the concept of personalized cardiovascular medicine. In the meantime, much work lies ahead in the identification, functional analysis, and integration of noncoding sequences that control vascular cell phenotypes.
Fig. 6.
Noncoding sequence control of vascular cell phenotypes. The differentiated phenotype of Weibel-Palade body (WP-b) containing ECs (top electron micrograph) and subjacent, Myofilament (Mf)-containing SMCs (bottom electron micrograph) is regulated through TFBS (such as ETS and CArG elements, respectively), directing expression of mRNA, miR, and lncRNA genes whose mature products serve structural and complex regulatory roles, the latter likely involving direct or indirect effects of each transcribed unit on other transcribed sequences in the nucleus or cytoplasm (small arrows). TFBS-SNPs (denoted with an X in each element) may interfere with normal target gene expression and hence perturb the differentiated vascular cell phenotype. Intercellular control of vascular cell phenotypes (large arrows) may be achieved through the action of RNA-containing extracellular vesicles (e.g., exosomes denoted as small circles released locally in the interstitial space or via circulation in the case of ECs)
Acknowledgments
Work in the Miano lab is supported by National Institutes of Health grants HL-117907 and HL-112793. Work in the Long lab is supported by National Institutes of Health grant HL-112686 and a Scientist Development Grant from the American Heart Association (3670036). The authors apologize to those authors whose important work was not cited due to space constraints.
Abbreviations
- CRISPR
Clustered regularly interspaced short palindromic repeats
- EC
Endothelial cell
- ENCODE
Encyclopedia of DNA Elements
- ETS
E26 transformation specific
- LncRNA
Long noncoding RNA
- LSS
Laminar shear stress
- Mir
MicroRNA
- MYOCD
Myocardin
- NAT
Natural antisense transcript
- NICD
Notch intracellular domain
- RACE
Rapid amplification of cDNA ends
- SMC
Smooth muscle cell
- SNP
Single nucleotide polymorphism
- SRF
Serum response factor
- TFBS
Transcription factor binding site
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84(3):767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Naik V, Leaf EM, Hu JH, Yang HY, Nguyen NB, Giachelli CM, Speer MY. Sources of cells that contribute to atherosclerotic intimal calcification: an in vivo genetic fate mapping study. Cardiovasc Res. 2012;94(3):545–554. doi: 10.1093/cvr/cvs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014;115(7):662–667. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 4.Tang Z, Wang A, Yuan F, Yan Z, Liu B, Chu JS, Helms JA, Li S. Differentiation of multipotent vascular stem cells contributes to vascular diseases. Nat Commun. 2012;3:875. doi: 10.1038/ncomms1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy E, Mooney CJ, Hakimjavadi R, Fitzpatrick E, Guha S, Collins LE, Loscher CE, Morrow D, Redmond EM, Cahill PA. Adult vascular smooth muscle cells in culture express neural stem cell markers typical of resident multipotent vascular stem cells. Cell Tissue Res. 2014;358(1):203–216. doi: 10.1007/s00441-014-1937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemenoff RA, Horita H, Ostriker AC, Furgeson SB, Simpson PA, VanPutten V, Crossno J, Offermanns S, Weiser-Evans MC. SDF-1alpha induction in mature smooth muscle cells by inactivation of PTEN is a critical mediator of exacerbated injury-induced neointima formation. Arterioscler Thromb Vasc Biol. 2011;31(6):1300–1308. doi: 10.1161/ATVBAHA.111.223701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen AT, Gomez D, Bell RD, Campbell JH, Clowes AW, Gabbiani G, Giachelli CM, Parmacek MS, Raines EW, Rusch NJ, Speer MY, Sturek M, Thyberg J, Towler DA, Weiser-Evans MC, Yan C, Miano JM, Owens GK. Smooth muscle cell plasticity: fact or fiction? Circ Res. 2013;112(1):17–22. doi: 10.1161/CIRCRESAHA.112.281048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herring BP, Hoggatt AM, Burlak C, Offermanns S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vascular Cell. 2014;6:21. doi: 10.1186/2045-824X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies PF, Civelek M, Fang Y, Fleming I. The atherosusceptible endothelium: endothelial phenotypes in complex haemodynamic shear stress regions in vivo. Cardiovasc Res. 2013;99(2):315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111(17):6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 15.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 16.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489(7414):83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 19.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27(6):1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 20.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 21.Minty A, Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986;6(6):2125–2136. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 23.Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90(9):1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- 24.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 25.Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, Jr, Miano JM. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson CC, Zhou Q, Long X, Miano JM. Identifying functional single nucleotide polymorphisms in the human CArGome. Physiol Genomics. 2011;43:1038–1048. doi: 10.1152/physiolgenomics.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Liu ZC, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 28.Mack CP, Owens GK. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ Res. 1999;84:852–861. doi: 10.1161/01.res.84.7.852. [DOI] [PubMed] [Google Scholar]
- 29.Touw K, Hoggatt AM, Simon G, Herring BP. Hprt -targeted transgenes provide new insight into smooth muscle-restricted promoter activity. Am J Physiol Cell Physiol. 2007;292:C1024–C1032. doi: 10.1152/ajpcell.00445.2006. [DOI] [PubMed] [Google Scholar]
- 30.Manabe I, Owens GK. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J Clin Invest. 2001;107:823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilly B, Olson EN, Beckerle MC. Identification of a CArG box-dependent enhancer within the cysteine-rich protein 1 gene that directs expression in arterial but not venous or visceral smooth muscle cells. Dev Biol. 2001;240:531–547. doi: 10.1006/dbio.2001.0507. [DOI] [PubMed] [Google Scholar]
- 32.Long X, Tharp DL, Georger MA, Slivano OJ, Lee MY, Wamhoff BR, Bowles DK, Miano JM. The smooth muscle cell-restricted KCNMB1 ion channel subunit is a direct transcriptional target of serum response factor and myocardin. J Biol Chem. 2009;284(48):33671–33682. doi: 10.1074/jbc.M109.050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long X, Slivano OJ, Cowan SL, Georger MA, Lee TH, Miano JM. Smooth muscle calponin: an unconventional CArG-dependent gene that antagonizes neointimal formation. Arterioscler Thromb Vasc Biol. 2011;31(10):2172–2180. doi: 10.1161/ATVBAHA.111.232785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanda V, Miano JM. Leiomodin 1: a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J Biol Chem. 2012;287(4):2459–2467. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miano JM, Ramanan N, Georger MA, de Mesy-Bentley KL, Emerson RL, Balza RO, Jr, Xiao Q, Weiler H, Ginty DD, Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci USA. 2004;101(49):17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS, Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14(1):64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 37.Cooper SJ, Trinklein ND, Nguyen L, Myers RM. Serum response factor binding sites differ in three human cell types. Genome Res. 2007;17(2):136–144. doi: 10.1101/gr.5875007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5(9):829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK. SRF utilizes distinct promoter and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol. 2011;31(4):861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang SX, Gras EG, Wycuff DR, Marriot SJ, Kadeer N, Yu W, Olson EN, Garry DJ, Parmacek MS, Schwartz RJ. Identification of direct serum response factor gene targets during DMSO induced P19 cardiac cell differentiation. J Biol Chem. 2005;280(19):19115–19126. doi: 10.1074/jbc.M413793200. [DOI] [PubMed] [Google Scholar]
- 41.Garrido-Martin EM, Blanco FJ, Roque M, Novensa L, Tarocchi M, Lang UE, Suzuki T, Friedman SL, Botella LM, Bernabeu C. Vascular injury triggers kruppel-like factor 6 mobilization and cooperation with specificity protein 1 to promote endothelial activation through upregulation of the activin receptor-like kinase 1 gene. Circ Res. 2013;112(1):113–127. doi: 10.1161/CIRCRESAHA.112.275586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147(1):32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28(9):943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 45.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y, Slivano OJ, Christie CK, Cheng AW, Miano JM. CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice. Arterioscler Thromb Vasc Biol. 2015;35:312–315. doi: 10.1161/ATVBAHA.114.305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Q, Taurin S, Sethakorn N, Long X, Imamura M, Wang DZ, Zimmer WE, Dulin NO, Miano JM. Myocardin-dependent activation of the CArG box-rich smooth muscle gamma actin gene: Preferential utilization of a single CArG element through functional association with the NKX3.1 homeodomain protein. J Biol Chem. 2009;284(47):32582–32590. doi: 10.1074/jbc.M109.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang DZ, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 49.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110(14):5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34(10):1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Wang DZ, Pipes GCT, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100(12):7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long X, Bell RD, Gerthoffer WT, Zlokovic BV, Miano JM. Myocardin is sufficient for a SMC-like contractile phenotype. Arterioscler Thromb Vasc Biol. 2008;28(8):1505–1510. doi: 10.1161/ATVBAHA.108.166066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raphel L, Talasila A, Cheung C, Sinha S. Myocardin overexpression is sufficient for promoting the development of a mature smooth muscle cell-like phenotype from human embryonic stem cells. PLoS One. 2012;7(8):e44052. doi: 10.1371/journal.pone.0044052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida T, Kawai-Kowase K, Owens GK. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential cells. Arterioscler Thromb Vasc Biol. 2004;24(9):1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
- 55.Kitchen CM, Cowan SL, Long X, Miano JM. Expression and promoter analysis of a highly restricted integrin alpha gene in vascular smooth muscle. Gene. 2013;513(1):82–89. doi: 10.1016/j.gene.2012.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parmacek MS. Myocardin: dominant driver of the smooth muscle cell contractile phenotype. Arterioscler Thromb Vasc Biol. 2008;28(8):1416–1417. doi: 10.1161/ATVBAHA.108.168930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boucher J, Gridley T, Liaw L. Molecular pathways of notch signaling in vascular smooth muscle cells. Front Physiol. 2012;3:81. doi: 10.3389/fphys.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 59.Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27(9):1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noseda M, Fu Y, Niessen K, Wong F, Chang L, McLean G, Karsan A. Smooth muscle alpha-actin is a direct target of Notch/CSL. Circ Res. 2006;98(12):1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 61.Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via RBP-J k-dependent pathway. J Biol Chem. 2006;281(39):28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- 62.Boucher JM, Peterson SM, Urs S, Zhang C, Liaw L. The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286(32):28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rensen SSM, Niessen PM, Long X, Doevendans PA, Miano JM, van Eys GJJM. Contribution of serum response factor and myocardin to transcriptional regulation of smoothelins. Cardiovasc Res. 2006;70:136–145. doi: 10.1016/j.cardiores.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 64.Briot A, Jaroszewicz A, Warren CM, Lu J, Touma M, Rudat C, Hofmann JJ, Airik R, Weinmaster G, Lyons K, Wang Y, Kispert A, Pellegrini M, Iruela-Arispe ML. Repression of Sox9 by Jag1 is continuously required to suppress the default chondrogenic fate of vascular smooth muscle cells. Dev Cell. 2014;31(6):707–721. doi: 10.1016/j.devcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long X, Creemers EE, Wang DZ, Olson EN, Miano JM. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proc Natl Acad Sci USA. 2007;104(42):16570–16575. doi: 10.1073/pnas.0708253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 67.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 68.Mao X, Debenedittis P, Sun Y, Chen J, Yuan K, Jiao K, Chen Y. Vascular smooth muscle cell Smad4 gene is important for mouse vascular development. Arterioscler Thromb Vasc Biol. 2012;32(9):2171–2177. doi: 10.1161/ATVBAHA.112.253872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125(22):4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 70.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 71.Claussnitzer M, Dankel SN, Klocke B, Grallert H, Glunk V, Berulava T, Lee H, Oskolkov N, Fadista J, Ehlers K, Wahl S, Hoffmann C, Qian K, Ronn T, Riess H, Muller-Nurasyid M, Bretschneider N, Schroeder T, Skurk T, Horsthemke B, Diagram+Consortium. Spieler D, Klingenspor M, Seifert M, Kern MJ, Mejhert N, Dahlman I, Hansson O, Hauck SM, Bluher M, Arner P, Groop L, Illig T, Suhre K, Hsu YH, Mellgren G, Hauner H, Laumen H. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156(1-2):343–358. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park C, Kim TM, Malik AB. Transcriptional regulation of endothelial cell and vascular development. Circ Res. 2013;112(10):1380–1400. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishikawa SI. A complex linkage in the developmental pathway of endothelial cell and hematopoietic cells. Curr Opin Cell Biol. 2001;13:673–678. doi: 10.1016/s0955-0674(00)00270-2. [DOI] [PubMed] [Google Scholar]
- 74.Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. 2013;33(6):1366–1375. doi: 10.1161/ATVBAHA.112.301167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai TN, Baban D, Ragoussis J, Huang Y, Han JD, Zeng L, Hu Y, Xu Q. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci USA. 2012;109(34):13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, Schachterle W, Pulijaal VR, Mathew S, Chasen ST, Xiang J, Rosenwaks Z, Shido K, Elemento O, Rabbany SY, Rafii S. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFbeta suppression. Cell. 2012;151(3):559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morita R, Suzuki M, Kasahara H, Shimizu N, Shichita T, Sekiya T, Kimura A, Sasaki K, Yasukawa H, Yoshimura A. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc Natl Acad Sci USA. 2015;112(1):160–165. doi: 10.1073/pnas.1413234112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlaeger TM, Qin Y, Fujiwara Y, Magram J, Sato TN. Vascular endothelial cell lineage-specific promoter in transgenic mice. Development. 1995;121:1089–1098. doi: 10.1242/dev.121.4.1089. [DOI] [PubMed] [Google Scholar]
- 79.Aird WC, Jahroudi N, Weiler-Guettler H, Rayburn HB, Rosenberg RD. Human von Willebrand factor gene sequences target expression to a subpopulation of endothelial cells in transgenic mice. Proc Natl Acad Sci USA. 1995;92(10):4567–4571. doi: 10.1073/pnas.92.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kappel A, Ronicke V, Damert A, Flamme I, Risau W, Breier G. Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood. 1999;93(12):4284–4292. [PubMed] [Google Scholar]
- 81.Gory S, Vernet M, Laurent M, Dejana E, Dalmon J, Huber P. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood. 1999;93(1):184–192. [PubMed] [Google Scholar]
- 82.Iljin K, Petrova TV, Veikkola T, Kumar V, Poutanen M, Alitalo K. A fluorescent Tie1 reporter allows monitoring of vascular development and endothelial cell isolation from transgenic mouse embryos. FASEB J. 2002;16(13):1764–1774. doi: 10.1096/fj.01-1043com. [DOI] [PubMed] [Google Scholar]
- 83.Seki T, Hong KH, Yun J, Kim SJ, Oh SP. Isolation of a regulatory region of activin receptor-like kinase 1 gene sufficient for arterial endothelium-specific expression. Circ Res. 2004;94(8):e72–e77. doi: 10.1161/01.RES.0000127048.81744.31. [DOI] [PubMed] [Google Scholar]
- 84.Mollica LR, Crawley JT, Liu K, Rance JB, Cockerill PN, Follows GA, Landry JR, Wells DJ, Lane DA. Role of a 5’-enhancer in the transcriptional regulation of the human endothelial cell protein C receptor gene. Blood. 2006;108(4):1251–1259. doi: 10.1182/blood-2006-02-001461. [DOI] [PubMed] [Google Scholar]
- 85.Okada Y, Yano K, Jin E, Funahashi N, Kitayama M, ADoi T, Spokes K, Beeler DL, Shih SC, Okada H, Danilov TA, Maynard E, Minami T, Oettgen P, Aird WC. A three-kilobase fragment of the human Robo4 promoter directs cell type-specific expression in endothelium. Circ Res. 2007;100(12):1712–1722. doi: 10.1161/01.RES.0000269779.10644.dc. [DOI] [PubMed] [Google Scholar]
- 86.Jin E, Liu J, Suehiro J, Yuan L, Okada Y, Nikolova-Krstevski V, Yano K, Janes L, Beeler D, Spokes KC, Li D, Regan E, Shih SC, Oettgen P, Minami T, Aird WC. Differential roles for ETS, CREB, and EGR binding sites in mediating VEGF receptor 1 expression in vivo. Blood. 2009;114(27):5557–5566. doi: 10.1182/blood-2009-05-220434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wythe JD, Dang LT, Devine WP, Boudreau E, Artap ST, He D, Schachterle W, Stainier DY, Oettgen P, Black BL, Bruneau BG, Fish JE. ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 2013;26(1):45–58. doi: 10.1016/j.devcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD, Adams RH. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465(7297):483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 89.Shu XZ, Zhang LN, Zhang R, Zhang CJ, He HP, Zhou H, Wang N, Zhang TC. Histone acetyltransferase p300 promotes MRTF-A-mediates transactivation of VE-cadherin gene in human umbilical vein endothelial cells. Gene. 2015;563(1):17–23. doi: 10.1016/j.gene.2015.02.076. [DOI] [PubMed] [Google Scholar]
- 90.Holtz ML, Misra RP. Endothelial-specific ablation of serum response factor causes hemorrhaging, yolk sac vascular failure, and embryonic lethality. BMC Dev Biol. 2008;8:65. doi: 10.1186/1471-213X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meadows SM, Myers CT, Krieg PA. Regulation of endothelial cell development by ETS transcription factors. Semin Cell Dev Biol. 2011;22(9):976–984. doi: 10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135(6):1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robinson AS, Materna SC, Barnes RM, De Val S, Xu SM, Black BL. An arterial-specific enhancer of the human endothelin converting enzyme1 (ECE1) gene is synergistically activated by Sox17, FoxC2, and Etv2. Dev Biol. 2014;395(2):379–389. doi: 10.1016/j.ydbio.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 95.Topper JN, Cai J, Falb D, Gimbrone MA., Jr Identification of vascular endothelial genes differentially responsive to fluid mechanical stimuli: cyclooxygenase-2, manganese superoxide dismutase, and endothelial cell nitric oxide synthase are selectively up-regulated by steady laminar shear stress. Proc Natl Acad Sci USA. 1996;93(19):10417–10422. doi: 10.1073/pnas.93.19.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100(5):1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 97.Huddleson JP, Ahmad N, Srinivasan S, Lingrel JB. Induction of KLF2 by fluid shear stress requires a novel promoter element activated by a phosphatidylinositol 3-kinase-dependent chromatin-remodeling pathway. J Biol Chem. 2005;280(24):23371–23379. doi: 10.1074/jbc.M413839200. [DOI] [PubMed] [Google Scholar]
- 98.Khachigian LM, Anderson KR, Halnon NJ, Gimbrone MA, Jr, Resnick N, Collins T. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol. 1997;17(10):2280–2286. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- 99.Zhang R, Min W, Sessa WC. Functional analysis of the human endothelial nitric oxide synthase promoter. Sp1 and GATA factors are necessary for basal transcription in endothelial cells. J Biol Chem. 1995;270(25):15320–15326. doi: 10.1074/jbc.270.25.15320. [DOI] [PubMed] [Google Scholar]
- 100.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199(10):1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fan P, Chen Z, Tian P, Liu W, Jiao Y, Xue Y, Bhattacharya A, Wu J, Lu M, Guo Y, Cui Y, Gu W, Gu W, Yue J. miRNA biogenesis enzyme Drosha is required for vascular smooth muscle cell survival. PLoS One. 2013;8(4):e60888. doi: 10.1371/journal.pone.0060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Z, Wu J, Yang C, Fan P, Balazs L, Jiao Y, Lu M, Gu W, Li C, Pfeffer LM, Tigyi G, Yue J. DiGeorge syndrome critical region 8 (DGCR8) protein-mediated microRNA biogenesis is essential for vascular smooth muscle cell development in mice. J Biol Chem. 2012;287(23):19018–19028. doi: 10.1074/jbc.M112.351791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105(37):14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 107.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100(11):1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 109.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284(6):3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104(4):476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, Su M, Han Y, Shi HJ, Wen JK. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12(1):56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, Li YJ. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24(10):1087–1093. doi: 10.1038/ajh.2011.116. [DOI] [PubMed] [Google Scholar]
- 114.Reddy MA, Jin W, Villeneuve L, Wang M, Lanting L, Todorov I, Kato M, Natarajan R. Pro-inflammatory role of microrna-200 in vascular smooth muscle cells from diabetic mice. Arterioscler Thromb Vasc Biol. 2012;32(3):721–729. doi: 10.1161/ATVBAHA.111.241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maegdefessel L, Spin JM, Raaz U, Eken SM, Toh R, Azuma J, Adam M, Nagakami F, Heymann HM, Chernugobova E, Jin H, Roy J, Hultgren R, Caidahl K, Schrepfer S, Hamsten A, Eriksson P, McConnell MV, Dalman RL, Tsao PS. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. 2014;5:5214. doi: 10.1038/ncomms6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Balderman JA, Lee HY, Mahoney CE, Handy DE, White K, Annis S, Lebeche D, Hajjar RJ, Loscalzo J, Leopold JA. Bone morphogenetic protein-2 decreases microRNA-30b and microRNA-30c to promote vascular smooth muscle cell calcification. J Am Heart Assoc. 2012;1(6):e003905. doi: 10.1161/JAHA.112.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 118.Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, Shi Z, Kilsdonk EP, Gui Y, Wang DZ, Zheng XL. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol. 2011;31(2):368–375. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20(2):205–210. doi: 10.1089/scd.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heidersbach A, Saxby C, Carver-Moore K, Huang Y, Ang YS, de Jong PJ, Ivey KN, Srivastava D. microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9(9):e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 123.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Torella D, Iaconetti C, Catalucci D, Ellison GM, Leone A, Waring CD, Bochicchio A, Vicinanza C, Aquila I, Curcio A, Condorelli G, Indolfi C. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ Res. 2011;109:880–893. doi: 10.1161/CIRCRESAHA.111.240150. [DOI] [PubMed] [Google Scholar]
- 125.Madsen CS, Regan CP, Owens GK. Interaction of CArG elements and a GC-rich repressor element in transcriptional regulation of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J Biol Chem. 1997;272(47):29842–29851. doi: 10.1074/jbc.272.47.29842. [DOI] [PubMed] [Google Scholar]
- 126.Talasila A, Yu H, Ackers-Johnson M, Bot M, van Berkel T, Bennett MR, Bot I, Sinha S. Myocardin regulates vascular response to injury through miR-24/-29a and platelet-derived growth factor receptor-beta. Arterioscler Thromb Vasc Biol. 2013;33(10):2355–2365. doi: 10.1161/ATVBAHA.112.301000. [DOI] [PubMed] [Google Scholar]
- 127.Ackers-Johnson M, Talasila A, Sage AP, Long X, Bot I, Morrell NW, Bennett MR, Miano JM, Sinha S. Myocardin regulates vascular smooth muscle cell inflammatory activation and disease. Arterioscler Thromb Vasc Biol. 2015;35(4):817–828. doi: 10.1161/ATVBAHA.114.305218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Long X, Miano JM. Transforming growth factor-β1 (TGF-β1) utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286(34):30119–30129. doi: 10.1074/jbc.M111.258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105(2):158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Turczynska KM, Sadegh MK, Hellstrand P, Sward K, Albinsson S. MicroRNAs are essential for stretch-induced vascular smooth muscle contractile differentiation via microRNA (miR)-145-dependent expression of L-type calcium channels. J Biol Chem. 2012;287(23):19199–19206. doi: 10.1074/jbc.M112.341073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M, Upton P, Xin M, van Rooij E, Olson EN, Morrell NW, MacLean MR, Baker AH. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res. 2012;111(3):290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 135.Yu X, Zhang L, Wen G, Zhao H, Luong LA, Chen Q, Huang Y, Zhu J, Ye S, Xu Q, Wang W, Xiao Q. Upregulated sirtuin 1 by miRNA-34a is required for smooth muscle cell differentiation from pluripotent stem cells. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, Ramratnam B, Comstock CE, Knudsen KE, Gomella L, Spaeth GL, Hark L, Katz LJ, Witkiewicz A, Rostami A, Jimenez SA, Hollingsworth MA, Yeh JJ, Shaw CA, McKenzie SE, Bray P, Nelson PT, Zupo S, Van Roosbroeck K, Keating MJ, Calin GA, Yeo C, Jimbo M, Cozzitorto J, Brody JR, Delgrosso K, Mattick JS, Fortina P, Rigoutsos I. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. 2015;112(10):E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, Steer BM, Ingram AJ, Gupta M, Al Omran M, Teoh H, Marsden PA, Verma S. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 2012;126(11 Suppl 1):S81–S90. doi: 10.1161/CIRCULATIONAHA.111.084186. [DOI] [PubMed] [Google Scholar]
- 139.Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 2010;285(13):9383–9389. doi: 10.1074/jbc.M109.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang Y, Yin H, Zheng XL. MicroRNA-1 inhibits myocardin-induced contractility of human vascular smooth muscle cells. J Cell Physiol. 2010;225(2):506–511. doi: 10.1002/jcp.22230. [DOI] [PubMed] [Google Scholar]
- 141.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226(4):1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 143.Kumar S, Kim CW, Simmons RD, Jo H. Role of flow-sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero-miRs. Arterioscler Thromb Vasc Biol. 2014;34(10):2206–2216. doi: 10.1161/ATVBAHA.114.303425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Neth P, Nazari-Jahantigh M, Schober A, Weber C. MicroRNAs in flow-dependent vascular remodelling. Cardiovasc Res. 2013;99(2):294–303. doi: 10.1093/cvr/cvt096. [DOI] [PubMed] [Google Scholar]
- 145.Chen K, Fan W, Wang X, Ke X, Wu G, Hu C. MicroRNA-101 mediates the suppressive effect of laminar shear stress on mTOR expression in vascular endothelial cells. Biochem Biophys Res Commun. 2012;427(1):138–142. doi: 10.1016/j.bbrc.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 146.Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2010;107(7):3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14(3):249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 148.Kohlstedt K, Trouvain C, Boettger T, Shi L, Fisslthaler B, Fleming I. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ Res. 2013;112(8):1150–1158. doi: 10.1161/CIRCRESAHA.113.301282. [DOI] [PubMed] [Google Scholar]
- 149.Climent-Salarich M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFbeta triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 150.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107(30):13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Njock MS, Cheng HS, Dang LT, Nazari-Jahantigh M, Lau AC, Boudreau E, Roufaiel M, Cybulsky MI, Schober A, Fish JE. Endothelial cells suppress monocyte activation through secretion of extracellular vesicles containing anti-inflammatory microRNAs. Blood. 2015 doi: 10.1182/blood-2014-11-611046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300(5):H1762–H1769. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr, Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, Li YS, Chien S, Garcia-Cardena G, Shyy JY. Flow-dependent regulation of Kruppel-like factor 2 is mediated by microRNA-92a. Circulation. 2011;124(5):633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fan W, Fang R, Wu X, Liu J, Feng M, Dai G, Chen G, Wu G. Shear-sensitive microRNA-34a modulates flow-dependent regulation of endothelial inflammation. J Cell Sci. 2015;128(1):70–80. doi: 10.1242/jcs.154252. [DOI] [PubMed] [Google Scholar]
- 155.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105(5):1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang S, Aurora AB, Johnson BA, Qi X, McNally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Harris TA, Yamakuchi M, Kondo M, Oettgen P, Lowenstein CJ. Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arterioscler Thromb Vasc Biol. 2010;30(10):1990–1997. doi: 10.1161/ATVBAHA.110.211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schmidt M, Paes K, De Maziere A, Smyczek T, Yang S, Gray A, French D, Kasman I, Klumperman J, Rice DS, Ye W. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development. 2007;134(16):2913–2923. doi: 10.1242/dev.002576. [DOI] [PubMed] [Google Scholar]
- 160.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135(24):3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 161.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 162.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299(1):E110–E116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, Ramakrishnan S. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120(11):4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cunningham A, Dokun AO. MicroRNAs as potential therapeutic targets in peripheral artery disease. Int J Diabetol Vasc Dis Res. 2014;2(6):67–70. [Google Scholar]
- 165.Shi L, Fisslthaler B, Zippel N, Fromel T, Hu J, Elgheznawy A, Heide H, Popp R, Fleming I. MicroRNA-223 antagonizes angiogenesis by targeting beta1 integrin and preventing growth factor signaling in endothelial cells. Circ Res. 2013;113(12):1320–1330. doi: 10.1161/CIRCRESAHA.113.301824. [DOI] [PubMed] [Google Scholar]
- 166.Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, McCahill A, Breen C, Mackenzie RM, Delles C, Mountford JC, Milligan G, Emanueli C, Baker AH. Role of microRNAs 99b, 181a, and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem Cells. 2012;30(4):643–654. doi: 10.1002/stem.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Di Bernardini E, Campagnolo P, Margariti A, Zampetaki A, Karamariti E, Hu Y, Xu Q. Endothelial lineage differentiation from induced pluripotent stem cells is regulated by microRNA-21 and transforming growth factor beta2 (TGF-beta2) pathways. J Biol Chem. 2014;289(6):3383–3393. doi: 10.1074/jbc.M113.495531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miller CL, Haas U, Diaz R, Leeper NJ, Kundu RK, Patlolla B, Assimes TL, Kaiser FJ, Perisic L, Hedin U, Maegdefessel L, Schunkert H, Erdmann J, Quertermous T, Sczakiel G. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10(3):e1004263. doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SPA, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 170.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9(6):e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43(Database issue):D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 175.Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE, Sims D, Lindsay MA. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10(9):637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V, Sjoberg M, Keane TM, Verma A, Ala U, Tay Y, Wu D, Seitzer N, Velasco-Herrera MD, Bothmer A, Fung J, Langellotto F, Rodig SJ, Elemento O, Shipp MA, Adams DJ, Chiarle R, Pandolfi PP. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O, Coito C, Hsiao J, Lawrence M, Wahlestedt C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 190.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 191.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 192.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 193.Zhang B, Gunawardane L, Niazi F, Jahanbani F, Chen X, Valadkhan S. A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol Cell Biol. 2014;34(12):2318–2329. doi: 10.1128/MCB.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343(6172):1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Han DKM, Liau G. Identification and characterization of developmentally regulated genes in vascular smooth muscle cells. Circ Res. 1992;71(3):711–719. doi: 10.1161/01.res.71.3.711. [DOI] [PubMed] [Google Scholar]
- 198.Han DK, Khaing ZZ, Pollock RA, Haudenschild CC, Liau G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J Clin Invest. 1996;97(5):1276–1285. doi: 10.1172/JCI118543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kim DK, Zhang L, Dzau VJ, Pratt RE. H19, a developmentally regulated gene, is reexpressed in rat vascular smooth muscle cells after injury. J Clin Invest. 1994;93:355–360. doi: 10.1172/JCI116967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ding Z, Wang X, Schnackenberg L, Khaidakov M, Liu S, Singla S, Dai Y, Mehta JL. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. Int J Cardiol. 2013;168(2):1378–1385. doi: 10.1016/j.ijcard.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 202.Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci USA. 2011;108(40):16759–16764. doi: 10.1073/pnas.1110904108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 204.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Motterle A, Pu X, Wood H, Xiao Q, Gor S, Ng FL, Chan K, Cross F, Shohreh B, Poston RN, Tucker AT, Caulfield MJ, Ye S. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. 2012;21(18):4021–4029. doi: 10.1093/hmg/dds224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464(7287):409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, Scherer SW, Nakabayashi K, Marsden PA. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279(36):37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 208.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113(3):266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li X, Jiang XY, Ge J, Wang J, Chen GJ, Xu L, Xie DY, Yuan TY, Zhang DS, Zhang H, Chen YH. Aberrantly expressed lncRNAs in primary varicose great saphenous veins. PLoS One. 2014;9(1):e86156. doi: 10.1371/journal.pone.0086156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Li L, Li X, The E, Wang LJ, Yuan TY, Wang SY, Feng J, Wang J, Liu Y, Wu YH, Ma XE, Ge J, Cui YY, Jiang XY. Low expression of lncRNA-GAS5 is implicated in human primary varicose great saphenous veins. PLoS One. 2015;10(3):e0120550. doi: 10.1371/journal.pone.0120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M, Oberhuber A, Love DC, Hanover JA, Cinquetti R, Karousou E, Viola M, D’Angelo ML, Hascall VC, De Luca G, Passi A. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J Biol Chem. 2014;289(42):28816–28826. doi: 10.1074/jbc.M114.597401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7(11):7643–7652. [PMC free article] [PubMed] [Google Scholar]
- 215.Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282(21):15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 216.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115(1):133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 218.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015 doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 221.Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, Kung H, Zhao B, Miao J. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10(6):957–971. doi: 10.4161/auto.28363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, Esteban CR, Campistol JM, Yeo GW, Izpisua Belmonte JC. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131(14):1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Li K, Chowdhury T, Vakeel P, Koceja C, Sampath V, Ramchandran R (2015) Delta-like4 mRNA is regulated by adjacent natural antisense transcripts. Vascular Cell 7(3) [DOI] [PMC free article] [PubMed]
- 225.Zhang Y, Yang L, Chen LL. Life without A tail: new formats of long noncoding RNAs. Int J Biochem Cell Biol. 2014;54:338–349. doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 226.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015 doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wright MW. A short guide to long non-coding RNA gene nomenclature. Hum Genomics. 2014;8:7. doi: 10.1186/1479-7364-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, Suh BB, Hinrichs AS, Clawson H, Zweig AS, Kirkup V, Fujita PA, Rhead B, Smith KE, Pohl A, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39(Database issue):D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhang M, Ren Y, Wang Y, Wang R, Zhou Q, Peng Y, Li Q, Yu M, Jiang Y. Regulation of smooth muscle contractility by competing endogenous mRNAs in intracranial aneurysms. J Neuropathol Exp Neurol. 2015;74(5):411–424. doi: 10.1097/NEN.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 230.Ning S, Zhao Z, Ye J, Wang P, Zhi H, Li R, Wang T, Li X. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinform. 2014;15:152. doi: 10.1186/1471-2105-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43(Database issue):D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, Higgs DR, Miska EA, Ponting CP. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 234.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kang K, Peng X, Zhang X, Wang Y, Zhang L, Gao L, Weng T, Zhang H, Ramchandran R, Raj JU, Gou D, Liu L. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288(35):25414–25427. doi: 10.1074/jbc.M113.460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59(11):2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Choe N, Kwon JS, Kim JR, Eom GH, Kim Y, Nam KI, Ahn Y, Kee HJ, Kook H. The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis. 2013;229(2):348–355. doi: 10.1016/j.atherosclerosis.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 238.Li S, Ran Y, Zhang D, Chen J, Li S, Zhu D. MicroRNA-138 plays a role in hypoxic pulmonary vascular remodelling by targeting Mst1. Biochem J. 2013;452(2):281–291. doi: 10.1042/BJ20120680. [DOI] [PubMed] [Google Scholar]
- 239.Wang YS, Wang HY, Liao YC, Tsai PC, Chen KC, Cheng HY, Lin RT, Juo SH. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc Res. 2012;95(4):517–526. doi: 10.1093/cvr/cvs223. [DOI] [PubMed] [Google Scholar]
- 240.Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Cote J, Simard MJ, Bonnet S. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208(3):535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Qiao W, Chen L, Zhang M. MicroRNA-205 regulates the calcification and osteoblastic differentiation of vascular smooth muscle cells. Cell Physiol Biochem. 2014;33(6):1945–1953. doi: 10.1159/000362971. [DOI] [PubMed] [Google Scholar]
- 242.Jalali S, Ramanathan GK, Parthasarathy PT, Aljubran S, Galam L, Yunus A, Garcia S, Cox RR, Jr, Lockey RF, Kolliputi N. Mir-206 regulates pulmonary artery smooth muscle cell proliferation and differentiation. PLoS One. 2012;7(10):e46808. doi: 10.1371/journal.pone.0046808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gou D, Ramchandran R, Peng X, Yao L, Kang K, Sarkar J, Wang Z, Zhou G, Raj JU. miR-210 has an antiapoptotic effect in pulmonary artery smooth muscle cells during hypoxia. Am J Physiol Lung Cell Mol Physiol. 2012;303(8):L682–L691. doi: 10.1152/ajplung.00344.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhao H, Wen G, Huang Y, Yu X, Chen Q, Afzal TA, Luong LA, Zhu J, Shu Y, Zhang L, Xiao Q. MicroRNA-22 regulates smooth muscle cell differentiation from stem cells by targeting methyl CpG-binding protein 2. Arterioscler Thromb Vasc Biol. 2015 doi: 10.1161/ATVBAHA.114.305212. [DOI] [PubMed] [Google Scholar]
- 245.Liu X, Cheng Y, Chen X, Yang J, Xu L, Zhang C. MicroRNA-31 regulated by the extracellular regulated kinase is involved in vascular smooth muscle cell growth via large tumor suppressor homolog 2. J Biol Chem. 2011;286(49):42371–42380. doi: 10.1074/jbc.M111.261065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang J, Yan CH, Li Y, Xu K, Tian XX, Peng CF, Tao J, Sun MY, Han YL. MicroRNA-31 controls phenotypic modulation of human vascular smooth muscle cells by regulating its target gene cellular repressor of E1A-stimulated genes. Exp Cell Res. 2013;319(8):1165–1175. doi: 10.1016/j.yexcr.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 247.Merlet E, Atassi F, Motiani RK, Mougenot N, Jacquet A, Nadaud S, Capiod T, Trebak M, Lompre AM, Marchand A. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovasc Res. 2013;98(3):458–468. doi: 10.1093/cvr/cvt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kim MH, Ham O, Lee SY, Choi E, Lee CY, Park JH, Lee J, Seo HH, Seung M, Choi E, Min PK, Hwang KC. MicroRNA-365 inhibits the proliferation of vascular smooth muscle cells by targeting cyclin D1. J Cell Biochem. 2014;115(10):1752–1761. doi: 10.1002/jcb.24841. [DOI] [PubMed] [Google Scholar]
- 249.Li P, Liu Y, Yi B, Wang G, You X, Zhao X, Summer R, Qin Y, Sun J. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc Res. 2013;99(1):185–193. doi: 10.1093/cvr/cvt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Li P, Zhu N, Yi B, Wang N, Chen M, You X, Zhao X, Solomides CC, Qin Y, Sun J. MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation. Circ Res. 2013;113(10):1117–1127. doi: 10.1161/CIRCRESAHA.113.301306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kim S, Hata A, Kang H. Down-regulation of miR-96 by bone morphogenetic protein signaling is critical for vascular smooth muscle cell phenotype modulation. J Cell Biochem. 2014;115(5):889–895. doi: 10.1002/jcb.24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cao H, Hu X, Zhang Q, Wang J, Li J, Liu B, Shao Y, Li X, Zhang J, Xin S. Upregulation of let-7a inhibits vascular smooth muscle cell proliferation in vitro and in vein graft intimal hyperplasia in rats. J Surg Res. 2014;192(1):223–233. doi: 10.1016/j.jss.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 253.Hassel D, Cheng P, White MP, Ivey KN, Kroll J, Augustin HG, Katus HA, Stainier DY, Srivastava D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res. 2012;111(11):1421–1433. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JP, Hoefer I, Pasterkamp G, Bode C, Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123(9):999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 255.Meng S, Cao J, Wang L, Zhou Q, Li Y, Shen C, Zhang X, Wang C. MicroRNA 107 partly inhibits endothelial progenitor cells differentiation via HIF-1beta. PLoS One. 2012;7(7):e40323. doi: 10.1371/journal.pone.0040323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20(4):368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(3):1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med. 2013;5(7):949–966. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Chatterjee V, Beard RS, Jr, Reynolds JJ, Haines R, Guo M, Rubin M, Guido J, Wu MH, Yuan SY. MicroRNA-147b regulates vascular endothelial barrier function by targeting ADAM15 expression. PLoS One. 2014;9(10):e110286. doi: 10.1371/journal.pone.0110286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chamorro-Jorganes A, Araldi E, Rotllan N, Cirera-Salinas D, Suarez Y. Autoregulation of glypican-1 by intronic microRNA-149 fine tunes the angiogenic response to FGF2 in human endothelial cells. J Cell Sci. 2014;127(Pt 6):1169–1178. doi: 10.1242/jcs.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Yin KJ, Olsen K, Hamblin M, Zhang J, Schwendeman SP, Chen YE. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J Biol Chem. 2012;287(32):27055–27064. doi: 10.1074/jbc.M112.364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wu Y, Huang A, Li T, Su X, Ding H, Li H, Qin X, Hou L, Zhao Q, Ge X, Fang T, Wang R, Gao C, Li J, Shao N. MiR-152 reduces human umbilical vein endothelial cell proliferation and migration by targeting ADAM17. FEBS Lett. 2014;588(12):2063–2069. doi: 10.1016/j.febslet.2014.04.037. [DOI] [PubMed] [Google Scholar]
- 264.Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension. 2012;60(6):1407–1414. doi: 10.1161/HYPERTENSIONAHA.112.197301. [DOI] [PubMed] [Google Scholar]
- 265.Pulkkinen KH, Yla-Herttuala S, Levonen AL. Heme oxygenase 1 is induced by miR-155 via reduced BACH1 translation in endothelial cells. Free Radic Biol Med. 2011;51(11):2124–2131. doi: 10.1016/j.freeradbiomed.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 266.Yin R, Wang R, Guo L, Zhang W, Lu Y. MiR-17-3p inhibits angiogenesis by downregulating flk-1 in the cell growth signal pathway. J Vasc Res. 2013;50(2):157–166. doi: 10.1159/000345697. [DOI] [PubMed] [Google Scholar]
- 267.Kazenwadel J, Michael MZ, Harvey NL. Prox1 expression is negatively regulated by miR-181 in endothelial cells. Blood. 2010;116(13):2395–2401. doi: 10.1182/blood-2009-12-256297. [DOI] [PubMed] [Google Scholar]
- 268.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Hunninghake GM, Vera MP, Registry M, Blackwell TS, Baron RM, Feinberg MW. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest. 2012;122(6):1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286(3):2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, Antonini A, Martelli F, Capogrossi MC. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011;18(10):1628–1639. doi: 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Luo Z, Wen G, Wang G, Pu X, Ye S, Xu Q, Wang W, Xiao Q. MicroRNA-200C and -150 play an important role in endothelial cell differentiation and vasculogenesis by targeting transcription repressor ZEB1. Stem Cells. 2013;31(9):1749–1762. doi: 10.1002/stem.1448. [DOI] [PubMed] [Google Scholar]
- 272.Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393(4):643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.van Mil A, Grundmann S, Goumans MJ, Lei Z, Oerlemans MI, Jaksani S, Doevendans PA, Sluijter JP. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc Res. 2012;93(4):655–665. doi: 10.1093/cvr/cvs003. [DOI] [PubMed] [Google Scholar]
- 275.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R, Federici M. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120(15):1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 276.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107(11):1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol. 2010;30(15):3902–3913. doi: 10.1128/MCB.01237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Nicoli S, Knyphausen CP, Zhu LJ, Lakshmanan A, Lawson ND. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev Cell. 2012;22(2):418–429. doi: 10.1016/j.devcel.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA. 2011;108(20):8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang J, Wang Y, Wang Y, Ma Y, Lan Y, Yang X. Transforming growth factor beta-regulated microRNA-29a promotes angiogenesis through targeting the phosphatase and tensin homolog in endothelium. J Biol Chem. 2013;288(15):10418–10426. doi: 10.1074/jbc.M112.444463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D, Patient R, Boshoff C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 2012;120(25):5063–5072. doi: 10.1182/blood-2012-04-423004. [DOI] [PubMed] [Google Scholar]
- 282.Ying C, Sui-Xin L, Kang-Ling X, Wen-Liang Z, Lei D, Yuan L, Fan Z, Chen Z. MicroRNA-492 reverses high glucose-induced insulin resistance in HUVEC cells through targeting resistin. Mol Cell Biochem. 2014;391(1–2):117–125. doi: 10.1007/s11010-014-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123(3):282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 284.Yoo JK, Kim J, Choi SJ, Noh HM, Kwon YD, Yoo H, Yi HS, Chung HM, Kim JK. Discovery and characterization of novel microRNAs during endothelial differentiation of human embryonic stem cells. Stem Cells Dev. 2012;21(11):2049–2057. doi: 10.1089/scd.2011.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tang YY, Wo LK, Chai H. Effects of noncoding RNA NRON gene regulation on human umbilical vein endothelial cells functions. Zhonghua xin xue guan bing za zhi. 2013;41(3):245–250. [PubMed] [Google Scholar]
- 286.Bianchessi V, Badi I, Bertolotti M, Nigro P, D’Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J Mol Cell Cardiol. 2015;81:62–70. doi: 10.1016/j.yjmcc.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 287.Wang J, Chen L, Li H, Yang J, Gong Z, Wang B, Zhao X. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Mol Cell Biochem. 2015 doi: 10.1007/s11010-015-2379-1. [DOI] [PubMed] [Google Scholar]