Abstract
STUDY HYPOTHESIS
Susceptibility to inherited cryptorchidism in the LE/orl rat may be associated with genetic loci that influence developmental patterning of the gubernaculum by the fetal testis.
STUDY FINDING
Cryptorchidism in the LE/orl rat is associated with a unique combination of homozygous minor alleles at multiple loci, and the encoded proteins are co-localized with androgen receptor (AR) and Leydig cells in fetal gubernaculum and testis, respectively.
WHAT IS KNOWN ALREADY
Prior studies have shown aberrant perinatal gubernacular migration, muscle patterning defects and reduced fetal testicular testosterone in the LE/orl strain. In addition, altered expression of androgen-responsive, cytoskeletal and muscle-related transcripts in the LE/orl fetal gubernaculum suggest a role for defective AR signaling in cryptorchidism susceptibility.
STUDY DESIGN, SAMPLES/MATERIALS, METHODS
The long-term LE/orl colony and short-term colonies of outbred Crl:LE and Crl:SD, and inbred WKY/Ncrl rats were maintained for studies. Animals were intercrossed (LE/orl X WKY/Ncrl), and obligate heterozygotes were reciprocally backcrossed to LE/orl rats to generate 54 F2 males used for genotyping and/or linkage analysis. At least five fetuses per gestational time point from two or more litters were used for quantitative real-time RT–PCR (qRT–PCR) and freshly harvested embryonic (E) day 17 gubernaculum was used to generate conditionally immortalized cell lines. We completed genotyping and gene expression analyses using genome-wide microsatellite markers and single nucleotide polymorphism (SNP) arrays, PCR amplification, direct sequencing, restriction enzyme digest with fragment analysis, whole genome sequencing (WGS), and qRT–PCR. Linkage analysis was performed in Haploview with multiple testing correction, and qRT–PCR data were analyzed using ANOVA after log transformation. Imaging was performed using custom and commercial antibodies directed at candidate proteins in gubernaculum and testis tissues, and gubernaculum cell lines.
MAIN RESULTS AND THE ROLE OF CHANCE
LE/orl rats showed reduced fertility and fecundity, and higher risk of perinatal death as compared with Crl:LE rats, but there were no differences in breeding outcomes between normal and unilaterally cryptorchid males. Linkage analysis identified multiple peaks, and with selective breeding of outbred Crl:LE and Crl:SD strains for alleles within two of the most significant (P < 0.003) peaks on chromosomes 6 and 16, we were able to generate a non-LE/orl cryptorchid rat. Associated loci contain potentially functional minor alleles (0.25–0.36 in tested rat strains) including an exonic deletion in Syne2, a large intronic insertion in Ncoa4 (an AR coactivator) and potentially deleterious variants in Solh/Capn15, Ankrd28, and Hsd17b2. Existing WGS data indicate that homozygosity for these combined alleles does not occur in any other sequenced rat strain. We observed a modifying effect of the Syne2del allele on expression of other candidate genes, particularly Ncoa4, and for muscle and hormone-responsive transcripts. The selected candidate genes/proteins are highly expressed, androgen-responsive and/or co-localized with developing muscle and AR in fetal gubernaculum, and co-localized with Leydig cells in fetal testis.
LIMITATIONS, REASONS FOR CAUTION
The present study identified multiple cryptorchidism-associated linkage peaks in the LE/orl rat, containing potentially causal alleles. These are strong candidate susceptibility loci, but further studies are needed to demonstrate functional relevance to the phenotype.
WIDER IMPLICATIONS OF THE FINDINGS
Association data from both human and rat models of spontaneous, nonsyndromic cryptorchidism support a polygenic etiology of the disease. Both the present study and a human genome-wide association study suggest that common variants with weak effects contribute to susceptibility, and may exist in genes encoding proteins that participate in AR signaling in the developing gubernaculum. These findings have potential implications for the gene-environment interaction in the etiology of cryptorchidism.
LARGE SCALE DATA
Sequences were deposited in the Rat Genome Database (RGD, http://rgd.mcw.edu/).
STUDY FUNDING AND COMPETING INTERESTS
This work was supported by: R01HD060769 from the Eunice Kennedy Shriver National Institute for Child Health and Human Development (NICHD), 2P20GM103446 and P20GM103464 from the National Institute of General Medical Sciences (NIGMS), and Nemours Biomedical Research. The authors have no competing interests to declare.
Keywords: cryptorchidism, genetics, gubernaculum, whole genome sequencing, linkage analysis, rat strains
Introduction
Nonsyndromic cryptorchidism, or undescended testis, is a common reproductive anomaly of unclear etiology, most likely involving multilocus genetic susceptibility, environmental exposures and/or maternal factors. Clustering of cryptorchidism has been reported in a number of families (reviewed in (Savion et al., 1984)); in most of these cases, multiple individuals in the same generation have been affected, and phenotypic variability has been observed, particularly with regard to laterality. Extended pedigrees have not usually been examined, but autosomal dominance with reduced penetrance is most often cited as the probable mode of inheritance. Case–control studies also support familial aggregation suggesting moderate genetic risk (Czeizel et al., 1981; Jones and Young, 1982; Elert et al., 2003; Schnack et al., 2008) and higher maternal transmission, supporting the possibility that maternal factor(s), or X-linked risk alleles, influence expression of the phenotype. In a large population study, co-occurrence rates of persistent cryptorchidism were 1.8% in unrelated boys, 2.4% in paternal half-brothers, 4.3% in maternal half-brothers, 7.5% in full brothers, and 16.7 and 26.7% in dizygotic and monozygotic twins (Jensen et al., 2010). The frequencies of sporadic and familial cryptorchidism suggest multifactorial inheritance due to interaction of multiple, individually low risk susceptibility loci and/or environmental factors. While suggestive evidence implicates a role for altered hormonal signaling in the etiology of the disease (Jain and Singal, 2013; Thankamony et al., 2014), neither candidate gene nor genome-wide association studies have defined strong association with any locus (Dalgaard et al., 2012; Barthold et al., 2015a, b), nor is there consistent association with exposure to endocrine disrupting chemicals (Virtanen and Adamsson, 2012).
Spontaneous non-syndromic cryptorchidism occurs commonly in some mammalian species (Amann and Veeramachaneni, 2007) and the patterns suggest multilocus inheritance in both rats and swine (Ikadai et al., 1988; Rothschild et al., 1988), but specific loci have yet to be defined. Isolated cryptorchidism has been reported in substrains of King-Holtzman, Wistar Kyoto (WKA), Long Evans (LE) and Buffalo rats (Dressler et al., 1983; Ikadai et al., 1988; Mouhadjer et al., 1989; Patkowski et al., 1992) but is not reported as a sporadic phenotype in outbred strains. In inbred cryptorchid rat strains, 60–85% of offspring are affected with testes located in the superficial inguinal pouch, one of the most common locations found in clinical cases (Moul and Belman, 1988; van Brakel et al., 2011). Breeding studies show incomplete penetrance and evidence for complex inheritance (Ikadai et al., 1988). Fertility is reduced in affected males but spermatogenesis can be restored with orchidopexy (Patkowski et al., 1992; Lugg et al., 1996; Zhou et al., 1998). In previous studies comparing the fetal gubernaculum of a representative strain, the LE-derived LE/orl cryptorchid rat, with the outbred parental LE strain (Crl:LE), we reported altered expression of cytoskeletal and muscle-related genes (Barthold et al., 2008), abnormal shape (Barthold et al., 2006) and muscle patterning defects (Barthold et al., 2014). There was also a significant reduction in testicular testosterone and altered expression of androgen-responsive gubernacular transcripts in LE/orl fetuses (Barthold et al., 2013). In the present studies, we used genetic, genomic and protein expression methodology to define candidate genes at genomic loci associated with the LE/orl phenotype. Our findings suggest polygenic cryptorchidism susceptibility in this strain, with plausible causal variants in genes encoding proteins that co-localize with the androgen receptor (AR) in fetal gubernaculum and with Leydig cells in fetal testis.
Materials and Methods
Ethical statement
Breeding colonies were maintained at the Nemours Biomedical Research Life Science Center in accordance with Guidelines for Laboratory Animal Care and Use following approval of protocols by the Institutional Animal Care and Use Committee. The Life Science Center is accredited by the Association for the Accreditation and Assessment of Laboratory Animal Care.
Animal care and breeding
The LE/orl colony, a kind gift of Dr. Andrew Freedman, has been maintained long-term in our laboratory under standard conditions as previously described (Barthold et al., 2006, 2008, 2014), using breeding protocols to minimize inbreeding and associated subfertility. Outbred Long Evans (Crl:LE) and Sprague Dawley (Crl:SD), and inbred Wistar Kyoto (WKY/Ncrl) rats (all from Charles River Laboratories) were purchased at age 2–3 months. Standard rat strain terminology is used throughout based on the compendium in the Rat Genome Database (RGD, http://rgd.mcw.edu/). Rats received food (Lab Diet Rat Chow5021; PMI Nutrition International) and water ad libitum and were housed in polycarbonate cages with pine shaving bedding in a room with a 12:12 light cycle and controlled temperature (70 ± 2°C) and humidity (35–70%). Purchased strains were allowed a period of adjustment prior to placement with males to generate timed pregnancies, and tissue recovered at the time of ear punch identification was used for DNA extraction and genotyping. Vaginal smears were obtained to document the presence of sperm and the morning of the day of detection (sperm positive) was designated embryonic day (E) 0. We analyzed 5-year breeding records of Crl:LE and LE/orl rats used in this and prior studies (Johnson et al., 2010; Barthold et al., 2013, 2014) to determine the mean interval required to achieve sperm positive status, the number of successful deliveries, surviving pups and/or fetuses, and individual fertility data for LE/orl and Crl:LE strains.
Using microsatellite data available at the initiation of this study, we chose the WKY/NCrl rat strain for intercross-backcross experiments to maximize the number of polymorphic markers by comparison with the parent Crl:LE strain as a proxy for LE/orl. We generated a total of 25 female and 3 male LE/orl-WKY/Ncrl F1 heterozygous pups that were maintained into adulthood, then reciprocally backcrossed with LE/orl rats. F2 males were examined at or after 3 weeks of age, euthanized by CO2 inhalation, and testes were removed from affected males and processed for DNA extraction immediately or frozen. Additional breeding of outbred Crl:LE and Crl:SD strains was performed to generate animals homozygous for specific alleles in genes of interest within linkage peaks.
Fetal tissue samples and primary cell lines
For developmental studies, we generated timed pregnancies using Crl:LE and LE/orl females. We developed a restriction enzyme assay for a Esr2-Syne2 tag single nucleotide polymorphism (SNP; Supplementary Table SI) and after validation via direct sequencing, we used this assay to genotype Crl:LE adults to allow selective breeding and to genotype fetuses of heterozygous matings using DNA extracted from ear clips and liver, respectively. After confirmation of pregnancy by palpation, female dams were euthanized by CO2 inhalation at E17, E19 or E21 between 12 and 3 pm. Fetuses were collected, euthanized by decapitation, and fetal gubernacula were microdissected as previously described (Barthold et al., 2013, 2014). Pairs of gubernacula were removed en bloc and fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) overnight at 4°C. Gubernacula were then incubated in 27% sucrose for a further 12 h at 4°C, subsequently embedded in optimal cutting temperature media (OCT, Sakura Finetek USA, Inc.) and frozen at −70°C. Sectioning of the embedded tissue was carried out using a Leica CM3050 cryostat (Barthold et al., 2015b). Cell lines were generated from E17 rat gubernacula dissociated overnight by incubation with 5 mg/ml collagenase I (Worthington) in Hanks' Balanced Salt Solution (HBSS, Invitrogen) at 4°C. The organs were triturated and the cells were plated onto collagen I-coated (Sigma, 5 μg/cm3) T25 flasks in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12, Life Technologies) supplemented with 18% fetal bovine serum (Hyclone), 10 μg/ml rat epidermal growth factor (Peprotech), 2.5 μg/ml rat basic fibroblast growth factor (Peprotech), 0.4 μg/ml dexamethasone (Sigma), 10 μM of the Rho kinase inhibitor Y27632 (R&D systems) and penicillin/streptomycin (Hyclone). Cells were maintained in a humidified incubator at 37°C with 5% CO2.
Linkage analysis
DNA was isolated using the DNAeasy extraction kit (Qiagen) and PCR primer pairs (Rat Mappairs©, Research Genetics) were used for microsatellite genotyping. Four-color multiplexed marker panels were typed using capillary electrophoresis fragment analysis (either the 310 or 3130xl Genetic Analyzer, Applied Biosystems). We determined that given a maximum intermarker distance of approximately 20 cM in our genome-wide screening set, simulation of linkage analysis indicated that 15–20 rats were sufficient to detect potential areas of linkage with our chosen marker panel. However, analysis of marker genotypes of an initial cohort of affected LE/orl-WKY F2 males (n = 23) using LINKAGE ((Lathrop et al., 1984); http://www.jurgott.org/linkage/LinkagePC.html) failed to show any potential areas of linkage. To increase power to detect associated genomic regions, provide finer mapping, and to facilitate copy number variation (CNV) analysis, we generated additional affected males (n = 54 total) and used a genome-wide Rat Mapping 10 K SNP kit (Affymetrix) for genotyping by a commercial service (Cogenics, Inc.). In both analyses, markers were chosen based on a preliminary analysis of each parental strain, and uninformative microsatellites and SNPs and inconsistent or heterozygous genotypes were excluded. All data were analyzed using transmission disequilibrium testing with empirical estimation of significance by permutation analysis (n = 10 000 to correct for multiple testing) as implemented in the parenTDT function of Haploview ((Barrett et al., 2005); https://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview).
Direct (sanger) sequencing and fragment analysis
Regions of interest identified by association studies and for genotyping purposes were PCR amplified using specific primers obtained from Integrated DNA Technologies (Supplementary Table SI). For DNA fragments requiring sequence data, we used the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems) and reactions were analyzed using the 3130xl Genetic Analyzer. In some cases, we used restriction enzyme digestion (obtained from New England Biolabs) and fragment analysis for routine genotyping, after validation with formal sequencing analysis. These samples were either digested with enzyme followed by gel electrophoresis, or directly run on a 3% Nusieve agarose (Lonza)/TAE (Fisher) gel, and visualized under UV using ethidium bromide (Sigma) staining.
Whole genome sequencing
Genomic DNA libraries from Crl:LE (heterozygous Syne2-Esr2 haplotype), LE/orl and a cryptorchid Crl:LE-SD/crl intercross male (SLM26) were generated at the Delaware Biotechnology Institute (DBI) using the TruSeq DNA Sample Preparation Kit (Illumina). The three samples were indexed across six lanes of Illumina HiSeq 2500 paired-end 150 bp sequencing at the DBI Sequencing and Genotyping Center. The unprocessed Illumina sequencing data have been submitted to the NCBI Sequence Read Archive (SRA), and all data are linked under BioProject ID PRJNA289481.
Whole genome sequencing data analysis
Genome resequencing data were analyzed using genomic variant analysis pipelines developed by the Center for Bioinformatics and Computational Biology Core facility at DBI to support clinical and biomedical research (Crowgey et al., 2015). Briefly, sequencing data were trimmed and filtered to remove sequencing adapters and low-quality (Q > 0.01) bp using CLC Genomics Workbench (CLC Bio; Aarhus, Denmark). The filtered data were initially aligned to the Rattus norvegicus reference genome (RGSC 5.0/Rnor5.0) using BWA-MEM (Li and Durbin, 2009; Li, 2014). GATK UnifiedGenotyper (McKenna et al., 2010) was used to call SNPs, and PinDel (Ye et al., 2009) and BreakDancer (Chen et al., 2009; Fan et al., 2014) were used to identify larger structural variants and copy number variations (CNVs) from alignment files (BAM) and produce variant call files (VCFs). Quality control metrics were analyzed throughout the process using VCFtools ((Danecek et al., 2011), http://vcftools.sourceforge.net), PSEQ (https://atgu.mgh.harvard.edu/plinkseq/pseq.shtml), and custom scripts. VCF files were annotated with genome features based upon the RGD Genome Browser (Shimoyama et al., 2015) Rnor5.0 annotation (version 2014-08-14). Variants were further annotated with quality and genotypic data, using the Quality and Genetic Modules previously described (Crowgey et al., 2015). Various measures of deleteriousness were determined using SnpEff/ANNOVAR (Cingolani et al., 2012), Polyphen2 (Adzhubei et al., 2010) (http://genetics.bwh.harvard.edu/pph2) and PROVEAN (Choi et al., 2012) (http://provean.jcvi.org/). JBrowse (Skinner et al., 2009; Westesson et al., 2013) (http://jbrowse.org/) was utilized for visualization and comparison with the publicly available annotations and variants collected in RGD. In addition, exonic variant data available in the Rnor3.4 and Rnor5.0 assemblies of the RGD Variant Visualizer (http://rgd.mcw.edu/rgdweb/front/config.html) were downloaded for comparison analysis and our whole genome sequencing (WGS) data were submitted for inclusion in this RGD tool. We filtered VCFs in linkage peaks for potentially functional variants (premature stop codons, non-synonymous, frame shift and splice site mutations) and prioritized chromosome (chr) 6 and 16 genotypes common to LE/orl and SLM26 and distinct in Crl:LE. For CNVs, we used the eXome-Hidden Markov Model (XHMM; (Fromer and Purcell, 2014)).
We cross-referenced our data with variant data available for 42 rat strains in the RGD Variant Visualizer and we examined syntenic regions in other species using the tools available in UCSC Genome Bioinformatics (http://genome.ucsc.edu/). We uploaded all protein-coding genes within top linkage peaks from the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/projects/mapview/, Annotation Release 104) into Ingenuity Pathway Analysis (IPA©) and performed comparison analyses in IPA with our existing transcriptome data (Barthold et al., 2008, 2013, 2014; Johnson et al., 2010). We further prioritized potentially deleterious variants based on the following criteria: (i) knockout mouse or syndromic cryptorchidism genes collated in Mouse Genome Informatics (MGI; http://www.informatics.jax.org/), Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim) and/or PubMed (http://www.ncbi.nlm.nih.gov/pubmed) databases; (ii) developmental or hormone responsive genes defined by gubernaculum transcriptome data (Barthold et al., 2008, 2013, 2014; Johnson et al., 2010), (iii) genes encoding known AR-interacting proteins (http://androgendb.mcgill.ca/) (Gottlieb et al., 2012), (iv) participants in pathways defined by our cryptorchidism genome-wide association study data analyses (Barthold et al., 2015a) and/or (v) PhyloP conservation score (Pollard et al., 2010) defined in the UCSC genome browser of the corresponding human syntenic nucleotide(s), with >1.5 defined as conserved (Dong et al., 2015). Conserved and/or deleterious variants within the most promising candidate genes were prioritized for further focused studies of mRNA and protein expression in fetal gubernacular tissues and cells.
RNA extraction and qRT–PCR
Total RNA purification, cDNA generation, qRT–PCR, and data analysis using the ddCT method were described previously (Barthold et al., 2013, 2014). We utilized existing samples of cDNA from Crl:LE and LE/orl E17 and E19 fetal gubernacula, and E17 gubernacula cultured for 24 h with dihydrotestosterone (DHT; 0, 10 or 30 nM) following a 24 h washout period (n ≥ 5 replicates/group from ≥2 litters) for analysis of transcripts of interest, including those identified as hormone-responsive in our previous genome-wide studies (Johnson et al., 2010; Barthold et al., 2013). We genotyped fetal liver DNA for the Syne2-Esr2 tag SNP to allow statistical analysis of chr6 genotype-specific levels of differentially expressed (Crl:LE versus LE/orl) and/or hormone-responsive transcripts. Expression of target genes was normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression and quantified using a fetal rat embryo total RNA preparation (Applied Biosystems/Ambion) as calibrator. We used the ABI PRISM 7900HT Sequence Detection System and pre-validated TaqMan gene expression assays (Applied Biosystems) to measure transcript levels of ankyrin domain-containing 28 (Ankrd28): Rn01450330_m1; androgen receptor (Ar): Rn00560747_m1; bone morphogenetic protein 3 (Bmp3): Rn00567346_m1; chordin-like 2 (Chrdl2): Rn01510694_m1; cytochrome P450, family 11, subfamily A, polypeptide 1 (Cyp11a1): Rn00568733_m1; cytochrome P450, family 17, subfamily A, polypeptide 1 (Cyp17a1): Rn00562601_m1; cytokine receptor like factor 1 (Crlf1): Rn01419973_m1; chemokine (C-X-C motif) ligand 12 (Cxcl12): Rn00573260_m1; estrogen receptor 1 (Esr1): Rn01430446_m1; glyceraldehyde-3-phosphate dehydrogenase (Gapdh): Rn99999916_s1; hyaluronan synthase (Has2): Rn00565774_m1; insulin-like 3 (Insl3): Rn00586632_m1; luteinizing hormone/choriogonadotropin receptor (Lhcgr): Rn00564309_m1; myosin heavy chain 3 (Myh3): Rn00561539_m1; myosin heavy chain 7 (Myh7): Rn00568328_m1; myogenin (Myog): Rn00567418_m1; nuclear receptor coactivator 4 (Ncoa4): Rn01450250_g1; poly (ADP-ribose) glycohydrolase (Parg): Rn00580158_m1; prepronociceptin (Pnoc): Rn00564560_m1; relaxin/insulin-like family peptide receptor 2 (Rxfp2): Rn01412901_m1; secreted frizzled-related protein 2 (Sfrp2): Rn01458837_m1; slit homolog 3 (Slit3): Rn00580013_m1; small optic lobes homolog/calpain 15 (Solh/Capn15): Rn01309818_m1; scavenger receptor class B, member 1 (Scarb1): Rn00580588_m1; steroid acute regulatory protein (Star): Rn00580695_m1; spectrin repeat containing, nuclear envelope 2 (Syne2-5′): Rn01473640_m1 (exon 17–18); Syne2- 3′: Rn01472358_m1 (exon 108–109); transforming growth factor beta 2 (Tgfb2): Rn00579674_m1; wingless-type MMTV integration site family, member 4 (Wnt4): Rn00584577_m1 and wingless-type MMTV integration site family, member 5a (Wnt5a): Rn00575260_m1.
Imaging
Dissociated E17 gubernacula grown on collagen 1-coated plates (Fisher) in selective media were used for imaging at passage 4–5 following initial dissociation and plating. Y27632 was removed from the media, and the following day cells were washed with PBS and fixed in 4% paraformaldehyde in PBS on ice for 45 min. The cells were permeabilized by addition of 0.1% Triton X100 for 15 min, washed with PBS, and stored in PBS with 0.5% sodium azide at 4°C. For imaging of sections, intact gubernacular blocks were fixed in 4% paraformaldehyde for 12 h, incubated overnight in 27% sucrose, embedded in OCT and stored at −70°C until sectioning at 10 μm using a Leica CM3050 cryostat. A rat-specific anti-peptide nesprin-2 antibody (N2abd-83) was commercially generated at ThermoFisher Scientific (Pierce Biotechnology, Inc.) using an amino (N-) terminal polypeptide sequence within the actin binding domain (ABD) of the protein (rat sequence: KEAFRIAEHELKIPKLLEP, amino acids 245–263). The antibody was generated in rabbits and subsequently affinity purified. Other antibodies used for genotype-specific immunostaining of Crl:LE and LE/orl sections and cells included AR (Santa Cruz Biotechnology, N-20/sc-816; 1:100), N3 (anti-human C-terminal nesprin 2 antibody, a kind gift from Dr. Qiuping Zhang; 1:500), SOLH/calpain 15 (Abcam, 39881; 1:500), ANKRD28 (Bethyl Labs, A300-974A-T; 1:250) and myosin heavy chain/all isoforms (Developmental Studies Hybridoma Bank,; A4.1025; 1:50). Samples were blocked for non-specific binding by incubation with PBS/3% BSA solution for one hour at room temperature (RT). They were then incubated overnight in PBS with combinations of antibodies as indicated in the figure legends. Following incubation with primary antibodies, samples were washed 3× with PBS for 5 min each and then incubated with secondary AlexaFluor labeled antibodies (Life Technologies) in PBS for 1 h at RT. Samples were then washed and imaged using an Olympus BX60 digital photomicroscope equipped with ImagePro v7.0 software (Media Cybernetics).
Leydig cell image analysis
We observed lower levels of testicular testosterone in LE/orl as compared with Crl:LE fetuses in a prior study (Barthold et al., 2014). After defining genotype-specific testicular testosterone levels, as noted for qRT–PCR experiments, we generated estimates of Leydig cell counts per testis (Scott et al., 2008). Fetal testes were removed at E17 and E19, placed in Bouin's fixative for 1 h at room temperature, imaged for volume measurement using an Olympus BX51 microscope equipped with ImagePro Plus v7.0 software (Media Cybernetics), and placed in 70% ethanol. Samples were then embedded in paraffin and sectioned in their entirety at 5 μM. For analysis, slides were deparaffinized and permeabilized in PBS/0.05% Tween-20 (PBST) for 10 min at room temperature. Antigen retrieval was performed by heating slides at 85°F for 40 min in sodium citrate solution (pH 6) in 0.05% Tween-20. Slides were allowed to come to room temperature and a random number generator was used to select 5 sections. Slides were incubated in PBST plus 0.1% Triton for 5 min, washed in PBST for 5 min and then blocked with 10% BSA in PBST for 30 min at room temperature. A Leydig cell-specific anti-3 beta hydroxysteroid dehydrogenase antibody (3βHSD; p-18, Santa Cruz #30820) was diluted in blocking buffer at 1:250 and applied to selected sections overnight at 4°C. After rinsing, slides were incubated with AlexaFluor DaG488 secondary antibody in PBS for 1 h at room temperature. Slides were then washed with PBS, counterstained for nuclei with 4′,6-diamidino-2-phenylindole (DAPI), imaged at 20× and montaged together using the EVOS auto fluorescence microscope (Life Technologies). Images were overlaid with a grid using ImageJ (Girish and Vijayalakshmi, 2004) (http://imagej.nih.gov/ij/). Nuclear and cytoplasmic Leydig and non-Leydig point counting was used to determine Leydig cell volume per testis. For analysis of candidate proteins in testis, we used 5 μM sections processed as noted above, with 3β-HSD and anti-alpha smooth muscle actin (αSMA) antibodies to localize Leydig cells and peritubular myoid cells, respectively, and additional antibodies as described for gubernaculum studies.
Statistical analysis
Breeding data were analyzed using Fisher's exact test for frequencies and t tests for quantitative data. qRT–PCR genotype-specific gene expression data were log-transformed and analyzed using ANOVA with least significant difference (LSD) post hoc analysis. All statistics were performed using IBM® SPSS Statistics 22 and P-values of <0.05 were considered statistically significant.
Results
Fertility, fecundity and neonatal survival are reduced in the LE/orl strain
Breeding data from Crl:LE and LE/orl rats collected during a 5-year period (Table I) show significant reductions in the number of successful matings, fetuses/litter and surviving pups/litter in the LE/orl substrain compared to Crl:LE. While sperm positive status was less likely in LE/orl as compared with Crl:LE females during mating periods, successful conception, as measured by the percent of pregnancies confirmed by maternal necropsy or delivery in females documented as sperm positive, was not significantly different. However, unexplained death (12%) was more likely in LE/orl rats, typically following symptoms of respiratory distress, and total neonatal demise occurred in 30% of LE/orl litters but was not observed in the Crl:LE strain. Sudden death may be attributable to the increased susceptibility of the LE/orl strain to reactive airway disease (Rodriguez et al., 2014). As we did not maintain long term Crl:LE colonies, LE/orl rats used for breeding were significantly older (Table I). However, the differences between strains persisted when we limited our analyses to matings initiated at ≤90 days of age for both genders, and we observed no significant differences in pregnancy rates in older versus younger LE/orl males (data not shown).
Table I.
Breeding data for Long Evans Crl:LE and LE/orl rats.
| Crl:LE | LE/orl | P-value | |
|---|---|---|---|
| Females | 136 | 215 | |
| Age (mean days ± SD) | 83.4 ± 29.6 | 118.7 ± 39.3 | <0.001 |
| Males | 20 | 85 | |
| Age (mean days ± SD) | 101.5 ± 40.4 | 113.1 ± 35.9 | <0.001 |
| Mating events | 194 | 436 | |
| Time to sperm+ (mean ± SD) | 3.5 ± 3.4 | 4.4 ± 3.8 | 0.001 |
| Successful pregnancy | 67% | 54% | 0.018 |
| No sperm detected | 14% | 26% | <0.001 |
| Sperm+, not pregnant | 17% | 14% | ns |
| Maternal deaths | 3% | 12% | 0.0045 |
| Fetuses/litter | 13.2 ± 3.2 | 9.3 ± 2.5 | <0.001 |
| Surviving pups/litter | 11.9 ± 4.7 | 6.0 ± 2.8 | <0.001 |
| Subtotal pup loss | 1/14 (7%) | 15/154 (10%) | 0.019 |
| Total pup loss | 0 | 46/154 (30%) | 0.012 |
The frequency of cryptorchidism in LE/orl male pups was 51% during the period studied (78% unilateral, left > right) and a total of 31% of testes were undescended (Table II). LE/orl males with bilateral descended testes had fewer affected offspring (44%) than affected male sires (55%) but this difference was not statistically significant. However, the lower incidence of cryptorchidism in LE/orl males in recent years compared with a historical incidence of 64% (38% of testes undescended) may reflect our use of male breeders with unilateral or bilateral descended testes to optimize the vitality of the colony. Our breeding data suggest that paternity potential is indistinguishable between LE/orl males with normally descended testes and those with unilateral cryptorchidism, but significantly decreased in bilaterally cryptorchid males (Table II), as noted in man (Lee and Houk, 2013). The data suggest that reduced fertility and fecundity in the LE/orl strain is due to a combination of irregular estrus cycles, maternal illness, altered sperm counts in LE/orl males, fetal resorption and/or perinatal mortality.
Table II.
Cryptorchidism versus pregnancy outcome in LE/orl males.
| Paternal phenotype |
P-value | ||||
|---|---|---|---|---|---|
| Descended | Undescended |
||||
| Left | Right | Bilateral | |||
| Number of pups | 150 | 82 | 41 | 34 | |
| 49% | 27% | 13% | 11% | ||
| Pregnancy outcome | |||||
| Pregnant | 63% | 61% | 58% | 6% | <0.0001; excluding bilateral: ns |
| No sperm detected | 22% | 27% | 28% | 72% | |
| Sperm+, not pregnant | 14% | 12% | 14% | 22% | |
| Offspring | |||||
| All survived | 59% | 62% | Ns | ||
| Subtotal pup loss | 14% | 6% | |||
| Total pup loss | 27% | 32% | |||
LE/orl cryptorchidism is associated with multiple genomic loci
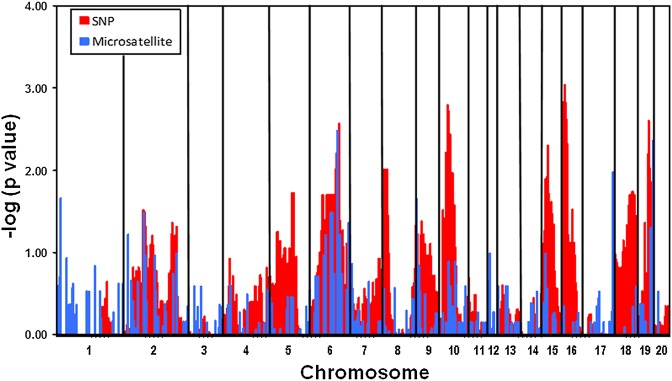
Inspection of intercross LE/orl X WKY/NCrl male progeny (n = 33) revealed a single affected cryptorchid rat (3%) suggesting either dominant inheritance or a susceptibility allele common to LE/orl and WKY/NCrl. Of 21 reciprocal backcrosses producing at least two litters, 16 delivered males with cryptorchidism; overall 54 of 292 (18%) F2 males were affected. There were no differences in occurrence of affected offspring based on the gender of the F1 and F2 parents. Results of linkage analyses using informative microsatellite (n = 242) and SNP (n = 3556) markers showed strongest signals at four regions on chromosomes 6, 10, 16 and 19 (P < 0.003; Fig. 1) and limited, less significant (P ≤ 0.01) signals on chromosomes 8 and 15. We noted overlap between microsatellite and SNP data for chromosomes 6 and 19, but not 10 and 16, for which limited polymorphic microsatellite data were available. The genomic coordinates for linkage peaks, which we defined as regions defined by markers with P-values < 0.01, are shown in Table III. We focused initially on a peak defined by both SNP and microsatellite data on chr6 and the most significant genomic peak, on chr16, by sequencing of the expressed and promoter regions of promising gene candidates. Esr2 (chr6) encodes estrogen receptor beta (ERβ), of potential interest in view of the inhibitory role of exogenous estrogen exposure on testicular descent (Staub et al., 2005) and the potential role of AR signaling (Handa et al., 2008; Rizza et al., 2014). We found a promoter variant in Esr2 (Supplementary Table SII) that flanks a conserved E–box element (maximum PhyloP score of 3.62) with functional activity (Cai et al., 2008); the reference (T) nucleotide is the minor allele, which is homozygous in LE/orl and heterozygous in Crl:LE, and the alternative nucleotide (C) is semi-conserved in other species. However, we noted low expression of Esr2/ERβ in fetal gubernaculum, as have other investigators (van Pelt et al., 1999; Staub et al., 2005). On chr16, we identified variants that are homozygous in LE/orl, absent in Crl:LE and heterozygous in Crl:SD. One of these is a 245-nt insertion, predicted to be a promoterless, partial long interspersed nucleotide element (LINE-1) sequence (www.repeatmasker.org/) in reverse orientation within intron 8–9 (chr16:8238108, Rnor5) of Ncoa4, which encodes an AR coactivator involved in muscle development (Siriett et al., 2006; Kollara and Brown, 2012). This variant does not appear to alter Ncoa4 splicing or mRNA sequence in this region in LE/orl gubernacula (data not shown). Another homozygous LE/orl allele is a non-conserved chr16 variant in the Insl3 3′UTR that is not associated with genotype-specific expression of testicular Insl3 (Barthold et al., 2014). While the variants we identified in other linkage peaks are heterozygous in Crl:LE rats (Supplementary Table SII), those on chr16 show complete divergence between LE/orl and the parent outbred LE strain.
Figure 1.
Linkage analysis of LE/orl cryptorchidism. LE/orl female rats were intercrossed with WKY/Ncrl rats, and F1 offspring were backcrossed with other LE/orl males and females. Affected males in the F2 generation were genotyped using a genome-wide microsatellite marker kit and a rat-specific single nucleotide polymorphism (SNP) array. Data were analyzed using transmission disequilibrium testing with empirical estimation of significance by permutation (n = 10 000 to correct for multiple testing) analysis as implemented in the parenTDT function of Haploview. Results are expressed as (–)log (P-value) with microsatellite and SNP results shown in blue and red, respectively.
Table III.
Major genomic linkage peaks associated with cryptorchidism in the orl rat.
| Chromosome | Baylor 3.4/rn4 assembly | RGSC 5.0/rn5 assembly | Lowest P-value | Gene candidates |
|---|---|---|---|---|
| 16 | 2186905-18078708 | 2542564-19213567 | 0.0009 | Ankrd28, Parg, Galnt15, Insl3* |
| 6 | 11855770-23900044 | 10602218-24231189 | 0.0016 | Syne2, Esr2 |
| 10 | 95430191-111097840 | 105326579-120281178 | 0.0027 | Solh/Capn15, Il9r, Slit3 |
| 19 | 47559752-49751347 | 60777664-62895213 | 0.0027 | Hsd17b2 |
| 5 | 33343313 | 37781361 | 0.005 | |
| 8 | 5589185-30926175 | 6916272-32275564 | 0.0097 |
Selective breeding for chromosome 6 and 16 alleles is sufficient to generate the LE/orl phenotype
Focusing on the functionally relevant genes in our linkage peaks, we bred Crl:LE rats heterozygous for alleles in Esr2 with Crl:SD males heterozygous for the Ncoa4 and Insl3 alleles to enrich for the LE/orl genotype. In the F1 generation of homozygous parents (F7 generation overall), we identified a male (SLM26; one of 11 siblings, and 1 of 48 males in this generation) with left-sided cryptorchidism, the first non-LE/orl male with the phenotype that we have observed in our rat studies. However, matings among this generation and with other homozygous rats failed to produce any additional cryptorchid males (of 64 male offspring) and was associated with a marked reduction in pregnancy rate (87% success, mean 15 pups/litter in heterozygous, and 30% success, mean 10 pups/litter in homozygous matings).
We generated WGS data for Crl:LE Esr2 mixed genotype, LE/orl and SLM26 male samples and prioritized conservative and/or potentially deleterious exonic variants in apparent linkage disequilibrium with the variants that we used for selective breeding to identify candidate risk genes. We achieved 14.6X, 16.7X, and 18.3X average per base coverage for the three samples respectively and identified a combined 7 238 358 SNPs, 1 582 721 indels (<15 bp), 312 519 long indels, and 90 526 other structural variants, which is similar to that reported for other rat strains (Atanur et al., 2013). We found no potentially deleterious, intragenic CNVs in our linkage peaks in LE/orl or SLM26 genomes. Following submission of our WGS data to the RGD Variant Visualizer, we directly compared LE/orl data with that of other sequenced rat strains and confirmed the strong overlap of our variant calls with those of other strains. We found no unique, potentially deleterious exonic variants within our linkage peaks that could account for the phenotype, suggesting that the combination of multiple common variants contributes to cryptorchidism susceptibility in this strain.
An exonic deletion in Syne2 is associated with LE/orl cryptorchidism
With improved annotation of the chr6 linkage peak in a newer rat genome assembly than was unavailable at completion of the linkage analysis, we identified multiple additional variants in Syne2, a gene poorly annotated in prior assemblies. We confirmed in individual samples that all coding Syne2 variants are in linkage disequilibrium (LD) with the Esr2 promoter SNP in LE-derived strains, including SLM26 (Supplementary Table SII). The most likely pathogenic Syne2 variants include a 24 bp deletion that produces loss of 8 amino acids (IAPSLATS) in the nesprin-2 protein (Syne2del), and potentially deleterious variants (S4178L and P4438L) within strongly predicted PEST (rich in proline, glutamic acid, serine, threonine) domains, motifs that predict regulation by proteolysis (Rechsteiner and Rogers, 1996) (PEST scores of 11.56 and 17.21, respectively; http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind).
WGS analysis revealed additional candidates, including potentially causal variants in Ankrd28, Parg, Solh (also known as Capn15), Slit3 and Hsd17b2 (Supplementary Table SII). Several additional candidates (hydroxysteroid (17-beta) dehydrogenase 2, Hsd17b2; polypeptide N-acetylgalactosaminyltransferase 15, Galnt15; and interleukin 9 receptor, Il9r) were not addressed further in the present study, as our existing data suggest that expression is low in gubernaculum (Barthold et al., 2008, 2014). In genotyping the strongest candidate variants in the transcrotal (TS) rat, another cryptorchid strain (tissue for genotyping kindly provided by Prof. John Hutson), we failed to find overlap with any chr6 or chr16 putative LE/orl causal variant, but note that the Solh/Capn15 variant is present and homozygous in the TS strain (Supplementary Table SII).
Altered gubernaculum transcript expression in Crl:LE fetuses homozygous for the Syne2 deletion
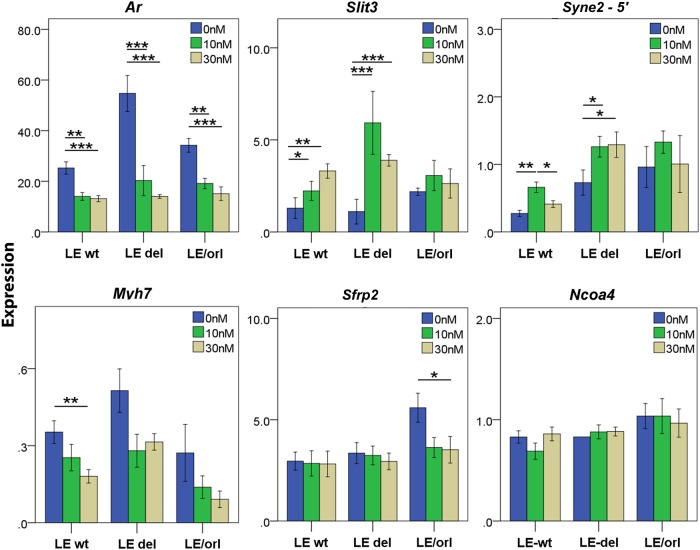
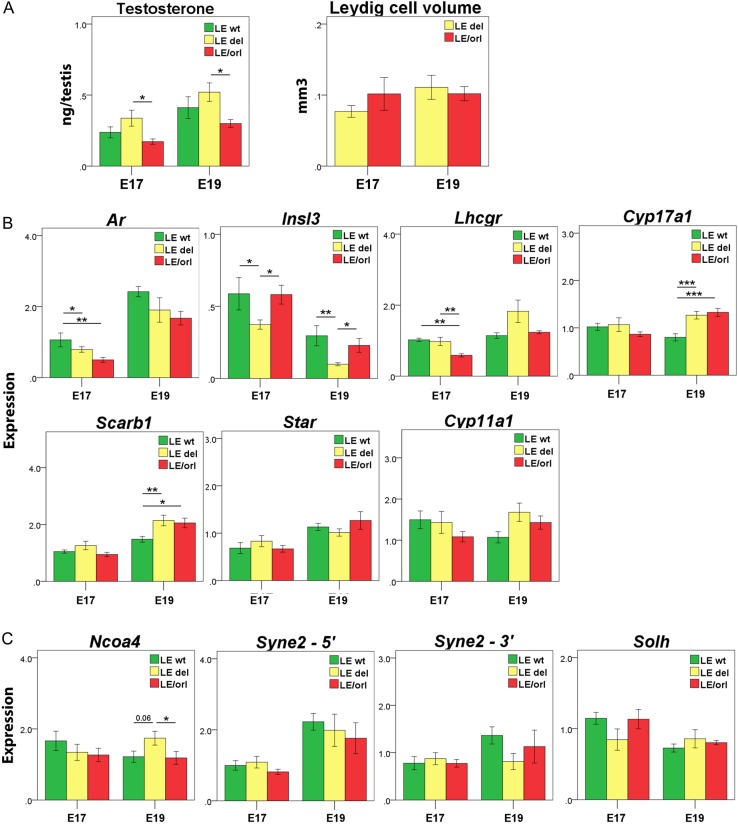
In previous studies, we identified altered expression of muscle-specific, hormone receptor and DHT- and/or INSL3-responsive (Johnson et al., 2010; Barthold et al., 2013) transcripts in LE/orl as compared with Crl:LE samples of unselected genotype (Barthold et al., 2014). We reanalyzed DHT response data to define expression patterns of samples from Crl:LE fetuses without (LE-Syne2wt) and homozygous for the Syne2 deletion allele (LE-Syne2del) and defined the genotype-specific response of a subset of LE/orl gene candidates to DHT. In prior studies, Slit3 and Myh7 transcript expression was DHT-responsive in Crl:LE but not LE/orl gubernacula, but the situation was reversed for Sfrp2. Syne2 and Slit3 are up-regulated in response to DHT in LE-Syne2wt and LE-Syne2del fetuses, but with differing patterns of expression (Fig. 2). Our data also suggest that Myh7 expression is more variable in LE-Syne2del than in LE-Syne2wt fetuses. We did not detect any genotype-specific differences in Sfrp2 and Ncoa4, which do not respond to DHT in Crl:LE fetuses (Fig. 2). Our microarray data suggested that INSL3 up-regulates Ankrd28 expression; qRT–PCR analysis showed a similar trend, but the results were not statistically significant (data not shown).
Figure 2.
Response of Ar and of LE/orl candidate gene transcripts to dihydrotestosterone (DHT). Transcript expression of androgen receptor (Ar), transcripts differentially regulated by DHT in cryptorchid and wild type (wt) fetuses, and LE/orl candidate genes in E17 gubernacula was analyzed based on fetal genotype: wt fetuses without (LE-Syne2wt), and wt (LE-Syne2del) and LE/orl fetuses with the Syne2 exonic deletion. Samples (n = 5–8 replicates/group) were exposed to DHT (0, 10 or 30 nM) for 24 h, RNA was extracted and transcript levels were measured by qRT–PCR using specific TaqMan® gene expression assays. Expression levels were calculated relative to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and an embryonic RNA control preparation. Ar, androgen receptor; Slit3, slit homolog 3; Syne2-5′, spectrin repeat containing, nuclear envelope 2 (exon 17–18 assay); Myh7, myosin heavy chain 7; Sfrp2, secreted frizzled-related protein 2 and Ncoa4, nuclear receptor coactivator 4. *P < 0.05, **P < 0.01 by ANOVA after log transformation.
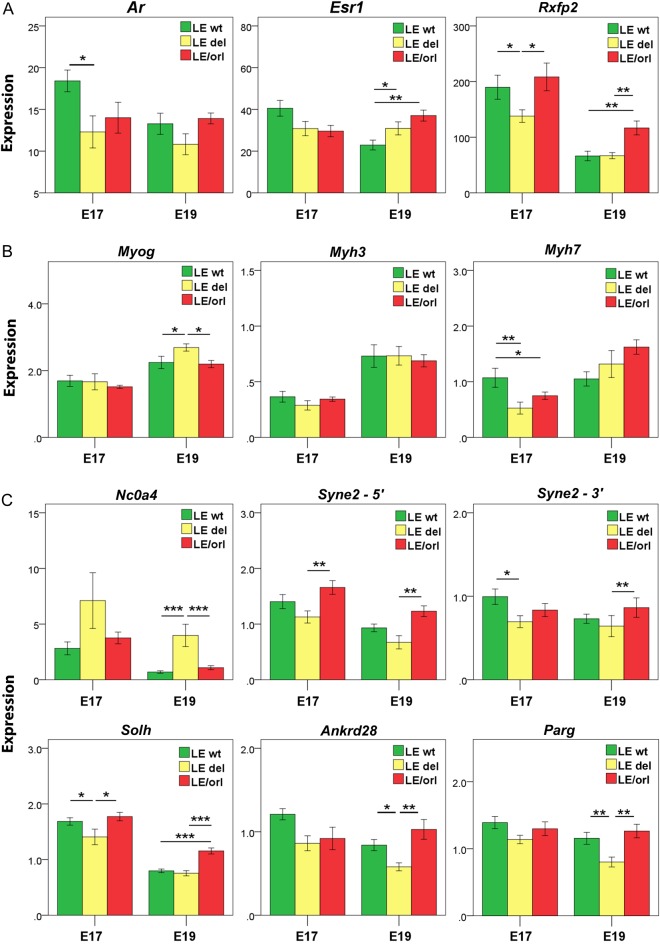
Syne2, Ankrd28, Ncoa4, Parg and Solh/Capn15 transcripts are expressed at higher levels in E17 than in E19 fetal gubernaculum and show Syne2 genotype-specific expression at one or both gestational days (Fig. 3). The majority of hormone receptor, muscle-specific and candidate gene transcripts were also differentially expressed in LE-Synedel as compared with LE-Syne2wt and/or LE/orl samples. Most notably, we observed a 2.5–4-fold increase in Ncoa4 expression at E17 and E19 in LE-Syne2del as compared with both LE/orl and LE-Syne2wt gubernaculum using an assay that spans the intron containing the Ncoa4 insertion in the LE/orl strain. This suggests that the Syne2 deletion allele modifies Ncoa4 expression in LE wt rats, but that significant up-regulation does not occur in LE/orl gubernacula, potentially due to the Ncoa4 variant or other factors that alter Ncoa4 expression in this strain. Further studies are needed to determine the relationship of Syne2 and Ncoa4, and their encoded proteins, with each other and with AR signaling pathways in the fetal gubernaculum.
Figure 3.
Genotype-specific transcript expression in fetal gubernaculum. Expression levels of (A) hormone receptors, (B) muscle-specific transcripts and (C) LE/orl candidate genes in E17 and E19 gubernacula (mean ± SEM) from wild type (wt) fetuses without (LE-Syne2wt), and wt (LE-Syne2del), and LE/orl fetuses with the Syne2 exonic deletion (n = 8–12 replicates per group). Expression levels were calculated relative to Gapdh and an embryonic RNA control preparation. (A) Ar, androgen receptor; Esr1, estrogen receptor 1; Rxfp2, relaxin/insulin-like family peptide receptor 2; (B) Myog, myogenin; Myh3, myosin heavy chain 3; Myh7, myosin heavy chain 7; (C) Ncoa4, nuclear receptor coactivator 4; Syne2-5′, spectrin repeat containing, nuclear envelope 2 (exon 17–18 assay); Syne2-3′, spectrin repeat containing, nuclear envelope 2 (exon 108–109 assay); Solh, small optic lobes homolog; Ankrd28, ankyrin domain-containing 28 and Parg, poly (ADP-ribose) glycohydrolase. *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA after log transformation.
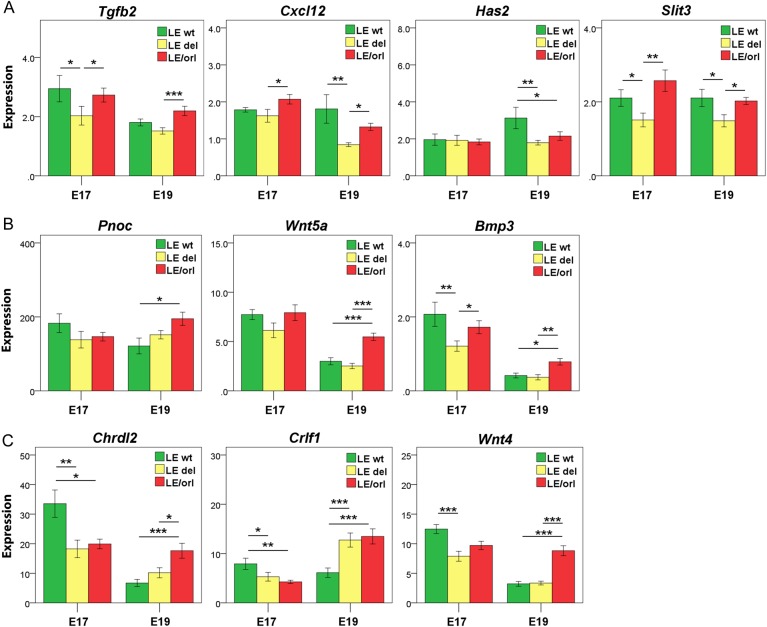
Since Ncoa4 is a known nuclear receptor coactivator (Kollara and Brown, 2012), we also measured levels of hormone-responsive genes (Johnson et al., 2010; Barthold et al., 2013) in LE-Syne2wt, LE-Syne2del and LE/orl gubernacula at E17 and E19, to identify additional modifying effects of the Syne2 deletion at the transcript level. Transcripts assayed included: Bmp4, Tgfb2 and Cxcl12 (down-regulated by DHT); Has2 and Slit3 (up-regulated by DHT); Pnoc, Wnt5a and Bmp3 (up-regulated by INSL3); Chrdl2 and Crlf1 (up-regulated by INSL3 and DHT); and Wnt4 (up-regulated by INSL3 and down-regulated by DHT). These data also show differential expression of all DHT-responsive genes E17 and/or E19 in LE-Syne2del as compared with LE-Syne2wt fetuses (Fig. 4).
Figure 4.
Genotype-specific expression of hormone-responsive transcripts in fetal gubernaculum. Expression levels of DHT- and/or INSL3-responsive transcripts in E17 and E19 gubernacula (mean ± SEM) collected from wild type (wt) fetuses without (LE-Syne2wt), and wt (LE-Syne2del), and LE/orl fetuses with the Syne2 exonic deletion (n = 8–12 replicates per group). Expression levels were calculated relative to Gapdh and an embryonic RNA control preparation. (A) Transcripts down-regulated (Tgfb2, transforming growth factor 2 and Cxcl12, chemokine (C-X-C motif) ligand 12) or up-regulated (Has2, hyaluronan synthase and Slit3, slit homolog 3) by DHT in wt fetal gubernaculum. (B) Transcripts up-regulated by INSL3 (Pnoc, prepronociceptin; Wnt5a, wingless-type MMTV integration site family, member 5a; Bmp3, bone morphogenetic protein 3) in wt fetal gubernaculum. (C) Transcripts up-regulated by both DHT and INSL3 (Chrdl2, chordin-like 2 and Crlf1, cytokine receptor like factor 1) or up-regulated by INSL3 and down-regulated by DHT (Wnt4, wingless-type MMTV integration site family, member 4) in wt fetal gubernaculum. *P < 0.05,**P < 0.01, ***P < 0.001 by ANOVA after log transformation.
Altered testicular transcript expression in Crl:LE fetuses homozygous for the Syne2 deletion
In our previous work, we identified reduced levels of testicular testosterone LE/orl as compared with Crl:LE fetuses (Barthold et al., 2014). When we calculated Syne2 genotype-specific testicular testosterone, mean levels were significantly different between LE/orl and LE-Syne2del, but not between LE/orl and LE-Syne2wt fetuses (Fig. 5A). However, despite lower testosterone levels, we did not observe a significant decrease in total Leydig cell volume in LE/orl as compared with LE-Syne2del testes (n = 5 replicates per group) in a limited morphometric analysis. To study this further, we measured levels of Leydig cell (Ar, Insl3 and Lhcgr), and steroidogenesis transcripts (Cyp17a1, Scarb1, Star and Cyp11a1) in LE-Syne2wt, LE-Syne2del and LE/orl testis at E17 and E19 (Fig. 5B). In these analyses, we noted differential expression of Ar, Insl3, Cyp17a1 and Scarb1 in LE-Syne2wt versus LE-Syne2del testes at E17 and/or E19. We also observed suggestive up-regulation of Ncoa4 expression in LE-Syne2del testes that was less robust than in gubernaculum, but no significant genotype-specific differences for Solh or Syne2 (Fig. 5C). These data are preliminary but suggest that nesprin-2 may have functional roles in both gubernaculum and Leydig cells, with the potential to regulate androgen production and action, respectively.
Figure 5.
Genotype-specific testicular testosterone and Leydig cell transcript expression. Expression levels in testis from wild type (wt) fetuses without (LE-Syne2wt), and wt (LE-Syne2del), and LE/orl fetuses with the spectrin repeat containing, nuclear envelope 2 (Syne2) exonic deletion (n = ≥5 replicates per group). (A) Testicular testosterone levels (ng/testis, mean ± SEM) were significantly higher in LE-Syne2del as compared with LE/orl testes, but stereoscopic Leydig cell volumes were not significantly different between these groups. (B) A subset of Leydig cell- and steroidogenesis-specific transcripts show differential expression in LE-Syne2wt and LE-Syne2del samples. (C) Expression of LE/orl candidate genes does not show genotype-specific differences in testis, except for Ncoa4, which shows modest up-regulation in LE-Syne2del fetal testis as compared with gubernaculum. Ar, androgen receptor; Insl3, insulin-like 3; Lhcgr, luteinizing hormone/choriogonadotropin receptor; Cyp17a1, cytochrome P450, family 17, subfamily A, polypeptide 1; Scarb1, scavenger receptor class B, member 1; Star, steroid acute regulatory protein; Cyp11a1, cytochrome P450, family 11, subfamily A, polypeptide 1; Ncoa4, nuclear receptor coactivator 4; Syne2-5′, spectrin repeat containing, nuclear envelope 2 (exon 17–18 assay); Syne2-3′, spectrin repeat containing, nuclear envelope 2 (exon 108–109 assay) and Solh, small optic lobes homolog. *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA after log transformation of data.
Cryptorchidism gene candidates encode proteins expressed in fetal gubernaculum and testis
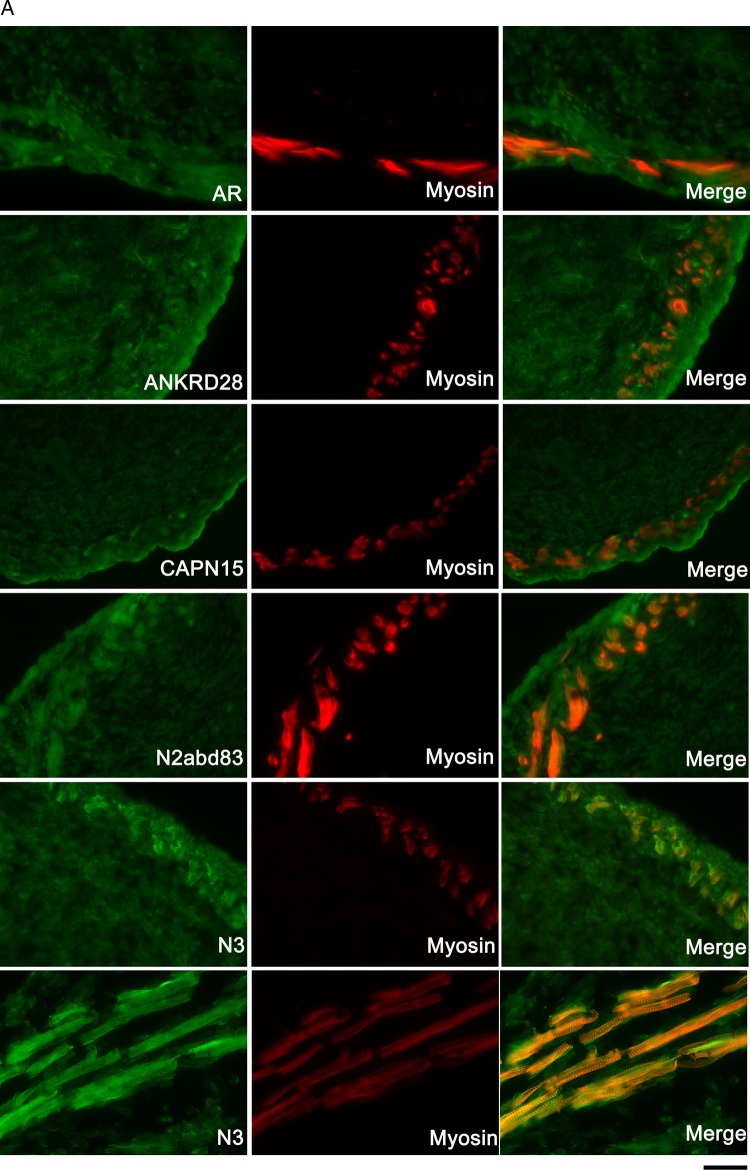
We studied expression patterns of AR and of proteins encoded by our cryptorchidism susceptibility loci (Fig. 6) and also found nuclear AR expression in the majority of mesenchymal cells within the core of the E17 gubernaculum, and in association with the outer layer of developing muscle. We also found enriched expression of ANKRD28, SOLH/calpain 15, and nesprin-2 in cells surrounding the developing cremaster (Fig. 6A). Imaging using a custom Nterminal antibody specific for the actin-binding domain of rat nesprin-2 (N2abd-83) showed expression in peripheral mesenchymal cells while N3, a well-characterized nesprin-2 C-terminal antibody, showed expression within muscle striations, consistent with known localization of this antibody to the Z-disk and I-band in striated muscle (Zhang et al., 2005).
Figure 6.
Gubernacular expression of proteins encoded by LE/orl cryptorchidism candidate genes. (A) Crl:LE E17 gubernaculum sections stained for AR (androgen receptor), ANKRD28 (ankyrin domain-containing 28), calpain 15 (CAPN15, also known as small optic lobes homolog, SOLH), nesprin-2 (N3abd83 (N-terminal), and N3 (C-terminal) antibodies) and myosin heavy chain (all isoforms; A4.1025) shows enhanced expression of ANKRD28, calpain 15 and the actin-binding domain specific nesprin-2 antibody (green) in the developing muscle layer localized by myosin (red). AR is seen in mesenchymal nuclei surrounding developing muscle, and the C-terminal nesprin-2-specific antibody (N3) is co-localized with myosin within striated muscle (×40, except for lowest 3 N3 images, ×100; scale bar: 40 μm for ×40 images and 100 μm for ×100 images). (B) Primary Crl:LE gubernacular cells grown on collagen 1-coated plates and immunostained for androgen AR, ANKRD28, CAPN15 and nesprin-2 (N3abd83 antibody) show co-localization of all proteins with AR in these cells (×40, scale bar: 40 μm).
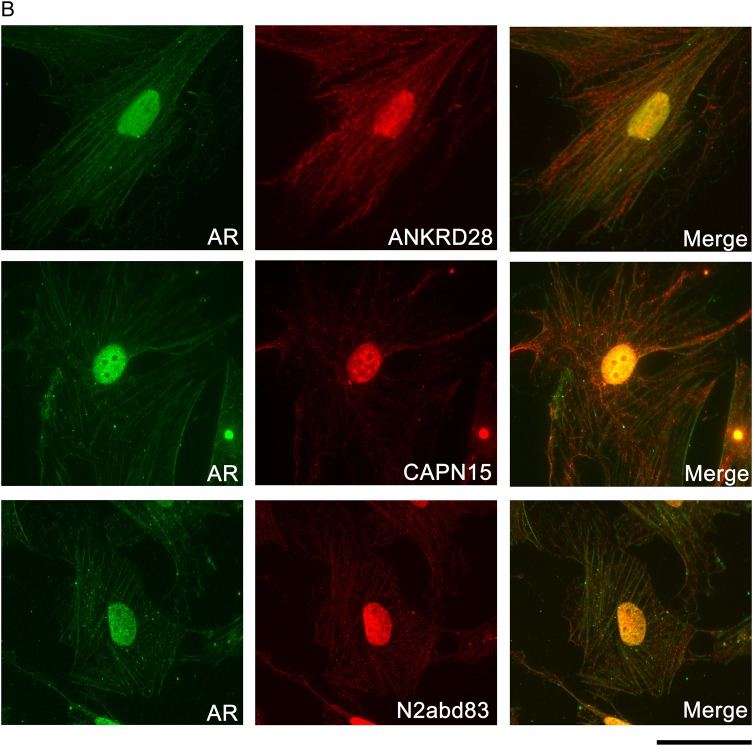
Our conditionally immortalized (Liu et al., 2012) primary gubernacular cell lines express myogenic markers including desmin, αSMA (also expressed in the striated fetal cremaster muscle) and integrin α7, but do not express paired box 7 (PAX7) or myosin heavy chain protein, indicative of committed and differentiated muscle, respectively (data not shown). These cells also consistently express AR, localized to both nucleus and cytoplasm, and we observed co-expression of AR with nesprin-2 (N2abd83), ANKRD28 and calpain 15 in both locations (Fig. 6B). In contrast, immunolocalization of nesprin-2 using the carboxy (C)-terminal N3 antibody (data not shown) revealed poor co-localization with AR, and prominent staining in nucleoli, structures that consistently show low AR expression in our cells (Fig. 6B). We did not observe qualitative differences in nesprin-2 expression based on strain or Syne2 genotype (data not shown).
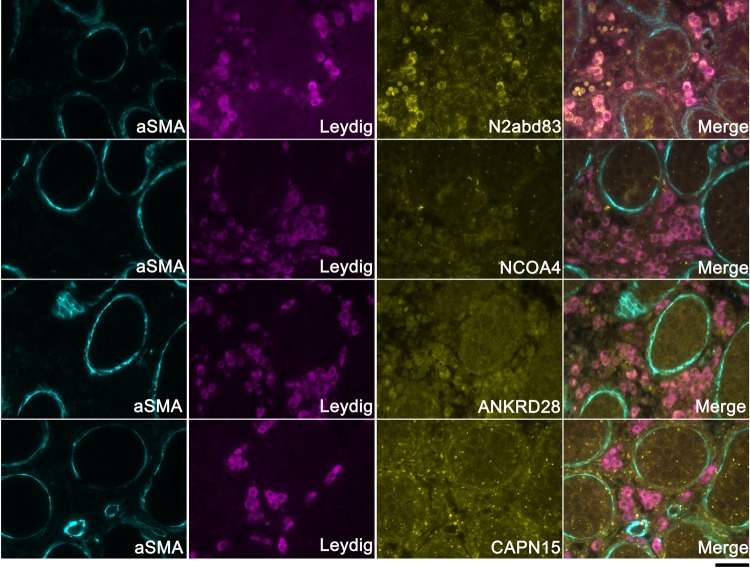
In testis images, 3βHSD in Leydig cells was strongly co-localized with nesprin-2, but less strongly with other candidate proteins, including NCOA4/ARA70 (androgen receptor associated 70), ANKRD28 and SOLH/calpain 15 at E17 (Fig. 7) and E19 (data not shown). Calpain 15 in particular appeared to be more widely expressed in other cells within the interstitial space and within tubules. Expression of nesprin-2 in Leydig cells is consistent with Syne2 genotype-specific differences in Ar, Insl3 and genes involved in steroidogenesis.
Figure 7.
Testicular expression of proteins encoded by LE/orl cryptorchidism candidate genes. Imaging of E19 testis suggests localization of ankyrin domain containing 28 (ANKRD28), nuclear receptor coactivator 4/androgen receptor associated 70 (NCOA4/ARA70) and nesprin-2 in Leydig cells with minimal if any intratubular expression of these proteins, and calpain 15 (CAPN15, also known as small optic lobes homolog, SOLH) in both interstitial and intratubular locations. Peritubular myoid and Leydig cells are localized using the specific antibodies alpha smooth muscle actin (aSMA) and 3-beta hydroxysteroid dehydrogenase (3βHSD), respectively (×40, scale bar: 40 μm).
Discussion
These data suggest that cryptorchidism in the LE/orl rat is a polygenic trait that is associated with variants in genes that are expressed in both gubernaculum and testis. Breeding studies of another cryptorchid rat strain (Gumbreck et al., 1984), and association studies in pigs (Elansary et al., 2015) and man (Barthold et al., 2015a) provide further evidence that cryptorchidism is inherited as a polygenic trait. Using high throughput rat genotyping arrays and recent major advances in variant mapping of the rat genome, we identified promising candidates, including Syne2, Ncoa4, Solh/Capn15 and Hsd17b2, within four cryptorchidism-associated linkage peaks. These contain conserved or semi-conserved exonic variants with minor allele frequencies ranging from 0.24–0.36 in previously genotyped rat strains, but the combination of alleles are unique to LE/orl. We validated these loci by generating a cryptorchid male via selective breeding of outbred strains for alleles at two of these loci, yet the very low incidence of cryptorchidism that we observed in this breeding protocol implies that additional alleles that were not accounted for contribute to risk. Previous rat sequencing data suggested that strain-specific haplotype blocks underlie disease susceptibility (Atanur et al., 2013), but our analysis did not identify any unique, potentially damaging variants within LE/orl linkage peaks. Moreover, we found that only the Solh/Capn15 variant is present in the transcrotal cryptorchid rat strain, which may explain subtle phenotypic differences between LE/orl and TS (JM Hutson, unpublished data). Based on the disparate genetic architecture of the parental strains (Atanur et al., 2013; Hermsen et al., 2015) from which reported colonies of cryptorchid rats emerged, it is likely that susceptibility is dependent upon genetic background and varies by strain. However, the increased propensity for left-sided disease in LE/orl rats mirrors that observed in other cryptorchid models; in Buffalo rats (Patkowski et al., 1992) and mice with inactivation of Wt1 (Kaftanovskaya et al., 2013), cryptorchidism is always left-sided.
The promising susceptibility genes that we identified in cryptorchidism-associated linkage peaks show functional overlap with our prior rat studies (Barthold et al., 2008, 2014) and with suggestive loci that we identified in a genome-wide association study of nonsyndromic cryptorchidism (Barthold et al., 2015a, b). In the LE/orl fetus, we found misexpression of cytoskeletal and hormone-responsive genes in fetal gubernaculum, abnormal muscle development and reduced testicular testosterone. Syne2, Ncoa4, Ankrd28 and Solh/Capn15 encode proteins that co-localize with nuclear AR expression in gubernaculum cells, with the cremaster myogenic zone in the periphery of the fetal gubernaculum, and with Leydig and other interstitial cells of the testis, suggesting that they may participate in cytoskeletal pathways, muscle development and hormone production and/or responses in the developing gubernaculum and testis. AR-positive mesenchymal cells are required for normal fetal gubernacular development and testicular descent, and transgenic deletion of Ar in these cells interferes with normal muscle development (Kaftanovskaya et al., 2012). Interestingly, SYNE2, ANKRD28 and PARG were recently identified as novel AR-interacting proteins in prostate cancer cells (Hsiao et al., 2015). A role for cryptorchidism risk genes participating at multiple levels in hormonal pathways required for gubernacular development and testicular descent is biologically plausible, but defining a role for these LE/orl cryptorchidism susceptibility candidates in muscle development, hormone production and/or AR signaling will require further studies.
The most potentially damaging allele we identified is a deletion within Syne2, which encodes nesprin-2, a large cytoskeletal protein with a myriad of isoforms that participates in muscle development and has been associated with Emery–Dreifuss muscular dystrophy in humans, although transgenic inactivation of the gene in mice does not produce an evident phenotype (Zhang et al., 2007; Randles et al., 2010; Autore et al., 2013; Duong et al., 2014). Nesprin-2 isoform expression changes during myogenesis, and the apparent differential expression pattern using N- and C-terminal antibodies in the developing cremaster muscle suggests that cell-specific isoforms may also exist in the developing gubernaculum. Nesprins contain tandem N-terminal calponin homology (CH) domains that are structurally similar to the actin-binding domains of actinins, known nuclear receptor coregulators (Huang et al., 2004; Khurana et al., 2011). Actin-binding and other cytoskeletal proteins are also calpain substrates (Franco and Huttenlocher, 2005) and interact with AR in muscle (Ting and Chang, 2008). Notably, Esr1/Syne1 and Esr2/Syne2 comprise adjacent gene pairs on rat chromosomes 1 and 6, respectively.
As the Syne2 product has structural similarities to other actin-binding proteins that participate in nuclear receptor signaling, we compared DHT-responsive transcript expression in LE-Syne2wt and LE-Syne2del fetal gubernaculum, and found differences that suggest that the Syne2 allele modifies expression of DHT response in wt rats. Most notably, expression levels of Ncoa4, a gene within our chr16 linkage peak that is highly expressed in the fetal gubernaculum and encodes a nuclear receptor coactivator (Kollara and Brown, 2012), were markedly increased in LE-Syne2del male gubernacula. If nesprin-2 participates in AR signaling, a compensatory response may exist in Crl:LE fetuses homozygous for the Syne2 deletion, as many proteins contribute to the AR interactome in androgen sensitive tissues. The LE/orl Ncoa4 intronic insertion does not appear to directly alter transcript levels; however, there is a shorter developmental isoform of NCOA4/ARA70 exists (Kollara and Brown, 2010) that is expressed in gubernaculum (data not shown), but its origin and regulation are poorly understood. Further studies will be required to define a functional role for the Ncoa4 variant, and to determine whether nesprin-2 and NCOA4/ARA70 function as AR coactivators in the gubernaculum. In the fetal testis, these proteins are expressed in Leydig cells, and the LE-Synedel genotype is associated with increased expression of Ncoa4, although this up-regulation is less striking than in the gubernaculum. However, testicular testosterone and expression of Ar, Insl3 and other Leydig cell-specific genes is altered in LE-Syne2del as compared with LE-Syne2wt and/or LE/orl males, suggesting a direct or indirect functional association between Syne2 and steroidogenesis.
Candidate variants in other linkage peaks encode proteins that could interact with each other and with AR. For example, interaction of spectrin and ankyrin domain-containing proteins has a role in organ development and in red blood cell shape (Bennett and Baines, 2001; Saito et al., 2015). The function of the LE/orl candidate ANKRD28 remains unknown, but the protein is known to be associated with focal adhesion formation and may therefore participate in cytoskeleton-dependent processes such as cell migration and muscle development (Kiyokawa and Matsuda, 2009; Tachibana et al., 2009). Solh/Capn15, an atypical calpain with multiple zinc-finger, potential nucleic acid binding domains, is well-expressed in fetal male reproductive tissues (http://gudmap.org) (Harding et al., 2011) and also shows nuclear and cytoplasmic localization in gubernacular cells. A potentially damaging variant could affect the protease function of calpain 15, and may be of functional importance if the additional, potentially deleterious variants in the predicted PEST domains of nesprin-2 are calpain target sites. Moreover, the 5 mb telomeric region of the chr10 linkage peak containing Solh/Capn15 is syntenic with human 16p13.3; deletion or translocation of this region is associated with the α-thalassemia mental retardation syndrome, ATX-16 (Holinski-Feder et al., 2000; Gibson et al., 2008). Although cryptorchidism is a component of this syndrome, the genetic etiology has not been determined.
We did not find any intragenic structural variants in our linkage peaks to consider further as potential causal alleles. The rat genome assembly is less complete, accurate and well-annotated than those of human and mouse, and future re-analysis of our data may provide additional insight into the genetic basis of cryptorchidism in this strain. However, the present data add further support for altered AR and cytoskeletal signaling in the pathogenesis of cryptorchidism susceptibility, and show overlap with suggestive GWAS signals that we have identified in our human studies of nonsyndromic cryptorchidism (Barthold et al., 2015a). Studies that define the AR interactome and the role of AR signaling in gubernacular development will help to better define the mechanism of action of specific candidate alleles, and determine how gubernaculum-specific perturbations in AR signaling may contribute to the risk of cryptorchidism.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors' roles
J.S.B., A.K.R., S.W.P., K.S.-C., S.M.M., R.E.A.: study design; J.S.B., A.K.R., S.W.P.: writing of manuscript; J.P.: animal care phenotyping and breeding; J.P., J.S.B., A.K.R., A.M., M.L.M., J.R., M.O.A.: data and/or sample collection and analysis; A.M., A.K.R.: imaging; J.P., A.M., Y.W., K.S.-C., S.M.M., R.E.A., M.D., M.L.M., J.R., M.O.A.: review/approval of manuscript.
Funding
This work was supported by: R01HD060769 from the Eunice Kennedy Shriver National Institute for Child Health and Human Development (NICHD), 2P20GM103446 and P20GM103464 from the National Institute of General Medical Sciences (NIGMS), and Nemours Biomedical Research.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We thank: Erin Crowgey for her guidance and assistance in performing bioinformatics analyses; Dr Quiping Zhang for helpful discussions related to nesprin-2; the CBCB Bioinformatics Core Facility, Bruce Kingham and the Sequencing and Genotyping Center at the University of Delaware, the Biomolecular Core Laboratory of Nemours Biomedical Research, and Delaware INBRE Core Access Award funding that allowed successful completion of this work.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RP, Veeramachaneni DN. Cryptorchidism in common eutherian mammals. Reproduction 2007;133:541–561. [DOI] [PubMed] [Google Scholar]
- Atanur SS, Diaz AG, Maratou K, Sarkis A, Rotival M, Game L, Tschannen MR, Kaisaki PJ, Otto GW, Ma MC et al. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 2013;154:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autore F, Pfuhl M, Quan X, Williams A, Roberts RG, Shanahan CM, Fraternali F. Large-scale modelling of the divergent spectrin repeats in nesprins: giant modular proteins. PLoS One 2013;8:e63633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- Barthold JS, Si X, Stabley D, Sol-Church K, Campion L, McCahan SM. Failure of shortening and inversion of the perinatal gubernaculum in the cryptorchid long-evans orl rat. J Urol 2006;176:1612–1617. [DOI] [PubMed] [Google Scholar]
- Barthold JS, McCahan SM, Singh AV, Knudsen TB, Si X, Campion L, Akins RE. Altered expression of muscle- and cytoskeleton-related genes in a rat strain with inherited cryptorchidism. J Androl 2008;29:352–366. [DOI] [PubMed] [Google Scholar]
- Barthold JS, Wang Y, Robbins A, Pike J, McDowell E, Johnson KJ, McCahan SM. Transcriptome analysis of the dihydrotestosterone-exposed fetal rat gubernaculum identifies common androgen and insulin-like 3 targets. Biol Reprod 2013;89:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold JS, Robbins A, Wang Y, Pugarelli J, Mateson A, Anand-Ivell R, Ivell R, McCahan SM, Akins RE Jr. Cryptorchidism in the orl rat is associated with muscle patterning defects in the fetal gubernaculum and altered hormonal signaling. Biol Reprod 2014;91:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold JS, Wang Y, Kolon TF, Kollin C, Nordenskjold A, Olivant Fisher A, Figueroa TE, BaniHani AH, Hagerty JA, Gonzalez R et al. Pathway analysis supports association of nonsyndromic cryptorchidism with genetic loci linked to cytoskeleton-dependent functions. Hum Reprod 2015a;30:2439–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold JS, Wang Y, Kolon TF, Kollin C, Nordenskjold A, Olivant Fisher A, Figueroa TE, BaniHani AH, Hagerty JA, Gonzalez R et al. Phenotype specific association of the TGFBR3 locus with nonsyndromic cryptorchidism. J Urol 2015b;193:1637–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev 2001;81:1353–1392. [DOI] [PubMed] [Google Scholar]
- Cai W, Rambaud J, Teboul M, Masse I, Benoit G, Gustafsson JA, Delaunay F, Laudet V, Pongratz I. Expression levels of estrogen receptor beta are modulated by components of the molecular clock. Mol Cell Biol 2008;28:784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009;6:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012;7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wangle L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowgey EL, Stabley DL, Chen C, Huang H, Robbins KM, Polson SW, Sol-Church K, Wu CH. An integrated approach for analyzing clinical genomic variant data from next-generation sequencing. J Biomol Tech 2015;26:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel A, Erodi E, Toth J. Genetics of undescended testis. J Urol 1981;126:528–529. [DOI] [PubMed] [Google Scholar]
- Dalgaard MD, Weinhold N, Edsgard D, Silver JD, Pers TH, Nielsen JE, Jorgensen N, Juul A, Gerds TA, Giwercman A et al. A genome-wide association study of men with symptoms of testicular dysgenesis syndrome and its network biology interpretation. J Med Genet 2012;49:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al. The variant call format and VCFtools. Bioinformatics 2011;27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler JB, Allison JE, Chung KW. Ectopic testes: a heritable mutation in the King-Holtzman rat: androgen-binding protein in testes and epididymides. Biol Reprod 1983;29:1313–1317. [DOI] [PubMed] [Google Scholar]
- Duong NT, Morris GE, Lamle T, Zhang Q, Sewry CA, Shanahan CM, Holt I. Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS One 2014;9:e94380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elansary M, Stinckens A, Ahariz N, Cambisano N, Coppieters W, Grindflek E, van Son M, Buys N, Georges M. On the use of the transmission disequilibrium test to detect pseudo-autosomal variants affecting traits with sex-limited expression. Anim Genet 2015;46:395–402. [DOI] [PubMed] [Google Scholar]
- Elert A, Jahn K, Heidenreich A, Hofmann R. The familial undescended testis. Klin Padiatr 2003;215:40–45. [DOI] [PubMed] [Google Scholar]
- Fan X, Abbott TE, Larson D, Chen K. BreakDancer—identification of genomic structural variation from paired-end read mapping. Curr Protoc Bioinformatics 2014;2014:15:15.1:15.6.1–15.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci 2005;118:3829–3838. [DOI] [PubMed] [Google Scholar]
- Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet 2014;81:7.23.1–7.23.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson WT, Harvard C, Qiao Y, Somerville MJ, Lewis ME, Rajcan-Separovic E. Phenotype-genotype characterization of alpha-thalassemia mental retardation syndrome due to isolated monosomy of 16p13.3. Am J Med Genet A 2008;146A:225–232. [DOI] [PubMed] [Google Scholar]
- Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J Cancer 2004;41:47. [PubMed] [Google Scholar]
- Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M. The androgen receptor gene mutations database: 2012 update. Hum Mutat 2012;33:887–894. [DOI] [PubMed] [Google Scholar]
- Gumbreck LG, Stanley AJ, Allison JE, Peeples EE. Ectopic testes in the Norway rat. J Exp Zool 1984;230:151–154. [DOI] [PubMed] [Google Scholar]
- Handa RJ, Pak TR, Kudwa AE, Lund TD, Hinds L. An alternate pathway for androgen regulation of brain function: activation of estrogen receptor beta by the metabolite of dihydrotestosterone, 5alpha-androstane-3beta,17beta-diol. Horm Behav 2008;53:741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y et al. The GUDMAP database—an online resource for genitourinary research. Development 2011;138:2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R, de Ligt J, Spee W, Blokzijl F, Schafer S, Adami E, Boymans S, Flink S, van Boxtel R, van der Weide RH et al. Genomic landscape of rat strain and substrain variation. BMC Genomics 2015;16:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinski-Feder E, Reyniers E, Uhrig S, Golla A, Wauters J, Kroisel P, Bossuyt P, Rost I, Jedele K, Zierler H et al. Familial mental retardation syndrome ATR-16 due to an inherited cryptic subtelomeric translocation, t(3;16)(q29;p13.3). Am J Hum Genet 2000;66:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao JJ, Ng BH, Smits MM, Martinez HD, Jasavala RJ, Hinkson IV, Fermin D, Eng JK, Nesvizhskii AI, Wright ME. Research resource: androgen receptor activity is regulated through the mobilization of cell surface receptor networks. Mol Endocrinol 2015;29:1195–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Huang CJ, Wang WM, Kang JC, Hsu WC. The enhancement of nuclear receptor transcriptional activation by a mouse actin-binding protein, alpha actinin 2. J Mol Endocrinol 2004;32:481–496. [DOI] [PubMed] [Google Scholar]
- Ikadai H, Ajisawa C, Taya K, Imamichi T. Suprainguinal ectopic scrota of TS inbred rats. J Reprod Fertil 1988;84:701–707. [DOI] [PubMed] [Google Scholar]
- Jain VG, Singal AK. Shorter anogenital distance correlates with undescended testis: a detailed genital anthropometric analysis in human newborns. Hum Reprod 2013;28:2343–2349. [DOI] [PubMed] [Google Scholar]
- Jensen MS, Toft G, Thulstrup AM, Henriksen TB, Olsen J, Christensen K, Bonde JP. Cryptorchidism concordance in monozygotic and dizygotic twin brothers, full brothers, and half-brothers. Fertil Steril 2010;93:124–129. [DOI] [PubMed] [Google Scholar]
- Johnson KJ, Robbins AK, Wang Y, McCahan SM, Chacko JK, Barthold JS. Insulin-like 3 exposure of the fetal rat gubernaculum modulates expression of genes involved in neural pathways. Biol Reprod 2010;83:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones IR, Young ID. Familial incidence of cryptorchidism. J Urol 1982;127:508–509. [DOI] [PubMed] [Google Scholar]
- Kaftanovskaya EM, Huang Z, Barbara AM, De Gendt K, Verhoeven G, Gorlov IP, Agoulnik AI. Cryptorchidism in mice with an androgen receptor ablation in gubernaculum testis. Mol Endocrinol 2012;26:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaftanovskaya EM, Neukirchner G, Huff V, Agoulnik AI. Left-sided cryptorchidism in mice with Wilms’ tumour 1 gene deletion in gubernaculum testis. J Pathol 2013;230:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Chakraborty S, Cheng X, Su YT, Kao HY. The actin-binding protein, actinin alpha 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J Biol Chem 2011;286:1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa E, Matsuda M. Regulation of focal adhesion and cell migration by ANKRD28-DOCK180 interaction. Cell Adh Migr 2009;3:281–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollara A, Brown TJ. Variable expression of nuclear receptor coactivator 4 (NcoA4) during mouse embryonic development. J Histochem Cytochem 2010;58:595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollara A, Brown TJ. Expression and function of nuclear receptor co-activator 4: evidence of a potential role independent of co-activator activity. Cell Mol Life Sci 2012;69:3895–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 1984;81:3443–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PA, Houk CP. Cryptorchidism. Curr Opin Endocrinol Diabetes Obes 2013;20:210–216. [DOI] [PubMed] [Google Scholar]
- Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 2014;30:2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 2012;180:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugg JA, Penson DF, Sadeghi F, Petrie B, Freedman AL, Gonzalez-Cadavid NF, Hikim AS, Rajfer J. Prevention of seminiferous tubular atrophy in a naturally cryptorchid rat model by early surgical intervention. J Androl 1996;17:726–732. [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhadjer N, Pointis G, Malassine A, Bedin M. Testicular steroid sulfatase in a cryptorchid rat strain. J Steroid Biochem 1989;34:555–558. [DOI] [PubMed] [Google Scholar]
- Moul JW, Belman AB. A review of surgical treatment of undescended testes with emphasis on anatomical position. J Urol 1988;140:125–128. [DOI] [PubMed] [Google Scholar]
- Patkowski D, Czernik J, Jelen M. The natural course of cryptorchidism in rats and the efficacy of orchidopexy or orchidectomy in its treatment before and after puberty. J Pediatr Surg 1992;27:870–873. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010;20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles KN, Lamle T, Sewry CA, Puckelwartz M, Furling D, Wehnert M, McNally EM, Morris GE. Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development. Dev Dyn 2010;239:998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci 1996;21:267–271. [PubMed] [Google Scholar]
- Rizza P, Barone I, Zito D, Giordano F, Lanzino M, De Amicis F, Mauro L, Sisci D, Catalano S, Dahlman Wright K et al. Estrogen receptor beta as a novel target of androgen receptor action in breast cancer cell lines. Breast Cancer Res 2014;16:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, Barthold JS, Kreiger PA, Armani MH, Wang J, Michelini KA, Wolfson MR, Boyce R, Barone CA, Zhu Y et al. The orl rat is more responsive to methacholine challenge than wild type. Pulm Pharmacol Ther 2014;29:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild MF, Christian LL, Blanchard W. Evidence for multigene control of cryptorchidism in swine. J Hered 1988;79:313–314. [PubMed] [Google Scholar]
- Saito M, Watanabe-Nakayama T, Machida S, Osada T, Afrin R, Ikai A. Spectrin-ankyrin interaction mechanics: a key force balance factor in the red blood cell membrane skeleton. Biophys Chem 2015;200–201:1–8. [DOI] [PubMed] [Google Scholar]
- Savion M, Nissenkorn I, Servadio C, Dickerman Z. Familial occurrence of undescended testes. Urology 1984;23:355–358. [DOI] [PubMed] [Google Scholar]
- Schnack TH, Zdravkovic S, Myrup C, Westergaard T, Wohlfahrt J, Melbye M. Familial aggregation of cryptorchidism--a nationwide cohort study. Am J Epidemiol 2008;167:1453–1457. [DOI] [PubMed] [Google Scholar]
- Scott HM, Hutchison GR, Jobling MS, McKinnell C, Drake AJ, Sharpe RM. Relationship between androgen action in the ‘male programming window,’ fetal sertoli cell number, and adult testis size in the rat. Endocrinology 2008;149:5280–5287. [DOI] [PubMed] [Google Scholar]
- Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang SJ et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res 2015;43:D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriett V, Nicholas G, Berry C, Watson T, Hennebry A, Thomas M, Ling N, Sharma M, Kambadur R. Myostatin negatively regulates the expression of the steroid receptor co-factor ARA70. J Cell Physiol 2006;206:255–263. [DOI] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res 2009;19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub C, Rauch M, Ferriere F, Trepos M, Dorval-Coiffec I, Saunders PT, Cobellis G, Flouriot G, Saligaut C, Jegou B. Expression of estrogen receptor ESR1 and its 46-kDa variant in the gubernaculum testis. Biol Reprod 2005;73:703–712. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Kiyokawa E, Hara S, Iemura S, Natsume T, Manabe T, Matsuda M. Ankyrin repeat domain 28 (ANKRD28), a novel binding partner of DOCK180, promotes cell migration by regulating focal adhesion formation. Exp Cell Res 2009;315:863–876. [DOI] [PubMed] [Google Scholar]
- Thankamony A, Lek N, Carroll D, Williams M, Dunger DB, Acerini CL, Ong KK, Hughes IA. Anogenital distance and penile length in infants with hypospadias or cryptorchidism: comparison with normative data. Environ Health Perspect 2014;122:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting HJ, Chang C. Actin associated proteins function as androgen receptor coregulators: an implication of androgen receptor's roles in skeletal muscle. J Steroid Biochem Mol Biol 2008;111:157–163. [DOI] [PubMed] [Google Scholar]
- van Brakel J, Dohle GR, de Muinck Keizer-Schrama SM, Hazebroek FW. Surgical findings in acquired undescended testis: an explanation for pubertal descent or non-descent? Eur J Pediatr Surg 2011;21:351–355. [DOI] [PubMed] [Google Scholar]
- van Pelt AM, de Rooij DG, van der Burg B, van der Saag PT, Gustafsson JA, Kuiper GG. Ontogeny of estrogen receptor-beta expression in rat testis. Endocrinology 1999;140:478–483. [DOI] [PubMed] [Google Scholar]
- Virtanen HE, Adamsson A. Cryptorchidism and endocrine disrupting chemicals. Mol Cell Endocrinol 2012;355:208–220. [DOI] [PubMed] [Google Scholar]
- Westesson O, Skinner M, Holmes I. Visualizing next-generation sequencing data with JBrowse. Brief Bioinform 2013;14:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci 2005;118:673–687. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007;16:2816–2833. [DOI] [PubMed] [Google Scholar]
- Zhou B, Hutson JM, Hasthorpe S. Efficacy of orchidopexy on spermatogenesis in the immature mutant ‘trans-scrotal’ rat as a cryptorchid model by quantitative cytological analysis. Br J Urol 1998;81:290–294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.