Abstract
Using eight inbred founder strains of the mouse Collaborative Cross (CC) project and their reciprocal F1 hybrids, we quantified variation in craniofacial morphology across mouse strains, explored genetic contributions to craniofacial variation that distinguish the founder strains, and tested whether specific or summary measures of craniofacial shape display stronger additive genetic contributions. This study thus provides critical information about phenotypic diversity among CC founder strains and about the genetic contributions to this phenotypic diversity, which is relevant to understanding the basis of variation in standard laboratory strains and natural populations. Craniofacial shape was quantified as a series of size‐adjusted linear dimensions (RDs) and by principal components (PC) analysis of morphological landmarks captured from computed tomography images from 62 of the 64 reciprocal crosses of the CC founder strains. We first identified aspects of skull morphology that vary between these phenotypically ‘normal’ founder strains and that are defining characteristics of these strains. We estimated the contributions of additive and various non‐additive genetic factors to phenotypic variation using diallel analyses of a subset of these strongly differing RDs and the first eight PCs of skull shape variation. We find little difference in the genetic contributions to RD measures and PC scores, suggesting fundamental similarities in the magnitude of genetic contributions to both specific and summary measures of craniofacial phenotypes. Our results indicate that there are stronger additive genetic effects associated with defining phenotypic characteristics of specific founder strains, suggesting these distinguishing measures are good candidates for use in genotype–phenotype association studies of CC mice. Our results add significantly to understanding of genotype–phenotype associations in the skull, which serve as a foundation for modeling the origins of medically and evolutionarily relevant variation.
Keywords: collaborative cross, craniofacial form, diallel, micro‐computed tomography, morphometrics, mouse models, normal variation
Introduction
The craniofacial skeleton has a complex genetic and developmental basis, reflecting a number of different functions, including cognition, ingestion of food, verbal communication, the housing of a variety of sense organs, and support and protection of the brain. The expression and interaction of many genes across many developmental, signaling, and structural pathways are necessary to coordinate the development of the integrated craniofacial complex (Chai & Maxson, 2006; Feng et al. 2009; Buchtová et al. 2010; Szabo‐Rogers et al. 2010). Recent work has shown how modulation of key regulatory genes, including SHH (Hu & Marcucio, 2009; Young et al. 2010; Chong et al. 2012), WNT (Brugmann et al. 2007; Alexander et al. 2014), BMP (Abzhanov et al. 2004, 2006; Wu et al. 2004, 2006) and FGF (Abzhanov & Tabin, 2004; Szabo‐Rogers et al. 2008; Griffin et al. 2013), produce integrated patterns of change in the shape of the head. Nevertheless, the genetic and developmental architecture underlying morphological variation of the skull remains largely unknown.
Craniofacial shape is moderately to highly heritable in a variety of species, including mice (Leamy, 1982a; Richtsmeier & McGrath, 1986), fish (Kimura et al. 2007), non‐human primates (Cheverud, 1996b; Roseman et al. 2010), and humans (Johannsdottir et al. 2005; Sherwood et al. 2008a; Martínez‐Abadías et al. 2009). Quantitative trait loci (QTL) analyses have provided evidence of genomic regions that influence craniofacial variation in non‐human primates (Sherwood et al. 2008b), dogs (Boyko et al. 2010; Schoenebeck et al. 2012; Schoenebeck & Ostrander, 2013), mice (Leamy et al. 1999, 2008; Klingenberg et al. 2001, 2004), and fish (Albertson et al. 2003; Kimura et al. 2007). Genome‐wide association studies of three‐dimensional facial shape (Liu et al. 2012; Paternoster et al. 2012) and associations based on ancestry informative markers (Claes et al. 2014) have been used to identify genomic loci associated with normal variation in adult human facial shape, some of which were previously associated with disease phenotypes.
Most QTL analyses of craniofacial phenotypes have focused on limited clinical samples in humans, small populations of non‐human primates or crosses of contrasting strains in mice or fish. The Collaborative Cross (CC) mice provide an opportunity to perform genotype–phenotype association studies on mice that represent a wide range of normal phenotypic variation with high genetic mapping resolution. The CC is a large panel of recombinant inbred strains derived from a genetically diverse selection of five common laboratory and three wild‐derived inbred mouse strains (Churchill et al. 2004; Chesler et al. 2008; Collaborative Cross Consortium, 2012). These recombinant inbred strains have a high and uniform level of mouse genetic diversity across the entire genome (Keane et al. 2011; Yang et al. 2011; Welsh et al. 2012). Over half of the genomic regions have at least six haplotypes (derived from eight founders), and very few regions have less than four (Collaborative Cross Consortium, 2012). As a precursor to future genotype–phenotype association studies of skull morphology in these recombinant strains, we quantify genetic contributions to normal variation across the craniofacial complex within the eight CC founder strains and associated F1 crosses.
Craniofacial form has been previously quantified with linear distances and angles (e.g. Johannsdottir et al. 2005; Roseman et al. 2010), including those representing specific features (e.g. orbital height) and more general measures (e.g. skull length), as well as summary values derived from principal component analyses of landmark coordinates (e.g. Klingenberg et al. 2001). Here, we identify phenotypic measures that differ strongly among the CC founder strains, measure the heritability of a subset of these traits, and explore the nature of the genetic contributions to craniofacial variation in F1 hybrids. To do this, we measured craniofacial variation from computed tomography images of the eight CC founder strains and F1 animals from 54 hybrid crosses. Using relative linear dimensions (RDs) and principal component (PC) analysis, we quantified the variation in ‘normal’ craniofacial shape across these Mus musculus lines, identifying groups of RDs across the skull that differ strongly among the founder strains and PCs that represent the majority of craniofacial shape variation. These measures, which display strong variation among the CC founder strains, are likely to be associated with genetic variation in descendant crosses.
Although we expect the PCs and RDs associated with variation among founder strains to both have a strong genetic basis, the nature of the genetic contributions to these different types of measures may differ. The skull is a combined product of developmental processes occurring across overlapping spatio‐temporal contexts (Hallgrímsson et al. 2009); thus, the inheritance of craniofacial shape characteristics may be exceedingly complex. Measures that are specific to discrete morphological and developmental regions, including RDs, may capture the influences of a particular spatio‐temporally defined process. Multivariate measures that summarize many aspects of phenotypic variation across the skull, like PCs, may reflect variation in numerous genetic and environmental factors, as well as the effects of developmental inputs acting at different and overlapping times and places. Among strongly differing RDs, we hypothesize that the strain‐specific additive genetic effects associated with RDs would be strongest when the RD represents a defining characteristic of that particular strain.
To test this hypothesis and as a basis for comparing genetic contributions to RDs and PCs, we completed a nearly full diallel analysis (including founder strains and F1 hybrids) on representatives of both phenotypic measurement types to decompose genetic variation into additive, maternal, sex, inbreeding, and other non‐additive components and to estimate the heritability of these factors. Previous diallel analyses of body weight (Lenarcic et al. 2012) and CD23 antigen density (Phillippi et al. 2014) in these genotypes identified significant overall and strain‐specific genetic contributions. We compare significance of factors and heritability estimates of factor groups for RDs and PCs, to identify any fundamental differences in the nature or strength of the genetic contributions between specific and summary measures of craniofacial phenotype. In addition to testing our hypothesis, we search our diallel results for patterns of difference in genetic contributions to variation in RDs between regions of the skull, which are associated with different developmental bases. Our results provide critical information about the range of and genetic contributions to ‘normal’ phenotypic diversity among CC founder strains, which will be valuable in the design and interpretation of experimental work related to craniofacial phenotypes.
Materials and methods
Basic morphometrics
Our sample consisted of mice bred as part of the CC project at the University of North Carolina under the approval of the University of North Carolina's Animal Care and Use Committee. Craniofacial measurements from 13–28 specimens of each of the eight CC founder strains (Table 1), bred from mice originally obtained from the Jackson Laboratory (www.jax.org), and 54 (of 56 possible) F1 crosses were included in this analysis (total n = 1211) (Table 2). These mice were housed at UNC for 8–12 weeks with standard chow and housing. Each cross is identified by two letters, the first being the founder strain of the dam and the second being the founder strain of the sire (Table 1) (Collaborative Cross Consortium, 2012). The two crosses representing the missing hybrids (gray in Table 1) are not productive (Chesler et al. 2008). Because all founder strains are inbred, specimens within each founder strain and F1 cross are isogenic. Micro‐computed tomography (μCT) images of heads were obtained in the 3D Morphometrics Centre at the University of Calgary with a Scanco vivaCT40 scanner (Scanco Medical, Brüttisellen, Switzerland) at 0.035–0.038 mm voxel dimensions at 55 kV and 72–145 μA. Three‐dimensional coordinates of 54 adult landmarks (8 midline, 46 bilateral) were collected by a single observer from minimum threshold defined bone surfaces within analyze 3D (www.mayo.edu/bir/) (Fig. 1). These landmarks display low repeatability error and were chosen to represent overall morphology of the skull at a moderate landmark density. All subsequent analyses were performed on the symmetric component of landmark coordinate variation.
Table 1.
Definitions of founder strains, including letters used to identify them in the supplementary tables, JAX strain abbreviation, and strain origin (Beck et al. 2000)
| Letter | Strain IDs | Strain origin |
|---|---|---|
| A | A/J | Lab Inbred (Castle's Mice) |
| B | C57BL/6J | Lab Inbred (C57 Related) |
| C | 129S1/SvlmJ | Lab Inbred (Castle's Mice) |
| D | NOD/ShiLtJ | Lab Inbred (Swiss Mice) |
| E | NZO/HlLtJ | Lab Inbred (Castle's Mice/New Zealand) |
| F | CAST/EiJ | Wild Derived Inbred (Thailand) |
| G | PWK/PhJ | Wild Derived Inbred (Prague) |
| H | WSB/EiJ | Wild Derived Inbred (MD, USA) |
Table 2.
Sample sizes for all crosses in our analysis. Founder crosses are shown along the diagonal in bold, missing crosses are shown with gray background. Strains are identified by shortened versions of the strain IDs defined in Table 1
| Paternal strain | ||||||||
|---|---|---|---|---|---|---|---|---|
| A/J | C57 | 129 | NOD | NZO | CAST | PWK | WSB | |
| Maternal strain | ||||||||
| A/J | 18 | 19 | 19 | 20 | 19 | 20 | 20 | 20 |
| C57 | 20 | 19 | 20 | 18 | 19 | 20 | 16 | 18 |
| 129 | 21 | 17 | 19 | 17 | 18 | 19 | 24 | 22 |
| NOD | 19 | 21 | 19 | 13 | 22 | 24 | 21 | 18 |
| NZO | 19 | 18 | 22 | 20 | 17 | 0 | 0 | 19 |
| CAST | 20 | 26 | 19 | 18 | 21 | 18 | 23 | 19 |
| PWK | 19 | 21 | 20 | 17 | 19 | 18 | 18 | 18 |
| WSB | 19 | 19 | 20 | 20 | 19 | 28 | 20 | 18 |
Figure 1.
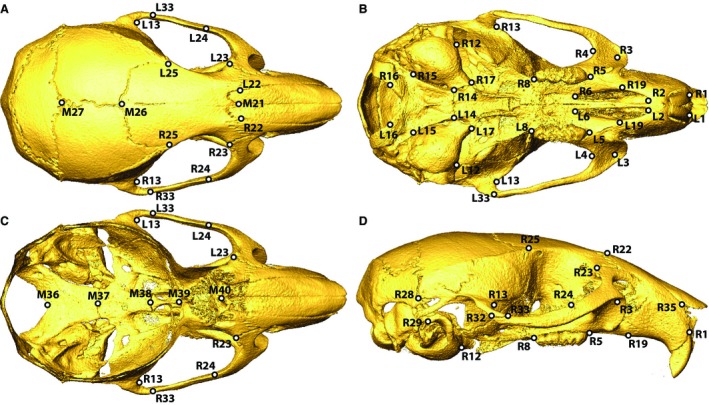
Landmark identification on the (A) superior, (B) inferior, (C) superior with calotte removed, and (D) lateral views of an adult mouse skull. Midline landmarks start with M, bilateral landmarks start with L&R.
Procrustes superimposition was performed in morphoj (Klingenberg, 2011) to transform the landmark coordinates of all specimens to a common scale and orientation. Residuals of a linear regression of the symmetric component of Procrustes coordinates on centroid size (CS) served as the basis of all further analysis to control for size‐associated shape variation. This allometric correction was considered appropriate because of the linear association between centroid size and a summary measure of shape across our entire sample (Fig. 2). The shape summary score represents the shape changes for which the regression on centroid scores accounts, allowing the visual identification of a linear or non‐linear relationship (Drake & Klingenberg, 2008). As this study is a precursor for future analysis of more derived CC mice, including recombinant inbred intercross strains for which each specimen will have different genotypes, we chose to correct for allometry using a single regression rather than strain‐specific allometric regressions. While an overall linear regression of shape variables on CS is largely appropriate, the overall linear relationship between shape and size does not precisely match the relationship for some specific genotypes. Therefore, there may be bias in the resulting residual values for these genotypes, so we must take some additional care in interpreting our results.
Figure 2.
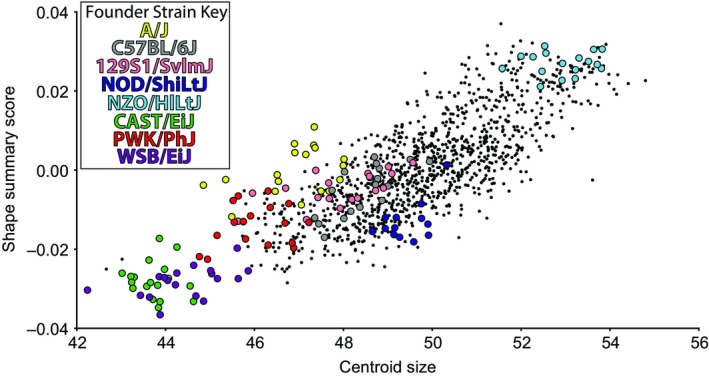
Linear relationship between centroid size of skull landmarks and a shape summary score derived from Procrustes coordinates in morphoj (Klingenberg, 2011) for each specimen. Founder strains are differentiated by colors and F1 cross‐specimens are black dots.
Principal components (PC) analysis was carried out to visualize and quantify the associations between strains along major axes of shape variation. Based on the strength of their associated eigenvalues, we selected PCs 1–8 for further analyses. To determine whether the founder strains have more extreme craniofacial shapes than their descendant F1 strains, we calculated the mean PC scores of each strain for PCs 1–8 and then calculated the Euclidean distance in this eight‐dimensional shape space between strain means and the grand mean PC scores. To test whether PC1 represents a common axis of intra‐genotype variation for all genotypes, we calculated the angle between the eigenvector of PC1 for all specimens and each PC1 eigenvector for PCAs of samples from individual genotypes. We repeated this for PCs 1–8. These angles are reported in radians.
Defining relative linear dimensions
We identified defining characteristics of the CC founder strains from relative linear dimensions (RDs) that quantify relative size of specific bones or morphological features (e.g. vault height). RDs were calculated as Euclidean distances between size‐adjusted landmark coordinates (regression residuals). These RDs are based on coordinates adjusted for size and so do not reflect the raw size of a specimen and can be interpreted as linear aspects of shape. For instance, if one mouse strain displays a larger value for an RD across the width of the foramen magnum, this suggests the strain displays a relatively wider foramen magnum than other strains and not that it displays an absolutely wider foramen magnum. Therefore, we define RD as ‘relative linear dimension’ rather than ‘linear distance’ to indicate that these measures relate to the shape component of morphological form.
All possible RDs were calculated for each specimen and RD means were calculated for each founder strain and F1 hybrid. Ratios of pairwise founder means were calculated for each RD, as in Euclidean distance matrix analysis (Lele & Richtsmeier, 1991). A given founder strain exhibited strong differences from other founder strains for an RD when the measure of that strain differed from the measures of three or more other strains by more than 8%, an arbitrary cut‐off value above which a manageable number of RDs displaying the largest inter‐strain differences were identified. Defining features of a given founder strain are RDs with strong differences for which the mean of that founder strain is always higher or always lower than the means of all other founder strains. 3D plots of strongly differing and defining features (Supporting Information Figs S1–S8) were used to identify aspects of shape for which a founder strain tends to differ from other founder strains. Because our dimensions are derived from the symmetrical component of landmark coordinate variation, the ratios for bilateral measures from the left and right side of the skull are identical. Therefore, bilateral measures are only reported for one side of the skull.
A subset of 34 strongly differing RDs was identified to represent relatively independent aspects of shape that tend to vary among CC founder strains (Fig. 3). Single RDs were chosen from groups of strongly differing RDs that appeared to represent the same type of shape variation within the skull (e.g. cranial vault height, zygomatic arch length). We preferred RDs identified as strongly differing for many of the CC founder strains or defining characteristics with strong difference ratios even if they were only identified for a smaller number of CC founder strains. We also preferred dimensions long enough to be less affected by measurement error but short enough to represent a single bone or small region of the head. We included three RDs that represented craniofacial morphology within regions in which strong differences among CC founder strains were not found. Although morphology across the skull is highly integrated (Moss & Young, 1960; Cheverud, 1982; Zelditch et al. 1992) and RDs that share a landmark endpoint are inherently associated, we consider the RDs in this subset to be relatively independent of one another because we typically chose single representatives from groups of strongly differing RDs running in parallel across particular regions of the skull. In combination with PCs 1–8, this subset of RDs are included in diallel analyses to test the nature of the genetic basis for variation in the F1 crosses.
Figure 3.
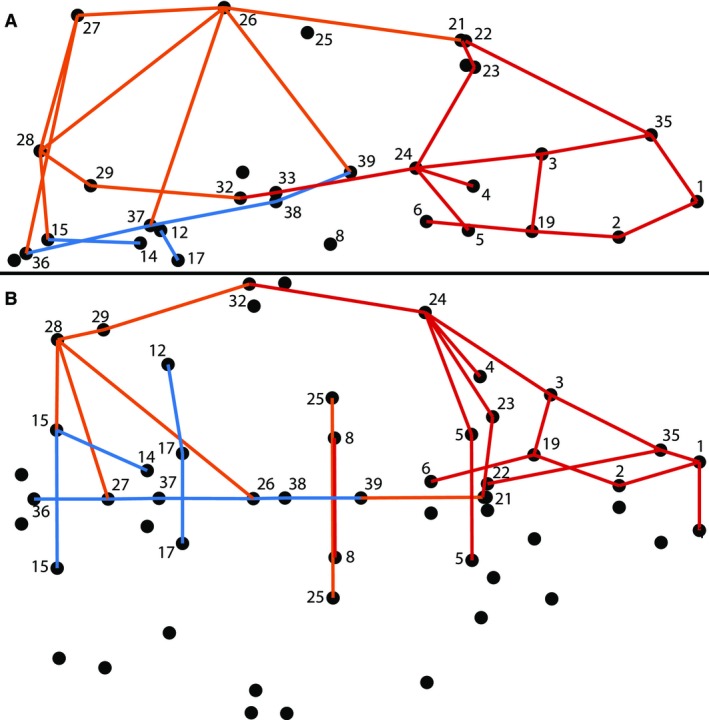
Subset relative linear dimension (RD) identification on the (A) lateral (anterior to left, superior up) and (B) inferior (anterior to left) views. Subset RDs representing the facial (red), cranial base (blue), and cranial vault (orange) regions of the skull are identified. Landmarks associated with these subset RDs are labeled.
Diallel analysis and heritability
We quantified the additive and various non‐additive genetic contributions to strongly differing aspects of shape across the craniofacial complex with separate diallel analyses for each of the 34 subset RDs and PCs 1–8. Diallel analysis was performed with the bayesdiallel v0.96 package (Lenarcic et al. 2012) in r (R Developmental Core Team, 2008) using a linear semi‐parametric model including additive, maternal, sex, inbreeding, and cross‐specific sources of variation, as well as sex interaction versions of the other factors. We transformed the distribution of measures for each RD and PC so they spanned 0–100 before diallel analysis, based on the expectations of the diallel package. Additive genetic factors conform to the expectation that an F1 phenotype is the average of founder phenotypes. Maternal effects are associated with the genotype of the dam of a litter, whereas sex effects are associated with the sex of the specimen. The inbreeding effect differs from dominance in that it is a deviation from additive effects calculated from all F1 crosses, rather than a deviation from expected founder means. Cross‐specific factors are non‐additive factors associated with specific founder strain interactions (Lenarcic et al. 2012).
Estimated highest posterior density intervals (α = 0.05) that did not include zero were used to identify diallel factors that contributed significantly to variation of each subset RD and PC. Heritability estimates were derived from the diallel results to measure how much phenotypic variation is associated with each type of genetic factor as well as groups of additive factors, non‐additive factors, and all factors (Lenarcic et al. 2012). Heritability estimates of the additive genetic factors are analogous to narrow‐sense heritability, whereas the total heritability estimate for all factors is analogous to broad‐sense heritability estimates, although covariates such as sex may already be accounted for before calculating broad‐sense heritability in other contexts (Kohn, 1991).
Because of their large size, detailed diallel results for each subset RD and PC are presented as supplementary tables (Supporting Information Tables S2 and S3). As a more manageable summary of these results, we calculated the frequency of significant contributions and average heritability estimates for each type of genetic factor. These summary results are presented for PCs, all subset RDs, and for RDs within three commonly identified regions of the skull – the cranial base, the cranial vault, and the face (Fig. 3). We fully recognize that the skull is highly integrated and these morphological regions are not truly independent (Hallgrímsson et al. 2009). However, this separation of RDs allows us to identify major differences in the nature of genetic contributions to shape variation between regions of the skull that are broadly associated with different cellular origins, mechanisms of ossification, and functions (Cheverud, 1982, 1996a; Lieberman et al. 2000; Hallgrímsson et al. 2007; Martínez‐Abadías et al. 2010).
To test for differences in the magnitude of additive contributions to PCs and RDs, we compared the distributions of estimated strain‐specific additive effects (absolute values standardized by SD) for subset RDs and PCs 1–8 using pair‐wise t‐tests. To test whether RDs representing defining features of a founder strain tend to have larger additive contributions to the phenotypes of descendant generations than strongly differing RDs that are not considered defining characteristics for a strain, or RDs not considered strongly differing for a strain, we made a similar comparison of estimated additive effect distributions using pair‐wise t‐tests.
Results
Basic morphometrics
There is a strong linear relationship between skull size (estimated by CS) and skull shape, represented by a shape summary score (Fig. 2). Founder strains differ substantially in size, with NZO/HILtJ mice having the largest skull sizes and the three wild‐derived strains CAST/EiJ, PWK/PhJ, WSB/EiJ having the smallest. Although centroid sizes of F1 crosses usually fall between these two extremes, the mean centroid size of all 54 F1 strains is larger than the average of associated founder strains, suggesting that F1 hybrids generally have larger skulls than expected from a pure additive model (Supporting Information Table S1).
After controlling for size, we conducted PC analysis to explore the association between craniofacial shapes of CC founder strains and F1 hybrids. We focused on the first eight PCs, which represent a cumulative 71% of craniofacial shape variance (Fig. 4A–D). The Euclidean distances between mean shapes of each strain (calculated using scores across PCs 1–8) and the grand mean shape of all specimens (~0 along all PCs) were calculated for all strains. Six of the eight CC founder strains are farther from the grand mean than all F1 strains, whereas WSB/EiJ and NZO/HlLtJ fall within the F1 range (Fig. 4E). This suggests that CC founder strains generally represent more extreme craniofacial shapes compared with the F1 hybrids.
Figure 4.
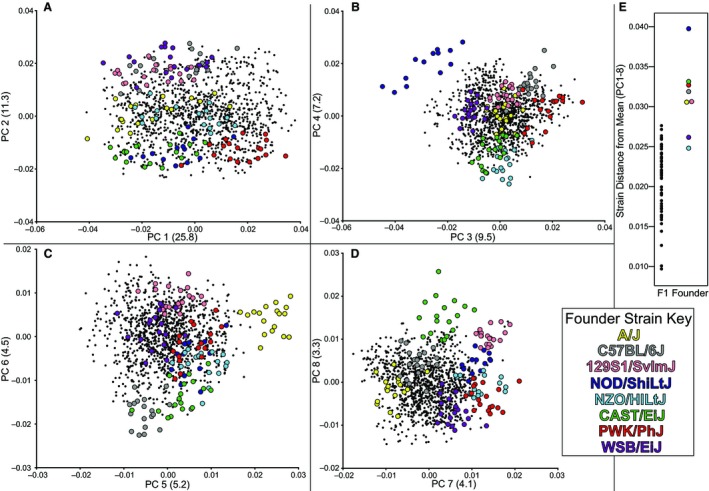
Specimens from founder strains (colored) and F1 strains (black dots) plotted across principal components (PC), with % of variance explained in parentheses, including (A) PC1 vs. PC2, (B) PC3 vs. PC4, (C) PC5 vs. PC6, (D) PC7 vs. PC8. (E) The Euclidean distance of strain mean PC scores and the grand mean across PCs 1–8.
No single PC axis separates all CC founder strains, but most founder strains do separate from F1 strains and other CC founder strains along at least one of the first eight PCs. Although variation along PC1 represents over one‐fourth of all craniofacial variation, it does not strongly separate founder genotypes (Fig. 4A) and appears to largely represent a common axis of intra‐genotype variation. This interpretation is supported by the fact that the angles between the eigenvector of PC1 for all specimens and the eigenvectors for PC1 of PCAs of specific genotypes (mean 0.608 radians, SD 0.203) are lower than the same angles calculated for each of PCs 2–8 (mean 1.414, SD 0.118). Nevertheless, PC1 is also associated with some inter‐genotype variation, including the fact that most CC founder strains trend towards the negative PC1 scores (Fig. 4A). PC2 serves to separate the CC founder genotypes into groups, including a group of the three with the most extreme phenotypes (Fig. 4E) towards the negative end of this axis (Fig. 4A). The other PCs, particularly in pairs, likewise provide separation of CC founder genotypes.
Defining features
We compared linear measures of shape across the skull to identify the characteristics that most strongly distinguish the different CC founder strains. Size‐adjusted RDs were identified for each CC founder strain that strongly differed (8% larger or smaller) from the mean of at least three other founder strains. From this list of strongly differing RDs, we identified defining characteristics of a CC founder strain (Fig. 5) as those that were always larger (red) or smaller (blue) than all other founder strains. Those strongly differing RDs that are not always larger or smaller than other founder strains (gray) are not considered defining characteristics of a given strain. Using wireframe visualizations (Figs S1–S8), we identified groups of defining and other differing RDs that differentiate a given CC founder strain from the other seven (Table 3). Certain features were identified as strongly differing for multiple CC founder strains, including distances along the zygomatic arches, cranial vault height (particularly between the coronal suture and basi‐occipital synchondrosis), lengths between points on the palate, and facial widths between the zygomatic process of the maxilla and the rest of the face. In some cases, there appears to be a trade‐off in relative size between different morphological areas. For instance, C57BL/6J and 129S1/SvlmJ tend to have a long or wide zygomatic process of the maxilla and a short jugal bone, whereas strain NOD/ShiLtJ displays the opposite pattern. In addition, strain NOD/ShiLtJ appears to compensate for a relatively long face and posterior cranial base/vault with a relatively short anterior cranial base/vault, or vice versa.
Figure 5.
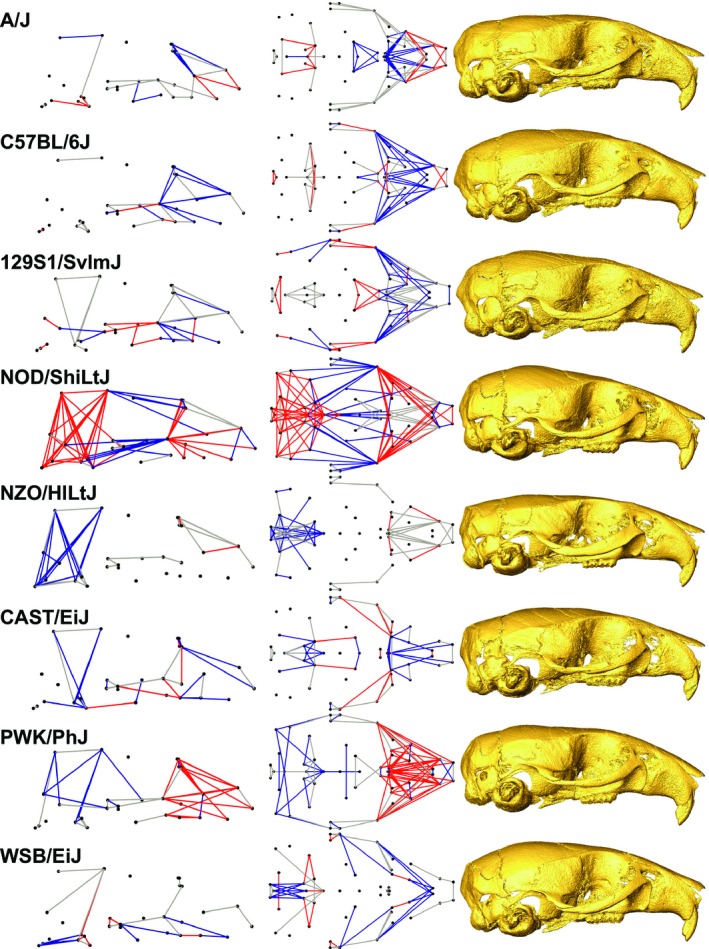
Comparison of founder strain (Table 1) features. Relative linear dimensions (RDs) identified as defining characteristics that are always larger (red) or shorter (blue) and strongly differing LDs that are not always larger or smaller (gray) are plotted from the lateral (left) and inferior (middle) views. μCT surface reconstructions of real specimens that are closest to mean strain shape are displayed for comparison, from the lateral view (right).
Table 3.
List of characteristics that help to differentiate a given founder strain from the other seven founder strains, derived from the highly variable relative linear dimensions (RDs) of a founder group whose mean value always tends to be higher or lower than the mean values for the other founder strains (Fig. 5)
| Founder strain Skull Region | Defining craniofacial features of founder strains (RDs after controlling for centroid size of the skull) |
|---|---|
| A/J | |
| Facial | Short fusion of nasal and maxillary bones |
| Short distance between posterior palatal foramen and posterior alveolus | |
| Narrow width between anterior alveolar ridges | |
| Zygomatic | Inferior and/or anterior landmark 3 |
| Cranial vault | Short midline parietal length at sagittal suture |
| Cranial base | Long medial petrous pyramid |
| C57BL/6J | |
| Facial | Short nasal bone and/or short fusion of nasal and maxillary bones |
| Anterior and/or lateral position of posterior palatal foramen | |
| Zygomatic | Short and/or narrow zygomatic process of the maxilla |
| Long jugal bone | |
| 129S1/SvlmJ | |
| Facial | Short distance between premaxillary‐maxillary suture and anterior palatal foramen |
| Long distance between posterior palatal foramen and posterior alvelous | |
| Narrow nasal inferior aperture | |
| Zygomatic | Short and/or narrow zygomatic process of maxilla |
| Long jugal bone | |
| Relatively posterior jugo‐temporal suture on zygomatic arch | |
| NOD/ShiLtJ | |
| Facial | Short height of premaxilla |
| Wide nasal aperture | |
| Zygomatic | Wide and/or long zygomatic process of maxilla |
| Short jugal and zygomatic process of temporal | |
| Cranial vault | Short frontal bones, as measured along sagittal suture |
| Long parietal bones, as measured along sagittal suture | |
| High posterior cranial vault | |
| Cranial base | Large extent of petrous temporal and basi‐occipital bones |
| Anterior sphenoid‐presphenoid junction | |
| NZO/HlLtJ | |
| Facial | Strong differences in facial length relative to other founder strains |
| Cranial vault | Short height of posterior cranial vault |
| CAST/EiJ | |
| Facial | Short superior nasal region |
| Narrow interorbital width at border of nasal and frontal bones | |
| Short premaxillary palate and anterior position of the first molar | |
| Cranial vault | Short posterior cranial vault |
| Cranial base | Anterior position of landmark 17 at the anterior midline petrous pyramid |
| Low sphenoid‐presphenoid junction | |
| PWK/PhJ | |
| Facial | Long upper facial region, including long fusion of premaxillary and nasal bones |
| Posterior position of posterior palatal foramen compared with premaxillary‐maxillary suture and the anterior alveolus | |
| Zygomatic | Long and/or wide zygomatic process of the maxilla |
| Cranial vault | Narrow anterior vault width at coronal suture |
| Narrow parietal bones | |
| Short cranial vault | |
| WSB/EiJ | |
| Facial | Short premaxillary palate |
| Long distance between premaxillary‐maxillary suture and anterior alveolus | |
| Zygomatic | Low and forward zygomatic arch |
| Cranial base | Short basi‐occipital |
Diallel analysis and heritability
We selected a subset of 34 strongly differing RDs that cross single bones or regions of the skull to represent the features that strongly vary between founder strains (Fig. 3). We completed separate diallel analyses for each subset RD and the first eight PCs to determine the contribution of additive and various non‐additive genetic sources to phenotypic variation. There was considerable variation in the number and types of factors that exert significant effects on these phenotypic values (Table S2). We calculated the frequency of significant effects for each factor type as a proxy for the importance of that factor type in determining craniofacial morphology across the head. The diallel analyses revealed a significant (~α = 0.05) overall effect of sex and inbreeding for 29% and 26%, respectively, of all subset RDs (Table 4). Strain‐specific additive effects were frequently (62%) significant across subset RDs, whereas strain‐specific maternal (17%), inbreeding (21%), and cross‐specific factors (19%) were significant at moderate frequencies. These results indicate that most factor types that do not include a sex interaction significantly contribute to RD variation at high or moderate frequencies. While strain‐specific additive effects are most frequently significant by far, the patterns of significance vary strongly among subset RDs and PCs.
Table 4.
The frequency of diallel factors found to be significant across analyses of all subset RDs (All), facial (Face), cranial base (Base), and cranial vault (Vault) subset RDs, as well as across the first eight PCs. Subset RDs defined by region in Fig. 2. Results of individual diallel analyses are available as Table S1
| Factor name | # Factors | % Sig All RDs | % Sig Face | % Sig Base | % Sig Vault | % Sig PCs |
|---|---|---|---|---|---|---|
| Overall sex | 1 | 29.4 | 25 | 43 | 27.3 | 50 |
| Overall inbreeding | 1 | 26.5 | 37.5 | 14 | 18.2 | 62.5 |
| Overall inbreeding/sex | 1 | 0 | 0 | 0 | 0 | 0 |
| Strain‐specific additive | 8 | 62.1 | 64.1 | 50 | 67 | 65.6 |
| Strain‐specific additive/sex | 8 | 5.9 | 8.6 | 0 | 5.7 | 12.5 |
| Strain‐specific maternal | 8 | 17.3 | 18.8 | 16.1 | 15.9 | 15.6 |
| Strain‐specific maternal/sex | 8 | 6.6 | 7 | 5.4 | 6.8 | 4.7 |
| Strain‐specific inbreeding | 8 | 21.3 | 11.7 | 28.6 | 30.7 | 18.8 |
| Strain‐specific inbreeding/sex | 8 | 7.4 | 10.2 | 3.6 | 5.7 | 12.5 |
| Cross‐specific symmetric | 28 | 19.2 | 21.7 | 16.8 | 17.2 | 21.9 |
| Cross‐specific symmetric/sex | 28 | 0 | 0 | 0 | 0 | 0 |
| Cross‐specific asymmetric | 28 | 7.6 | 9.2 | 6.1 | 6.2 | 12 |
| Cross‐specific asymmetric/sex | 28 | 0.3 | 0.4 | 0 | 0.3 | 1.8 |
As an initial test for whether developmentally and morphologically associated characters tend to have more similar patterns of genetic contributions, we grouped RDs by cranial region (cranial base, cranial vault, face). There are not many obvious differences in the frequency of significant contributions between RDs in these regions (Table 4). However, the cranial base has a lower frequency of significant additive effects and a higher frequency of significant mean sex effects. Additionally, the face has a higher overall frequency of significance for inbreeding, whereas base and vault have higher strain‐specific inbreeding frequencies. While our summary measures of significance frequencies generally indicate that the genetic contribution to craniofacial phenotype is complex and highly variable across the skull, these few differences between skull regions may indicate differences in the genetic architecture underlying the development of these specific regions.
The average frequencies of significance for most diallel factors across PCs 1–8 were similar to the average frequencies for subset RDs, with some slightly higher values for cross‐specific factors (though not for strain‐specific inbreeding). However, the frequency of significance for overall inbreeding is double the average for RDs (62.5 vs. 26.5%) and almost double for overall sex effects (50 vs. 29.4%). Overall, similar types of factors contribute to subset RDs and PCs variation, although PCs 1‐8 may better reflect morphological variation associated with inbreeding and sex effects.
We tested whether additive contributions to PC scores of F1 hybrids are generally stronger than those associated with subset RDs by comparing the distributions of estimated strain‐specific additive effect values. A t‐test revealed no significant difference between PCs and RDs (Fig. 6A), suggesting that PC scores and subset RD values are associated with additive effects of similar magnitude. Among subset RDs, we tested the hypothesis that the defining characteristics of founder strains would be associated with the strongest additive contributions to F1 craniofacial phenotypes. Because a given subset RD might be a defining characteristic for some founder strains and not a defining characteristic for others, each datapoint in this analysis is the strain‐specific additive contribution to a specific RD. Pairwise t‐tests revealed significant differences in strain‐specific additive effects within our RD subset between defining characters and both (i) other strongly differing RDs and (ii) RDs that do not strongly differ between founder strains (Fig. 6B). This supports our hypothesis that extreme characteristics of a founder strain should be more strongly associated with the genotype of that strain than are randomly chosen characteristics.
Figure 6.
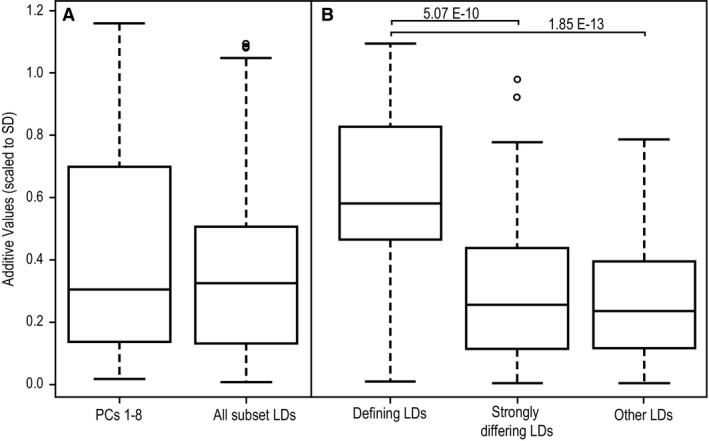
Distributions of diallel estimated strain‐specific additive effects (absolute values scaled to SD) for relative linear dimensions (RDs) identified as founder strain‐specific defining characteristics or strongly differing RDs of founder strains, as well as other strain‐specific additive values. Boxplots illustrate median and quartile values. Significant pair‐wise differences are labeled with the P‐values from pairwise t‐tests.
We calculated heritability estimates to quantify the proportion of phenotypic variation that each diallel factor is associated with for each subset RD and PCs 1–8. The average total heritability estimate is 0.68 for RDs and 0.81 for PCs 1–8, suggesting that most craniofacial variation is determined by genetic factors (Table 5; Fig. 7). The difference in total heritability between PCs and RDs is almost entirely accounted for by increased strain‐specific additive effects for the PCs. PC heritability values for all other factors are close to or within the range of variation between average RD heritability estimates across the three major skull regions, suggesting a similar magnitude of phenotypic effects associated with the contribution of other factors to variation in both measurement types. Across RD and PC measurements, heritability estimates for additive genetic effects are more than twice as high as the sum of all non‐additive effects (e.g. maternal, inbreeding, cross‐specific), though a few RDs do have non‐additive heritability values similar to or larger than additive values (Table S3). Of all the PCs, PC1 displays the lowest total heritability and additive heritability values (Fig. 7E). The fact that PC1 largely represents common intra‐genotype variation may explain this low value. By definition, additive genetic factors (and non‐additive factors) are associated with measured differences between genotypes, so we would expect associated heritability estimates to be low when variation is largely shared across genotypes.
Table 5.
The average estimated heritability values for strain and cross‐specific diallel factors across All, facial (Face), cranial base (Base), and cranial vault (Vault) subset RDs, as well as the first eight PCs. Total heritability explained by additive, non‐additive, and all factors are reported in bold. Subset RDs defined by region in Fig. 2. Heritability estimates for individual phenotypic measures are available as Table S2
| All RDs | Face | Base | Vault | PCs 1‐8 | |
|---|---|---|---|---|---|
| Sum additive factors | 0.458 | 0.486 | 0.333 | 0.498 | 0.62 |
| Strain‐specific additive | 0.453 | 0.48 | 0.329 | 0.493 | 0.616 |
| Strain‐specific additive/sex | 0.005 | 0.006 | 0.004 | 0.005 | 0.004 |
| Sum non‐additive factors | 0.224 | 0.211 | 0.271 | 0.214 | 0.192 |
| Strain‐specific maternal | 0.016 | 0.021 | 0.01 | 0.012 | 0.015 |
| Strain‐specific maternal/sex | 0.008 | 0.007 | 0.01 | 0.008 | 0.004 |
| Strain‐specific inbreeding | 0.044 | 0.028 | 0.063 | 0.057 | 0.030 |
| Strain‐specific inbreeding/sex | 0.004 | 0.005 | 0.003 | 0.003 | 0.004 |
| Cross‐specific symmetric | 0.135 | 0.131 | 0.167 | 0.121 | 0.123 |
| Cross‐specific symmetric/sex | 0.004 | 0.004 | 0.005 | 0.004 | 0.003 |
| Cross‐specific asymmetric | 0.011 | 0.013 | 0.011 | 0.008 | 0.010 |
| Cross‐specific asymmetric/sex | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| Sum all factors | 0.682 | 0.697 | 0.604 | 0.712 | 0.812 |
Figure 7.
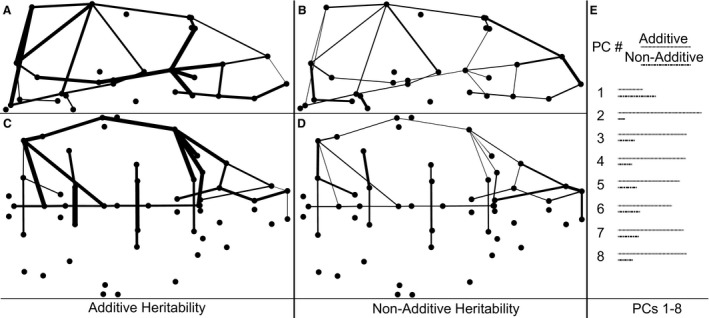
Heritability estimates for strain‐ and cross‐specific additive and non‐additive factors of phenotypic measures. Subset relative linear dimensions are illustrated as lines with widths proportional to estimated additive e (A,C) and non‐additive (B,D) heritability, from the lateral (top) and inferior (bottom) views. (E) Lines with length proportional to heritability estimates for additive and non‐additive factors associated with each of the first eight principal components (PCs).
Heritability estimates suggest that additive factors account for approximately 75% of total genetic contributions to phenotypic variation (sum additive factors/sum all factors) reflected in PC scores (n = 8) and 70% of subset RD variation in the face (n = 16) and cranial vault (n = 11), but only 55% of RD variation of the cranial base (n = 7) (Table 5). This suggests that additive genetic factors contribute most to PC scores, but least to RD variation of the cranial base (Fig. 7) (Table S3).
Discussion
Defining features of CC founder strains
As candidates for future genotype–phenotype analysis, we identified characteristics of each CC founder strain that differ strongly from several other founder strains and those that distinguish each CC founder strain from all others. These features are represented by size‐adjusted RDs between sparse homologous landmarks identified across the skull. A few clusters of strongly differing and defining RDs were identified for several CC founder strains, suggesting some regions of the skull are more likely to display strong variation across Mus musculus. These include (i) the zygomatic arch and associated processes, (ii) cranial vault height, and (iii) the nasal region of the face. Other defining characteristics of note include a relatively short basi‐occipital region for WSB/EiJ and a potential trade‐off between the contributions of different regions to overall skull length, with NOD/ShiLtJ having a relatively long face and posterior cranium but a relatively short anterior cranium (Fig. 5).
These three regions that display high inter‐genotype variation in our sample have also been identified as differing between wild mouse taxa in Europe and Western Asia. Scaled linear measurements of relative skull breadth, the lateral extent of the zygomatic arch, and relative rostral breadth (Macholan, 1996), as well as unscaled linear measurements of bi‐incisor breadth, cranial base breadth at the mastoid processes, and length of the foramen formed by the zygomatic arch (Gerasimov et al. 1990), were identified as features that distinguish Mus taxa. Geometric morphometric analyses of 2D landmarks, which ignore measures of skull height, were used to identify relative warps that differentiate taxa. The warps that differentiate Mus musculus musculus and M. m. domesticus populations in Denmark are associated with variation in relative cranial base width, length of the rostrum, and anterior–posterior location of the molar row (Auffray et al. 1996), whereas relative zygomatic arch length, relative width of cranial base and vault, and length of rostrum differentiate mouse taxa around the Eastern Mediterranean (Macholán et al. 2008). A 3D geometric morphometrics analysis found that M. m. musculus and M. m. domesticus differ in relative anterior cranial vault height and length, anterior facial width, and the angle of the nasal region (Pallares et al. 2014).
Previous studies commonly identify the zygomatic arch and nasal (rostral) regions as strongly varying between wild mouse populations, and the 3D morphometrics study that includes skull height variation identifies cranial vault height as strongly varying. However, we have not identified cranial base and vault widths as strongly varying across our founder strains. Although our results are not identical to previous studies of wild and wild‐derived mouse taxa, the parallels suggest that the regions of variation noted between our inbred populations also vary across wild mouse populations.
A thorough cranial shape analysis of C57BL/6, wild derived inbred Mus spretus, and associated inbred recombinant congenic strains identified landmarks on the top of the cranial vault, the zygomatic arch of the maxilla, and the posterior cranial base as strongly varying between genotypes. A CVA analysis of crosses indicated that variation between genotypes involved a change in the relative length of parietal and frontal bones, cranial vault height, zygomatic arch elevation at the maxilla, and angle of the nasal bones (Burgio et al. 2009), which closely matches the regions of strong variation that we noted in our sample.
Much of the variation we have identified in the zygomatic arch appears to be associated with a trade‐off in the relative length of the zygomatic process of the maxilla and the length of the jugal bone. For instance, C57BL/6J and 129S1/SvlmJ tend to have a long or wide zygomatic process of the maxilla and a short jugal bone, whereas NOD/ShiLtJ displays the opposite pattern. Therefore, the relative contribution of a single bone to zygomatic arch length may not be as canalized as the typical form of the whole arch. Given its association with many strongly differing RDs, the relative location of superior border of the jugal and zygomatic process of the maxilla (landmark 24) is central to the variation measured in the zygomatic region. Visualization of specimen Procrustes coordinates by genotype (results not shown) indicates that this landmark varies between founder genotypes primarily along the anterior–posterior axis, further indicating a high degree of variation in the relative length of the bones making up the zygomatic arch.
The strong variation in relative height of the cranial vault is the most visually obvious of the identified features, particularly between NOD/ShiLtJ and NZO/HlLtJ (Fig. 5). However, the main reason that NZO/HlLtJ is identified as having a relatively short cranial vault may be an indirect result of landmark coordinate scaling. Before scaling, NZO/HlLtJ mice have by far the largest skulls of all CC founder strains (Fig. 2), with an absolutely longer and wider facial region, and a longer cranial base, but with a posterior cranial vault more similar in size to other CC founder strains. Previous work has suggested that mouse genotypes with the largest skulls tend to be relatively long and narrow (Leamy, 1982b), which unscaled measurements of strain NZO/HlLtJ generally support. However, after Procrustes superimposition and control for overall allometry, the relative size of the face and cranial base are more similar to other CC founder strains, whereas the posterior vault is relatively shorter in height.
While the allometric relationship between size and shape are linear across the range of all specimens within our analysis (Fig. 2), there may be a more precise allometric relationship associated with each individual genotype. The example of relative vault size for NZO/HlLtJ indicates that vault size may not scale linearly with the rest of the skull in mice with the largest overall size, although further tests will be necessary to confirm this. Given the wide range of sizes that our sample encompasses and the importance of allometry for understanding craniofacial form, a separate study of allometry is being completed for this dataset. In practical terms, our use of a sample‐wide regression of shape variables on CS is largely appropriate but may not take into account differences in allometry that exist between genotypes. In addition, the regions and directions of strong variation noted in this analysis of shape may not be the same as they would be in an analysis carried out on unscaled landmark coordinates.
There is strong variation in RDs of the superior face around the nasal region, suggesting strong variation in the association of the nasal and premaxillary bones. Many of these RDs are associated with landmark 35, which is at the most anterior point along the suture between these two bones. Because the length of this suture does not necessarily covary with nasal and premaxillary bone size or shape, this strong facial variation may be a secondary effect of the relative orientations of these bones rather than their form. Three of the RDs with the lowest additive heritability meet at landmark 35 (Fig. 7), suggesting that significant differences in nasal–premaxillary suture length are not passed in an additive fashion to offspring. This example illustrates the care necessary in making inferences about development from even simple morphometric measures.
These areas of the skull displaying strong variation between CC strains do not correspond to the broader regions (i.e. cranial base, cranial vault, and face) that have been identified as morphological modules based on developmental and functional similarities (Cheverud, 1982, 1996a; Lieberman et al. 2000; Hallgrímsson et al. 2007; Martínez‐Abadías et al. 2010). Not all RDs within these wider modules vary in the same manner and there is noted variation in significance of diallel results between RDs within a single region. This highlights the fact that these general modules are not completely integrated units within which all aspects of morphology are driven by the same genetic and developmental factors. Although we found grouping RDs into these larger units useful in our search for overall patterns within our data, it is important to keep the complex interactions that underlie adult craniofacial form (Hallgrímsson et al. 2009) in mind when searching for the genetic or developmental bases for phenotypic variation.
Founder strains vs. F1 hybrids
Our results suggest that inbred CC founder strains have smaller skulls and more extreme craniofacial shapes than descendant F1 hybrids. Hybrid mice typically display larger body weights than inbred strains (e.g. Ingram et al. 1982; Kurnianto et al. 1999) and have longer craniofacial linear distances, except for certain skull width measures (Leamy, 1982b). Our results provide further evidence for this inbreeding effect on mouse skull size. In addition, our PC analysis suggests that CC founder strains (except perhaps WSB/EiJ and NZO/HlLtJ) have craniofacial shapes more divergent from overall mean shape than F1 strains. Previous work similarly indicates that hybrid mice display lower phenotypic variance within and between genotypes than their inbred parents do (Leamy, 1982a). The fact that F1 craniofacial shapes are less variable and tend to be closer to overall mean shape than their founder strains might support the additive expectation that a hybrid will generally display craniofacial morphology roughly average between its two inbred founder strains.
Regardless, the more extreme morphology of CC founder strains is not entirely additive in nature. Diallel analysis reveals that five of the first eight PCs (62.5%) are associated with a significant overall (rather than strain‐specific) inbreeding effect, suggesting that the inbred CC founder strains differ significantly from the additive expectation (derived from F1 cross morphology) in a common way. A significant overall inbreeding effect is found, on average, for only 26.5% of subset RDs, suggesting that PC analysis is better at separating CC founder from F1 specimens. This is not surprising, given that PCs 1–8 represent the strongest axes of covariation of all skull landmarks across all genotypes, whereas the RDs analyzed represent shape dimensions that vary among founder groups rather than being shared across groups. An RD would only be expected to be better than these first PCs at separating founder from F1 specimens if that particular aspect of shape represented a major site of variation between the groups. It is logical that our strongly differing RDs, which were chosen to differentiate founder strains from one another, rather than founders from F1s, are not as good as the first eight PCs at distinguishing these groups.
Additive vs. non‐additive genetic variation
Diallel analyses of a subset of 34 RDs that differ strongly among CC founder strains and 8 PCs that, in combination, differentiate CC founder strains were carried out to determine what genetic factors contribute to shape across the craniofacial complex (Table 4). Heritability estimates derived from these analyses quantify the proportion of phenotypic variance explained by each group of strain and cross‐specific factors (Fig. 7; Table 5). Although the contributions of genetic factors are not the same across different phenotypic measures (Table S2), general patterns emerge. Strain‐specific additive contributions to phenotypic variation are the most frequently significant and are, on average, associated with almost 50% of the phenotypic variance of RDs and about 60% of the phenotypic variance for PCs. Non‐additive factors are significant at lower frequencies than additive factors, except overall inbreeding effects for PCs (Table 4). The average total overall variance explained by all strain‐ and cross‐specific non‐additive factors is less than half that explained by strain‐specific additive factors, except in the case of cranial base RDs, for which these heritability values are more similar (Table 5). Overall, our results show that additive genetic factors play the largest role in determining variation in specific (RDs) and summary (PCs) measures of craniofacial shape. This prominence of additive genetic effects over non‐additive effects is frequently assumed by genetic models used for bioinformatics. So, the fact that additive factors explain most of the phenotypic variance is not a surprise. However, the significant influence of non‐additive factors requires a closer look.
Our heritability estimates indicate that, while half of phenotypic variation is accounted for by additive genetic factors, 20–25% of phenotypic variation is associated with non‐additive factors. This suggests that non‐additive factors contribute approximately one‐third of the total genetic variance, a larger proportion than might be expected (Hill et al. 2008). Even after removing heritability associated with sex, a factor frequently controlled for in other analyses, approximately 20% of phenotypic variation is still associated with non‐additive factors.
Comparable levels of additive and dominance effects associated with QTLs (Lander & Botstein, 1989) were found in previous analyses of mouse mandible (Cheverud et al. 1997; Klingenberg et al. 2001) and mouse skull (Leamy et al. 1999) measures. Although the values from these studies are not directly comparable to ours, they show that specific allelic changes in identified QTLs lead to similar magnitudes of non‐additive (dominance) and additive effects, supporting the idea that non‐additive factors play a major role in determining mouse craniofacial form. Understanding the strength and nature of non‐additive effects that contribute to complex traits such as skull shape (e.g. dominance, epistasis, variance heterogeneity) is important because most methods for gene discovery assume additive models of genetic variance. Such models are often blind to non‐additive genetic variation and this may contribute to the missing heritability problem for complex morphological traits (Carlborg & Haley, 2004; Hemani et al. 2013; Nelson et al. 2013).
Overall inbreeding effects are frequently noted for PC scores and less frequently for RD values (Table 4). Although intriguing, this significant non‐additive effect is unlikely to appear within wild outbred populations of mice or within the wider human population, suggesting that a portion of the strong non‐additive contribution to craniofacial variation noted in this study may be specific to the nature of our sample. Even within the first generation of a random bred population of the CC founder strains, the frequency of inbred mice is relatively low, leading the strain‐specific inbreeding factor to have a low heritability estimate compared with the frequency of significance for this factor. It has previously been argued that even if non‐additive effects such as dominance and epistasis influence a given trait, outbred populations, which are expected to have extreme allele frequencies, are likely to display additive genetic effects that account for most of total genetic variance (Hill et al. 2008). If true, this implies that the relatively high non‐additive genetic variance associated with craniofacial shape in our F1 hybrids, which should not display rare alleles within any cross, may not be representative of outbred mouse populations or human populations. This does not mean that non‐additive genetic factors cannot have significant effects on complex traits, only that non‐uniform allele frequencies in natural populations may tend to hide these effects.
Inbreeding can be a significant non‐additive source of variation that disappears in outbred populations. If the inbreeding effect contributes significantly to the variation of a phenotypic measure, we should be careful in applying the results of inbred strain experiments related to that measure as a way to understand the basis for variation in natural populations. This includes interpretations related to evolutionary and biomedically relevant variation in those populations. Based on our results, inbreeding is more likely to be a significant contributor to variation in PC scores than to variation in RDs, suggesting additional care is necessary when applying results related to PC scores. However, the definition of all PCs is sample‐dependent and PC scores are not discrete homologous phenotypes that are shared between populations, so direct comparison of PC score variation between separate studies is already highly problematic. Future analyses based on more advanced intercross generations from the CC will help to resolve the extent to which inbreeding effects might skew estimates of non‐additive genetic variation in this sample.
Other diallel analysis results
The lower average strain‐specific additive heritability for cranial base RDs opposes an existing hypothesis that cranial base measures should be more heritable because they finish developing before other parts of the skull and should be less impacted by environmental variation (Kohn, 1991; Roseman et al. 2010). However, because subset RDs were chosen specifically for high inter‐founder variation, the low heritability could indicate that among the most variable regional craniofacial characteristics, there is greater canalization of the cranial base than of other regions of the skull. Previous work in humans and primates did not find significant differences between the heritability of cranial base and other regions of the skull (Martínez‐Abadías et al. 2009; Roseman et al. 2010), although these studies were on relatively closely related populations and did not focus specifically on strongly differing RDs.
Overall sex effects are frequently significant for PCs 1–8 and moderately significant for RDs. As with the overall inbreeding effect, the PCs appear to separate specimens by sex better then individual RD measures. However, the RDs that have a significant sex effect represent the aspects of craniofacial shape with strong sexual dimorphism. Based on these RDs, females tend to have relatively smaller cranial base bones, larger cranial vaults, and a relatively wider face (Table S2). Moderate frequencies of significance for strain‐specific maternal effects (~20%) indicate that there are some phenotypic differences associated with whether specific strains are dam or sire within reciprocal crosses. Countering our expectation that the largest strain, NZO/HlLtJ, would always have a strong maternal effect based on their large overall body size, this strain does not frequently have a significant maternal effect on our scaled phenotypic measures.
The frequency of significance for cross‐specific symmetric factors is comparable to strain‐specific inbreeding and maternal effects (Table 4). Approximately 20% of all pairwise strain combinations display a phenotypic effect that cannot be explained by other factors, suggesting there are significant epistatic effects associated with background genotypes. Heritability estimates (Table 5) indicate that 10% of the phenotypic variation across all genotypes in our diallel sample can be explained by these strain‐specific symmetric factors. This value is higher than inbreeding heritability primarily because of the relatively low number of inbred mice within the full diallel population. The frequency of significance for asymmetric cross‐specific factors is lower, suggesting strain‐specific epistatic effects are not as frequently influenced by the maternal identity of a given founder strain. In combination, these results suggest there are some parent of origin‐based cross effects on craniofacial shape. Previous diallel analyses using the same CC founder genotypes also indicate some significant cross‐specific symmetric and asymmetric effects on body weight and white blood cell count, although the frequency of significance is lower (Lenarcic et al. 2012). Diallel analysis of a few immune system phenotypes indicated very few, if any, of these cross‐specific effects (Phillippi et al. 2014).
Comparing craniofacial measures
We found no significant difference in the strength of additive genetic effects between PCs 1–8 and subset RDs (Fig. 6), suggesting that these summary and specific measures of craniofacial phenotype are associated with similar magnitudes of additive effects. However, the mean heritability estimate of additive factors is higher for PCs than for subset RDs across all three craniofacial regions (Table 5), suggesting that additive factors contribute more strongly to PC scores than to RDs. This does not necessarily mean that PC scores will be associated with strong peaks in a QTL analysis. It is possible that a large proportion of genetic variation associated with PCs is based on a few major craniofacial regulatory genes or highly pleiotropic genes acting across the head. However, it is also possible that this strong genetic contribution to values along a major axis of craniofacial shape variation is a result of the expression and epigenetic interactions of a very large number of genes that could not be easily identified with a QTL analysis. Although it is unlikely that a phenotypic measure displaying very low heritability will be strongly associated with specific allelic variation, high heritability does not guarantee that such an association will be identified.
The results of this study support our hypothesis that additive genetic effects associated with RDs are strongest when the RD represents a defining characteristic of that particular strain. We found significant differences in the distributions of strain‐specific additive effects between defining characteristics and other subset RDs. This suggests that RDs associated with extreme founder phenotypic values may be the strongest candidate phenotypes for use in genotype–phenotype analysis. For instance, choosing defining characteristics of a founder strain for use within a standard QTL analysis based on an additive model will be beneficial because they are more likely to display strong additive effects associated with that strain genotype. We argue that choosing defining characteristics within an analysis increases the chances of identifying strong genotype–phenotype associations compared with randomly choosing RDs or choosing RDs that, in combination, appear to represent overall craniofacial shape. However, as with genetic contributions to PC values, this assumes that allelic variation between founder strains in a relatively small number of genes is associated with a large proportion of the genetic contribution to RD values. Actual genotype–phenotype association analysis with these candidate PC and RD measures will be required to pursue this issue further.
Concluding remarks
In this study, we quantified the nature of normal craniofacial variation among a wide range of mouse strains and estimated the relative contributions of additive and non‐additive genetic factors to phenotypic variation using diallel analyses. We found evidence of smaller skull sizes in inbred founder strains and evidence of an overall non‐additive inbreeding effect on skull shape. Common regions of strong shape variation between the founder strains were identified, including the zygomatic arch, cranial vault height, and the nasal region. We find little difference in the genetic contributions to RD measures and PC scores or to RDs associated with different skull regions, except for a lower additive contribution to cranial base RD variation, which might be associated with increased canalization. However, stronger estimated strain‐specific additive effects associated with defining characteristics of founder strains suggest that these RDs may be good candidates for use in future genotype–phenotype association studies of CC and descendant mice. Our results illustrate the nature of normal craniofacial variation across a wide range of inbred mouse lines and illuminate the heritable bases of this variation, serving as a foundation for predictions about the aspects of shape that will be most strongly associated with genetic variation between founder strains in future genetic analyses.
Author contributions
Concept/design: CJP, DKL, FPMV, RM, RS, BH. Sample collection: FPMV. Data analysis/interpretation: CJP, DKL, BH. Drafting of manuscript: CJP. Critical revision of manuscript: CJP, DKL, FPMV, RS, RM, BH. Approval for publication: CJP, DKL, FPMV, RS, RM, BH.
Supporting information
Figs S1–S8. OpenGL objects that display strongly differing relative linear dimensions (gray), defining characteristics that are always relatively shorter (red), and those that are always longer (blue) for a given founder strain. After downloading and unzipping this file, open the index.html file from the directory of a parental strain within your browser to view a 3D representation of these relative linear dimensions for that parental strain.
Table S1. F1 hybrid centroid sizes (CS) are all larger than expected by a purely additive model.
Table S2. Genetic factors with significant positive (‘Pos’, red) or negative (‘Neg’, blue) effects are identified for each subset relative linear dimension (RD) and for principal components (PC) 1–8.
Table S3. Heritability estimates for each group of strain and cross‐specific factors are presented for each subset relative linear dimension (RD) and for principal components (PC) 1–8.
Acknowledgements
Thanks to Wei Liu and Campbell Rolian for micro‐CT scanning and landmark acquisition. We also thank Darla Miller and Ginger Shaw for the generation of all mice used in this study. We gratefully acknowledge funding from NIH (1U01DE024440, R01‐DE19638, R01‐DE021708) and NSERC (Grant #238992‐12), as well as the CIHR Training Program in Genetics, Child Development and Health through the Alberta Children's Hospital Research Institute for Child and Maternal Health for support to CJP.
References
- Abzhanov A, Tabin CJ (2004) Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev Biol 273, 134–148. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, et al. (2004) Bmp4 and morphological variation of beaks in Darwin's finches. Science 305, 1462–1465. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Kuo WP, Hartmann C, et al. (2006) The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature 442, 563–567. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD (2003) Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc Natl Acad Sci U S A 100, 5252–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Piloto S, Le Pabic P, et al. (2014) Wnt signaling interacts with Bmp and Edn1 to regulate dorsal‐ventral patterning and growth of the craniofacial skeleton. PLoS Genet 10, e1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray J‐C, Alibert P, Latieule C, et al. (1996) Relative warp analysis of skull shape across the hybrid zone of the house mouse (Mus musculus) in Denmark. J Zool 240, 441–455. [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, et al. (2000) Genealogies of mouse inbred strains. Nat Genet 24, 23–25. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, et al. (2010) A simple genetic architecture underlies morphological variation in dogs. PLoS Biol 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, et al. (2007) Wnt signaling mediates regional specification in the vertebrate face. Development 134, 3283–3295. [DOI] [PubMed] [Google Scholar]
- Buchtová M, Kuo WP, Nimmagadda S, et al. (2010) Whole genome microarray analysis of chicken embryo facial prominences. Dev Dyn 239, 574–591. [DOI] [PubMed] [Google Scholar]
- Burgio G, Baylac M, Heyer E, et al. (2009) Genetic analysis of skull shape variation and morphological integration in the mouse using interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species. Evolution 63, 2668–2686. [DOI] [PubMed] [Google Scholar]
- Carlborg Ö, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5, 618–625. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE Jr (2006) Recent advances in craniofacial morphogenesis. Dev Dyn 235, 2353–2375. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, et al. (2008) The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome 19, 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM (1982) Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution 36, 499–516. [DOI] [PubMed] [Google Scholar]
- Cheverud JM (1996a) Developmental integration and the evolution of pleiotropy. Am Zool 36, 44–50. [Google Scholar]
- Cheverud JM (1996b) Quantitative genetic analysis of cranial morphology in the cotton‐top (Saguinus oedipus) and saddle‐back (S. fuscicollis) tamarins. J Evol Biol 9, 5–42. [Google Scholar]
- Cheverud JM, Routman EJ, Irschick DJ (1997) Pleiotropic effects of individual gene loci on mandibular morphology. Evolution 51, 2006–2016. [DOI] [PubMed] [Google Scholar]
- Chong HJ, Young NM, Hu D, et al. (2012) Signaling by SHH rescues facial defects following blockade in the brain. Dev Dyn 241, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, et al. (2004) The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36, 1133–1137. [DOI] [PubMed] [Google Scholar]
- Claes P, Liberton DK, Daniels K, et al. (2014) Modeling 3D facial shape from DNA. PLoS Genet 10, e1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium (2012) The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AG, Klingenberg CP (2008) The pace of morphological change: historical transformation of skull shape in St Bernard dogs. Proc R Soc B Biol Sci 275, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, et al. (2009) Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS ONE 4, e8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimov S, Nikolov H, Mihailova V, et al. (1990) Morphometric stepwise discriminant analysis of the five genetically determined European taxa of the genus Mus . Biol J Linn Soc 41, 47–64. [Google Scholar]
- Griffin JN, Compagnucci C, Hu D, et al. (2013) Fgf8 dosage determines midfacial integration and polarity within the nasal and optic capsules. Dev Biol 374, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Lieberman DE, Liu W, et al. (2007) Epigenetic interactions and the structure of phenotypic variation in the cranium. Evol Dev 9, 76–91. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Jamniczky H, Young NM, et al. (2009) Deciphering the palimpsest: studying the relationship between morphological integration and phenotypic covariation. Evol Biol 36, 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Knott S, Haley C (2013) An evolutionary perspective on epistasis and the missing heritability. PLoS Genet 9, e1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM (2008) Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4, e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS (2009) A SHH‐responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development 136, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DK, Reynolds MA, Les EP (1982) The relationship of genotype, sex, body weight, and growth parameters to lifespan in inbred and hybrid mice. Mech Ageing Dev 20, 253–266. [DOI] [PubMed] [Google Scholar]
- Johannsdottir B, Thorarinsson F, Thordarson A, et al. (2005) Heritability of craniofacial characteristics between parents and offspring estimated from lateral cephalograms. Am J Orthod Dentofac Orthop 127, 200–207. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Shimada A, Sakai N, et al. (2007) Genetic analysis of craniofacial traits in the medaka. Genetics 177, 2379–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP (2011) MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour 11, 353–357. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Routman EJ, et al. (2001) Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157, 785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Cheverud JM (2004) Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics 166, 1909–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn L (1991) The role of genetics in craniofacial morphology and growth. Annu Rev Anthropol 20, 261–278. [Google Scholar]
- Kurnianto E, Shinjo A, Suga D, et al. (1999) Diallel cross analysis of body weight in subspecies of mice. Exp Anim 48, 277–283. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy L (1982a) Morphometric studies in inbred and hybrid house mice II. Patterns in the variances. J Hered 73, 267–272. [DOI] [PubMed] [Google Scholar]
- Leamy L (1982b) Morphometric studies in inbred and hybrid house mice. I. Patterns in the mean values. J Hered 73, 171–176. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Routman EJ, Cheverud JM (1999) Quantitative trait loci for early‐and late‐developing skull characters in mice: a test of the genetic independence model of morphological integration. Am Nat 153, 201–214. [DOI] [PubMed] [Google Scholar]
- Leamy L, Klingenberg C, Sherratt E, et al. (2008) A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity 101, 518–526. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT (1991) Euclidean distance matrix analysis: a coordinate‐free approach for comparing biological shapes using landmark data. Am J Phys Anthropol 86, 415–427. [DOI] [PubMed] [Google Scholar]
- Lenarcic AB, Svenson KL, Churchill GA, et al. (2012) A general Bayesian approach to analyzing diallel crosses of inbred strains. Genetics 190, 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman DE, Ross CF, Ravosa MJ (2000) The primate cranial base: ontogeny, function, and integration. Am J Phys Anthropol 113(Suppl. 31), 117–169. [DOI] [PubMed] [Google Scholar]
- Liu F, van der Lijn F, Schurmann C, et al. (2012) A genome‐wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet 8, e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholan M (1996) Morphometric analysis of European house mice. Acta Theriol 41, 255–276. [Google Scholar]
- Macholán M, Mikula O, Vohralík V (2008) Geographic phenetic variation of two eastern‐Mediterranean non‐commensal mouse species, Mus macedonicus and M. cypriacus (Rodentia: Muridae) based on traditional and geometric approaches to morphometrics. Zool Anz ‐A J Comp Zool 247, 67–80. [Google Scholar]
- Martínez‐Abadías N, Esparza M, Sjøvold T, et al. (2009) Heritability of human cranial dimensions: comparing the evolvability of different cranial regions. J Anat 214, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Abadías N, Percival C, Aldridge K, et al. (2010) Beyond the closed suture in Apert mouse models: evidence of primary effects of FGFR2 signaling on facial shape at P0. Dev Dyn 239, 3058–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss ML, Young RW (1960) A functional approach to craniology. Am J Phys Anthropol 18, 281–292. [DOI] [PubMed] [Google Scholar]
- Nelson RM, Pettersson ME, Carlborg Ö (2013) A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet 29, 669–676. [DOI] [PubMed] [Google Scholar]
- Pallares LF, Harr B, Turner LM, et al. (2014) Use of a natural hybrid zone for genomewide association mapping of craniofacial traits in the house mouse. Mol Ecol 23, 5756–5770. [DOI] [PubMed] [Google Scholar]
- Paternoster L, Zhurov AI, Toma AM, et al. (2012) Genome‐wide association study of three‐dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am J Hum Genet 90, 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippi J, Xie Y, Miller DR, et al. (2014) Using the emerging Collaborative Cross to probe the immune system. Genes Immun 15, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Developmental Core Team (2008) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, Available from: http://www.R-project.org. [Google Scholar]
- Richtsmeier JT, McGrath JW (1986) Quantitative genetics of cranial nonmetric traits in randombred mice: heritability and etiology. Am J Phys Anthropol 69, 51–58. [DOI] [PubMed] [Google Scholar]
- Roseman CC, Willmore KE, Rogers J, et al. (2010) Genetic and environmental contributions to variation in baboon cranial morphology. Am J Phys Anthropol 143, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Ostrander EA (2013) The genetics of canine skull shape variation. Genetics 193, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Hutchinson SA, Byers A, et al. (2012) Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet 8, e1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RJ, Duren DL, Demerath EW, et al. (2008a) Quantitative genetics of modern human cranial variation. J Hum Evol 54, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RJ, Duren DL, Havill LM, et al. (2008b) A genomewide linkage scan for quantitative trait loci influencing the craniofacial complex in baboons (Papio hamadryas spp.). Genetics 180, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo‐Rogers HL, Geetha‐Loganathan P, Nimmagadda S, et al. (2008) FGF signals from the nasal pit are necessary for normal facial morphogenesis. Dev Biol 318, 289–302. [DOI] [PubMed] [Google Scholar]
- Szabo‐Rogers HL, Smithers LE, Yakob W, et al. (2010) New directions in craniofacial morphogenesis. Dev Biol 341, 84–94. [DOI] [PubMed] [Google Scholar]
- Welsh CE, Miller DR, Manly KF, et al. (2012) Status and access to the Collaborative Cross population. Mamm Genome 23, 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang T‐X, Suksaweang S, et al. (2004) Molecular shaping of the beak. Science 305, 1465–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang T‐X, Shen J‐Y, et al. (2006) Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dyn 235, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, et al. (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Chong HJ, Hu D, et al. (2010) Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development 137, 3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch ML, Bookstein FL, Lundrigan BL (1992) Ontogeny of integrated skull growth in the cotton rat Sigmodon fulviventer . Evolution 46, 1164–1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1–S8. OpenGL objects that display strongly differing relative linear dimensions (gray), defining characteristics that are always relatively shorter (red), and those that are always longer (blue) for a given founder strain. After downloading and unzipping this file, open the index.html file from the directory of a parental strain within your browser to view a 3D representation of these relative linear dimensions for that parental strain.
Table S1. F1 hybrid centroid sizes (CS) are all larger than expected by a purely additive model.
Table S2. Genetic factors with significant positive (‘Pos’, red) or negative (‘Neg’, blue) effects are identified for each subset relative linear dimension (RD) and for principal components (PC) 1–8.
Table S3. Heritability estimates for each group of strain and cross‐specific factors are presented for each subset relative linear dimension (RD) and for principal components (PC) 1–8.


