Abstract
Background: A quantity of case-control studies have been performed to address the association between the three interleukin-6 (IL-6) polymorphisms (-572G/C, -597G/A and -174G/C) and the risk of HBV related liver diseases. However, previous research results are inconsistent. We conducted this meta-analysis to clarify the correlation between these IL-6 polymorphisms and HBV related liver diseases. Methods: We searched in PubMed, EMBASE, Cochrane Library as well as Chinese databases including China National Knowledge Infrastructure (CNKI) and WanFang database for all the relevant studies up to April 15, 2015. The data were extracted by two independent authors. Odds ratios (ORs) and 95% confidence intervals (95% CI) were calculated. Results: A total of 10 studies consisting of 3879 cases and 2812 controls were included in this metaanalysis. For IL-6 polymorphism -572G/C, an association with increased chronic hepatitis B (CHB) risk was observed under in allelic, homozygous, heterozygous, dominant and recessive model. However, IL-6 polymorphisms (-572G/C) were not related to Inactive Carrier (IC), Liver Cirrhosis (LC) and Hepatocellular Carcinoma (HCC) risk in this study. We also found that IL-6 polymorphisms (-597G/A) were related to CHB in allelic, heterozygous, recessive model. For IL-6 polymorphism -174G/C, we did not find any association with CHB risk. Conclusion: The present meta-analysis indicated that IL-6 polymorphisms -572G/C and -597G/A significantly associate with CHB risk, but might not be significantly related to the progressive HBV such as LC and HCC. IL-6 polymorphisms -174G/C might not significantly associate with HBV related liver diseases.
Keywords: Interleukin-6, polymorphisms, hepatitis B virus, liver diseases, meta-analysis
Introduction
HBV infection is the most common cause of chronic liver disease around the world, the total number of HBV-infected population is 350 to 400 million and the number of deaths summed up to 250,000 [1], which continues to be a significant public health problem. The HBV-related liver disease contains self-limiting acute hepatitis, chronic hepatitis (CHB), fulminant hepatic failure (FHF), liver cirrhosis (LC) and hepatocellular carcinoma (HCC), the latter three are always considered to be the direct factors of HBV-infected patient’s death. However, about 95% of HBV infection individuals can successfully clear HBV, and only 5-10% will develop CHB. Among the CHB individuals, 20-30% will develop liver cirrhosis and 5% further progress to hepatocellular carcinoma [2]. The mechanism for persistent and progressive HBV infection is still unclear, which could be affected by complex factors including virus subtype, host condition, environmental and genetic elements. The current mainstream idea is that host immune factors and genetic factors may play important roles in the process [3].
Interleukin-6 (IL-6) is a multifunctional potent pleiotropic inflammatory cytokine that is considered a key growth-promoting and anti-apoptotic factor [4]. It plays a central role in the regulation of immune response. Previous studies have found that serum level of IL6 was increased in HBV-infected patients [5-7]. So high IL-6 levels might reflect more active hepatic necroinflammation and be associated with the presentation and severity in chronic HBV infection.
A number of studied about single nucleotide polymorphisms (SNPs) identified in the IL-6 gene, especially within the non-coding promoter sequence, involving -572G/C, -597G/A, and -174G/C, has been shown to influence the transcription and expression of IL-6. Zhao et al. have revealed four tag SNPs of IL-6 to be associated with susceptibility to chronic HBV infection in Chinese population [8]. Another study indicated that IL-6 (-572G/C) GC genotype shared a positive association with hepatitis among controls, and a negative association with cirrhosis and consequent HCC development among carriers in India [9]. However, in a Brazilian population, there are no significant differences in the polymorphism of IL-6 (-174G/C) between the chronic HBV-infected patient group and the self-limited infection group [10]. So far, large quantities of studies have confirmed the hypothesis that several IL-6 SNPs were potentially correlated with HBV infection risk, but the results were rather controversial and unconvincing, and no meta-analysis has examined the association between IL-6 polymorphism and HBV related liver diseases. Therefore, we have extensively reviewed literatures and conducted a meta-analysis to provide more credible evidence by systematically summarizing existed data.
Methods
Search strategy
We performed a systemic search in PubMed, EMBASE, Cochrane Library as well as Chinese databases including China National Knowledge Infrastructure (CNKI) and WanFang database for all the relevant studies utilizing the following search terms: “interleukin-6” or “IL-6”, “polymorphism” or “SNPs”, “Hepatitis B virus” or “HBV” (the latest research was updated to April 15, 2015). We only utilized data from fully published papers and not from meetings or conference abstracts. Additionally, articles in the reference list were manually searched for potentially relevant studies. When more than one of the same patient population was included in several publications, only the most recent or complete study was used. Search and literature retrieval were completed independently by two of the authors (Lei Chang and Tian Lan). Disagreements of the search result were settled by discussion among all the authors.
Inclusion and exclusion criteria
Studies were included in this meta-analysis if they met the following criteria: (1) the study assessed the association between HBV related disease and IL-6 polymorphisms (-572G/C, -597G/A and -174G/C); (2) chronic HBV infection cases included CHB, LC, HCC and IC (individuals who did not have any clinical symptoms and had persistently normal liver transaminase levels, serum albumin, and total serum bilirubin for at least 6 months); (3) case-control studies; (4) data in the studies were adequate to calculate odds ratio (OR) and 95% confidence interval (95% CI); (5) the genotype was tested in controls to ensure its fitting with the Hardy-Weinberg equilibrium (HWE). The major reasons for exclusion from our studies were: (1) family-based or sibling-based association studies; (2) the study without control group; (3) duplicated publication; (4) significant deviation of the genotype distribution in the control group from the HWE; (5) literature with insufficient data for evaluating OR and 95% CI.
Data extraction
Two investigators (Lei Chang and Yufeng Yuan) independently extracted the data from all eligible studies according to the inclusion and exclusion criteria above. The following information was collected from each study: author, year of publication, experimental method, ethnicity of research population, numbers of chronic HBV infection cases (including chronic hepatitis B, liver cirrhosis, hepatocellular carcinoma, and inactive carriers) and healthy controls, numbers of individual genotypes, distribution of haplotypes (-572/-597/-174), and the Hardy-Weinberg equilibrium (HWE) results.
Statistical analysis
We measure the strength of the association between IL-6 SNPs (-572G/C, -597G/A and -174G/C) and HBV related disease by pooled OR with its corresponding 95% CI. For -572G/C, the pooled ORs were estimated for allelic comparison (G vs C), homozygote comparison (GG vs CC), heterozygote comparison (GG vs CC), dominant model (GG + GC vs CC), and recessive model (GC + CC vs GG) respectively. For -597G/A, because of fewer researches the pooled ORs were estimated only for allelic comparison (G vs A), heterozygote comparison (GG vs GA) and recessive model (GA + AA vs GG). For -174G/C, the pooled ORs were estimated only for allelic comparison (G vs C), heterozygote comparison (GG vs GC) and recessive model (GC + CC vs GG). The P value of the pooled OR was considered significant if less than 0.05, which was examined by Z test. Heterogeneity across studies was determined by Chi-square test based Q statistic test and I2 statistic and the presence of heterogeneity was confirmed if the result was PQ < 0.05 or I2 ≥ 50%. In the condition of existence of heterogeneity, a random-effect model was utilized [11,12]; otherwise the fixed-effect model was employed to pool the results [13]. Additionally, in order to evaluate the stability of results, a sensitivity analysis was conducted. Both Begg’s test and Egger’s test were performed to test whether publication bias existed or not. All the analyses that have been mentioned were completed by STATA v.12.0.
Results
Characteristics of included studies
As shown in the Figure 1, 198 articles were found with the search strategy and then 135 duplicates are removed. After reading the titles or the abstracts, 35 records with improper titles, 6 articles were master thesis also excluded. Meanwhile, 2 articles based on the same population 4 articles studied on other polymorphisms. Eventually, 10 studies met the criteria were included in the meta-analysis of which 5 articles were in English and other 5 in Chinese [5,9,10,14-20]. The characteristics of included articles were showed in Table 1. For -572G/C polymorphism of IL-6, we included 8 articles including 3727 cases and 2709 controls (for subgroup analysis of IC; 426 cases and 777 controls; for subgroup analysis of CHB: 1413 cases and 2350 controls; for subgroup analysis of LC: 691 cases and 1408 controls; for subgroup analysis of HCC: 1197 cases and 2185 controls). 3 articles about -597G/A consisting of 578 cases and 450 controls (for subgroup analysis of CHB: 394 cases and 450 controls; for other subgroups, as only one studies was included, the analysis result tended to be less reliable, of which detailed data were not showed in this study) and 3 articles of -174G/C involving 263 cases and 203 controls were included as well (for subgroup analysis of CHB: 203 cases and 203 controls; for other subgroups, as only one studies was included, the analysis result tended to be less reliable, of which detailed data were not showed in this study). All these studies were case-control study.
Figure 1.
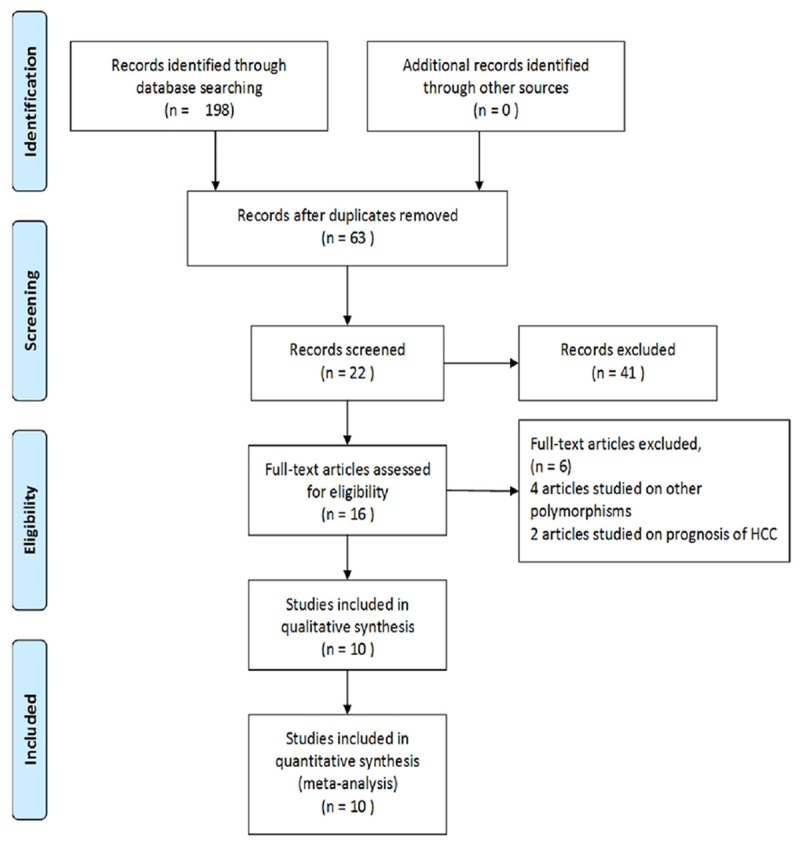
Flow chart of study selection.
Table 1.
Characteristics of studies included in the meta-analysis
| SNPs | Study | Year | Country | Genotyping methods | Genotype distribution | HWE | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||
| Controls | IC | CHB | LC | HCC | ||||||||||||||||
| -572G/C | GG | GC | CC | GG | GC | CC | GG | GC | CC | GG | GC | CC | GG | GC | CC | |||||
| Saxena, R. | 2014 | India | PCR-RFLP | 42 | 78 | 33 | 6 | 46 | 9 | 8 | 41 | 16 | 19 | 38 | 6 | 16 | 25 | 20 | 0.775 | |
| Lu, Y. | 2014 | China | PCR-RFLP | 16 | 96 | 100 | 7 | 78 | 134 | 0.279 | ||||||||||
| Tang, S. | 2013 | China | RT-PCR | 11 | 78 | 176 | 1 | 7 | 17 | 11 | 87 | 194 | 6 | 46 | 101 | 7 | 51 | 90 | 0.53 | |
| Liu, S. | 2012 | China | TaqMan | 15 | 132 | 271 | 6 | 74 | 206 | 13 | 136 | 237 | 0.827 | |||||||
| Qiu, X. Q. | 2011 | China | TaqMan | 13 | 105 | 241 | 23 | 107 | 210 | 12 | 110 | 259 | 0.71 | |||||||
| Dai, Y. | 2009 | China | PCR-RFLP | 23 | 116 | 73 | 7 | 87 | 66 | 0.073 | ||||||||||
| Xing, P. | 2007 | China | PCR | 8 | 54 | 38 | 7 | 59 | 45 | 0.062 | ||||||||||
| Park, B. L. | 2003 | Korea | PCR-RFLP | 62 | 371 | 557 | 17 | 88 | 175 | 32 | 169 | 274 | 12 | 92 | 117 | 0.983 | ||||
| -597G/A | GG | GA | AA | GG | GA | AA | GG | GA | AA | GG | GA | AA | GG | GA | AA | |||||
| Saxena, R. | 2014 | India | PCR-RFLP | 75 | 55 | 8 | 18 | 45 | 1 | 47 | 15 | 2 | 40 | 18 | 3 | 28 | 26 | 5 | 0.614 | |
| Lu, Y. | 2014 | China | PCR-RFLP | 211 | 1 | 0 | 219 | 0 | 0 | 0.973 | ||||||||||
| Xing, P. | 2007 | China | PCR | 99 | 1 | 0 | 109 | 2 | 0 | 0.960 | ||||||||||
| -174G/C | GG | GC | CC | GG | GC | CC | GG | GC | CC | GG | GC | CC | GG | GC | CC | |||||
| Ribeiro, C. S. | 2007 | Brazil | PCR | 20 | 18 | 2 | 15 | 14 | 1 | 0.416 | ||||||||||
| Xing, P. | 2007 | China | PCR | 98 | 2 | 0 | 107 | 4 | 0 | 0.920 | ||||||||||
| Chen, L. | 2006 | China | PCR | 62 | 1 | 0 | 59 | 1 | 0 | 62 | 0 | 0 | 0.949 | |||||||
Quantitative synthesis
Table 2 showed the main meta-analysis results of relationships between IL-6 -572G/C polymorphisms and HBV related disease for all population. Subgroup analysis by disease showed that IL-6 -572G/C exhibited the obvious association with CHB risk in allelic (Figure 2A), homozygous (Figure 2B), heterozygous (Figure 2C), recessive (Figure 2D) and dominant model (Figure 2E) (G vs C: OR = 0.786, 95% CI 0.701-0.881, PA = 0.000; GG vs CC: OR = 0.587, 95% CI 0.428-0.806, PA = 0.001; GG vs GC: OR = 0.681, 95% CI 0.497-0.934, PA = 0.017; GG+GC vs CC: OR = 0.768, 95% CI 0.666-0.886, PA = 0.000; GC+CC vs GG: OR = 1.630, 95% CI 1.202-2.210, PA = 0.002). However, there was no association between IL-6 -572G/C polymorphisms and IC (G vs C: OR = 1.121, 95% CI = 0.907-1.385, PA = 0.291; GG vs CC: OR = 1.347, 95% CI = 0.773-2.349, PA =0.293; GG vs GC: OR = 0.752, 95% CI = 0.454-1.246, PA = 0.269; GG + GC vs CC: OR = 1.262, 95% CI = 0.960-1.659, PA =0.095; GC + CC vs GG: OR = 1.145, 95% CI = 0.704-1.861, PA = 0.585), LC (G vs C: OR = 1.022, 95% CI = 0.880-1.187, PA = 0.777; GG vs CC: OR = 1.181, 95% CI = 0.811-1.719, PA = 0.385; GG vs GC: OR = 1.044, 95% CI = 0.730-1.494, PA = 0.813; GG + GC vs CC: OR = 1.011, 95% CI = 0.836-1.223, PA = 0.907; GC + CC vs GG: OR= 0.926, 95% CI = 0.657-1.304, PA = 0.658) and HCC (G vs C: OR = 1.038, 95% CI = 0.915-1.178, PA = 0.564; GG vs CC: OR = 0.891, 95% CI = 0.628-1.623, PA = 0.517; GG vs GC: OR = 0.911, 95% CI = 0.644-1.288, PA = 0.598; GG + GC vs CC: OR = 1.075, 95% CI = 0.922-1.253, PA = 0.357; GC + CC vs GG: OR = 1.083, 95% CI = 0.777-1.508, PA = 0.639) (Figure 2A-E).
Table 2.
The meta-analysis results of association between IL-6 -572G/C polymorphisms and HBV-related liver diseases
| SNPs | Contrast model | CHB | HCC | ||||||||||
|
|
|
||||||||||||
| Test for heterogeneity | OR (95% CI) | PA | Publication bias (Egger’s test) | Test for heterogeneity | OR (95% CI) | PA | Publication bias (Egger’s test) | ||||||
|
|
|
|
|||||||||||
| I2 (%) | P | t | P | I2 (%) | P | t | P | ||||||
|
| |||||||||||||
| -572G/C | G vs C | 1.0 | 0.417 | 0.786 (0.701-0.881) | 0.000 | -0.49 | 0.787 | 0.0 | 0.527 | 1.038 (0.915-1.178) | 0.564 | -1.41 | 0.539 |
| GG vs CC | 11.2 | 0.344 | 0.587 (0.428-0.806) | 0.001 | -1.59 | 0.135 | 0.0 | 0.867 | 0.891 (0.628-1.623) | 0.517 | 1.24 | 0.548 | |
| GG vs GC | 17.7 | 0.295 | 0.681 (0.497-0.934) | 0.017 | -2.37 | 0.105 | 0.00 | 0.941 | 0.911 (0.644-1.288) | 0.598 | 0.50 | 0.775 | |
| GG + GC vs CC | 0.0 | 0.525 | 0.768 (0.666-0.886) | 0.000 | 0.36 | 0.758 | 24.0 | 0.262 | 1.075 (0.922-1.253) | 0.357 | -1.96 | 0.334 | |
| GC + CC vs GG | 15.0 | 0.316 | 1.630 (1.202-2.210) | 0.002 | 4.02 | 0.268 | 0.0 | 0.992 | 1.083 (0.777-1.508) | 0.639 | -1.25 | 0.148 | |
|
| |||||||||||||
| SNPs | Contrast model | IC | LC | ||||||||||
|
|
|
||||||||||||
| Test for heterogeneity | OR (95% CI) | PA | Publication bias (Egger’s test) | Test for heterogeneity | OR (95% CI) | PA | Publication bias (Egger’s test) | ||||||
|
|
|
|
|||||||||||
| I2 (%) | P | t | P | I2 (%) | P | t | P | ||||||
|
| |||||||||||||
| -572G/C | G vs C | 47.7 | 0.148 | 1.121 (0.907-1.385) | 0.291 | -2.20 | 0.500 | 0.0 | 0.376 | 1.022 (0.880-1.187) | 0.777 | 1.96 | 0.434 |
| GG vs CC | 51.2 | 0.129 | 1.347 (0.773-2.349) | 0.293 | -2.38 | 0.515 | 19.0 | 0.291 | 1.181 (0.811-1.719) | 0.385 | 2.29 | 0.594 | |
| GG vs GC | 81.5 | 0.005 | 0.752 (0.454-1.246) | 0.269 | -1.24 | 0.796 | 0.0 | 0.866 | 1.044 (0.730-1.494) | 0.813 | -0.84 | 0.395 | |
| GG + GC vs CC | 0.0 | 0.678 | 1.262 (0.960-1.659) | 0.095 | -0.02 | 0.992 | 54.7 | 0.110 | 1.011 (0.836-1.223) | 0.907 | 3.80 | 0.287 | |
| GC + CC vs GG | 81.2 | 0.005 | 1.145 (0.704-1.861) | 0.585 | 1.59 | 0.872 | 0.0 | 0.953 | 0.926 (0.657-1.304) | 0.658 | 0.34 | 0.586 | |
Figure 2.
Forest plot of IL-6 -572G/C polymorphisms associated with hepatitis B virus related liver diseases risk for subgroup analysis of IC, CHB, LC, and HCC as compared with healthy control (HC). A: Allelic model, G vs C; B: Homozygous model, GG vs CC; C: Heterozygous modle, GG vs GC; D: Recessive model, GC + GG vs CC; E: Dominant model, GC + CC vs GG. The results indicated that IL-6 -572G/C exhibited the obvious association with CHB risk in all models.
Table 3 showed the main meta-analysis results of relationships between IL-6 -597G/A, IL-6 -174G/C polymorphisms and CHB for all population. The results indicated that IL-6 -597G/A exhibited the obvious association with CHB risk in allelic (Figure 3A), heterozygous, (Figure 3B) dominant model (Figure 3C) (G vs A: OR = 1.887, 95% CI 1.113-3.202, PA = 0.019; GG vs GA: OR = 2.209, 95% CI 1.117-3.923, PA = 0.021; GA + AA vs GG: OR = 0.470, 95% CI 0.257-0.861, PA = 0.014). As only one study of other models was included, the analysis result tended to be less reliable, of which detailed data were not showed in this study. Meanwhile, we did not find any association between IL-6 -174G/C polymorphisms and CHB risk (Figure 4A-C).
Table 3.
The meta-analysis results of association between IL-6 -597G/A, -174G/C polymorphisms and CHB risk
| SNPs | Contrast model | CHB | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Test for heterogeneity | OR (95% CI) | PA | Publication bias (Egger’s test) | ||||
|
|
|
||||||
| I2 (%) | P | t | P | ||||
| -597G/A | G vs A | 0 | 0.570 | 1.887 (1.113-3.202) | 0.019 | 3.54 | 0.175 |
| GG vs GA | 0 | 0.521 | 2.209 (1.117-3.923) | 0.021 | 3.17 | 0.195 | |
| GA + AA vs GG | 0 | 0.515 | 0.470 (0.257-0.861) | 0.014 | 0.51 | 0.698 | |
| -174G/C | G vs C | 0 | 0.630 | 0.992 (0.511-1.927) | 0.982 | 0.85 | 0.552 |
| GG vs GC | 0 | 0.644 | 0.920 (0.414-2.045) | 0.838 | 0.23 | 0.853 | |
| GC + CC vs GG | 0 | 0.638 | 1.058 (0.482-2.324) | 0.888 | 1.22 | 0.438 | |
Figure 3.
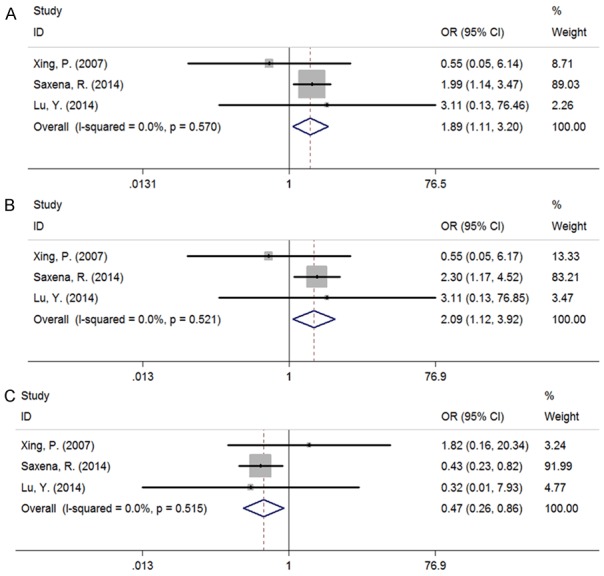
Forest plot of IL-6 -597G/A polymorphisms associated with CHB risk as compared with healthy control (HC). A: Allelic model, G vs A; B: Heterozygous model, GG vs GA; C: Dominant model, GA + AA vs GG; The results indicated that IL-6 -597G/A exhibited the obvious association with CHB risk in allelic, heterozygous, dominant model.
Figure 4.
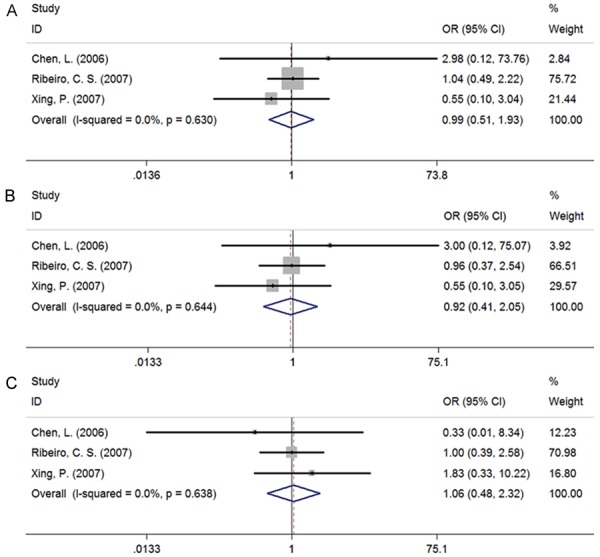
Forest plot of IL-6 -174G/C polymorphisms associated with CHB risk as compared with healthy control (HC). A: Allelic model, G vs C; B: Heterozygous model, GG vs GC; C: Dominant model, GC + CC vs GG; There was no any association between IL-6 -174G/C polymorphisms and CHB risk.
Publication bias and sensitivity analysis
The Begg’s test and Egger’s test were applied to assess the publication bias of included literature.The shapes of funnel plot were symmetrical, which have not implied the existence of publication bias. All the Egger’s test results of three polymorphisms were demonstrated in Tables 2 and 3, all P values were greater than 0.05 and thus there was no obvious publication bias in the meta-analysis.
Sensitivity analysis was performed to evaluate the stability of the result. Each data set was omitted individually to investigate the impact of a single study on the pooled ORs. The exclusion of any single study did not alter the overall conclusion, indicating that results were reliable.
Discussion
Human IL-6 genes located on chromosome 7p21 and total length of 5 KB, including four introns and five exons, which composed of 184 amino acids encoding protein IL-6 [21]. IL-6 was considered to be the starting stage of an important factor, when the virus infection happened, IL-6 inducied a variety of cell synthesis and secretion of a variety of acute phase protein, and promoted T, B lymphocyte proliferation, differentiation and produced immune globulin. IL-6 played a very important role in virus infection of inflammation [22]. Current researchers found that transcription factor such as NF-κB, Fos/Jun and glucocorticoid receptors can combine with the IL-6 promoter sequences and then regulation of expression of IL-6 gene at the transcriptional level [23]. IL-6 promoter polymorphism could lead to interindividual differences in gene transcription and expression, which affects the susceptibility to HBV infections and the progression of HBV-related diseases.
In current meta-analysis, we have investigated the relationships between three interlenkin-6 polymorphisms and HBV-related diseases risk. The results demonstrated that -572G/C in the promoter region of IL-6 gene was associated with an increased risk of HBV infection in heterozygous and dominant model (GG vs GC: OR = 0.832, 95% CI = 0.694-0.999, PA = 0.048; GC + CC vs GG: OR = 1.207, 95% CI = 1.014-1.437, PA = 0.034). Furthermore, In the subgroup analysis by HBV-related disease, an significant risk was found among CHB in all genetic models(G vs C: OR = 0.768, 95% CI 0.701-0.881, PA = 0.000; GG vs CC: OR = 0.587, 95% CI 0.428-0.806, PA = 0.001; GG vs GC: OR = 0.681, 95% CI 0.497-0.934, PA = 0.017; GG + GC vs CC: OR = 0.768, 95% CI 0.666-0.886, PA = 0.000; GC + CC vs GG: OR = 1.630, 95% CI 1.202-2.210, PA = 0.002), but no statistical association has been observed among other HBV-related diseases (LC, HCC and IC), indicating that the risk of HBV infection caused by -572G/C polymorphism may contribute to the difference of latter’s distribution in CHB patients. Therefore, we can conclude that to some extent, the single nucleotide polymorphism of -572G/C in IL-6 promoter region is inclined to promote the development of CHB.
For -597G/A, a total of 3 case-control studies [9,14,19] were analyzed to provide a comprehensive assessment of the association between its polymorphism and CHB. We noticed that a significant correlation exists under the allelic, heterozygous, dominant model (G vs A: OR = 1.887, 95% CI 1.113-3.202, PA = 0.019; GG vs GA: OR = 2.209, 95% CI 1.117-3.923, PA = 0.021; GA + AA vs GG: OR = 0.470, 95% CI 0.257-0.861, PA = 0.014). Simultaneously, there was no heterogeneity in the three genetic models, of which the result data possess a high centrality and credibility. Similarly, we have analyzed the data from three included studies [10,14,20] about the SNP of -174G/C in IL-6 promoter region and the result of meta-analysis illustrated that IL-6 -174G/C could not play a part in the susceptibility to CHB.
Nowadays, the number of studies about the association between the single nucleotide polymorphisms (-572G/C, -597G/A and -174G/C) and HBV infection risk is not enough. And in today’s era of evidence-based medicine, the system review or meta-analysis is insufficient. In addition, the research findings are conflicting and don’t achieve a consensus, which makes this meta-analysis indispensable. Simultaneously, this is the first meta-analysis concerning the association between IL-6 Polymorphisms and HBV-related Diseases.
However, the shortcomings of this study should be taken into consideration. Firstly, due to lack of related data, the quantity of involved studies about the SNP of -597G/A and -174G/C was not adequate. Only three studies were included respectively, although there was no heterogeneity in the results. Secondly, a few of the study controls were not from healthy populations. Selection bias could have occurred, which may have confounded the results. Third, the meta-analysis contains no prospective study to confirm the correlation between three IL-6 polymorphisms and CHB risk, which can provide a higher reliability.
In general, our meta-analysis illuminated that the IL-6 -572G/C and IL-6 -597G/A might be associated with CHB risk, but the IL-6 -174G/C did not play a part in the susceptibility to CHB. Nevertheless, because of the aforementioned limitations, more evidence of prospective, multi-centric and multi-populational trials are needed to further explore the association between IL-6 polymorphisms and HBV infection risk.
Disclosure of conflict of interest
None.
References
- 1.Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144–150. doi: 10.1111/j.1572-0241.2003.07179.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641–644. doi: 10.3748/wjg.v9.i4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res. 2001;52:113–116. doi: 10.1016/s0166-3542(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 4.Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–368. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 5.Park BL, Lee HS, Kim YJ, Kim JY, Jung JH, Kim LH, Shin HD. Association between interleukin 6 promoter variants and chronic hepatitis B progression. Exp Mol Med. 2003;35:76–82. doi: 10.1038/emm.2003.11. [DOI] [PubMed] [Google Scholar]
- 6.Park Y, Park JY, Han KH, Kim HS. Serum cytokine levels in chronic hepatitis B patients receiving peginterferon alpha-2a therapy. Hepatobiliary Pancreat Dis Int. 2012;11:499–506. doi: 10.1016/s1499-3872(12)60214-8. [DOI] [PubMed] [Google Scholar]
- 7.Kuo TM, Hu CP, Chen YL, Hong MH, Jeng KS, Liang CC, Chen ML, Chang C. HBV replication is significantly reduced by IL-6. J Biomed Sci. 2009;16:41. doi: 10.1186/1423-0127-16-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao XM, Gao YF, Zhou Q, Pan FM, Li X. Relationship between interleukin-6 polymorphism and susceptibility to chronic hepatitis B virus infection. World J Gastroenterol. 2013;19:6888–6893. doi: 10.3748/wjg.v19.i40.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena R, Chawla YK, Verma I, Kaur J. IL-6(-572/-597) polymorphism and expression in HBV disease chronicity in an Indian population. Am J Hum Biol. 2014;26:549–555. doi: 10.1002/ajhb.22562. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro CS, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorp- hism with hepatitis B infection evolution in adult patients. Mem Inst Oswaldo Cruz. 2007;102:435–440. doi: 10.1590/s0074-02762007005000043. [DOI] [PubMed] [Google Scholar]
- 11.Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava K, Srivastava A, Mittal B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010;55:495–499. doi: 10.1038/jhg.2010.54. [DOI] [PubMed] [Google Scholar]
- 13.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 14.Xing PX, Zou MJ, Xing QT, et al. Relationship between proinflammatory cytokine gene polymorphisms and diseases of HBV infection. Journal of Shandong University (Health Science) 2007;45:1229–33. [Google Scholar]
- 15.Dai Y, Liu XL, Chai QB. Relationship between the -572G/C polymorphism in IL-6 gene promoter and hepatitis virus B infection in Chinese Han population. World Chinese Journal of Digestology. 2009;17:1522–26. [Google Scholar]
- 16.Qiu XQ, Bei CH, Yu HP, Zeng XY, Zhong QA. Study on the relationship between single-nucleotide polymorphisms in IL-6, IL-10 genes and HBV-related hepatocellular carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:510–513. [PubMed] [Google Scholar]
- 17.Liu S, Qiu XQ, Zeng XY, Bai H, Bei CH, Yang Y. Relationship between IL6 -572G/C polymorphism and hepatocellular carcinoma in men. Zhonghua Gan Zang Bing Za Zhi. 2012;20:463–467. doi: 10.3760/cma.j.issn.1007-3418.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Tang S, Liu Z, Zhang Y, He Y, Pan D, Liu Y, Liu Q, Zhang Z, Yuan Y. Rather than Rs1800796 polymorphism, expression of interleukin-6 is associated with disease progression of chronic HBV infection in a Chinese Han population. Dis Markers. 2013;35:799–805. doi: 10.1155/2013/508023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Peng J, Wang C, Zhu Y, Wang F, Sun Z. IL-6 promoter functional polymorphism -572C/G affects spontaneous clearance of hepatitis B virus infection. Clin Lab. 2014;60:1903–1907. doi: 10.7754/clin.lab.2014.140311. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Chen ZX, Pan C, et al. The relationship between interleukin-6 polymorphisms and chronic HBV infection. Chinese Journal of Integrated Traditional and Western Medicine on Liver Diseases. 2006;16:82–6. [Google Scholar]
- 21.Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res Ther. 2006;8(Suppl 2):S2. doi: 10.1186/ar1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fielding CA, Mcloughlin RM, Mcleod L, Colmont CS, Najdovska M, Grail D, Ernst M, Jones SA, Topley N, Jenkins BJ. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J Immunol. 2008;181:2189–2195. doi: 10.4049/jimmunol.181.3.2189. [DOI] [PubMed] [Google Scholar]
- 23.Hosel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, Langenkamp A, Falk C, Büning H, Rose-John S, Protzer U. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]



