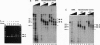Abstract
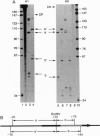
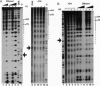
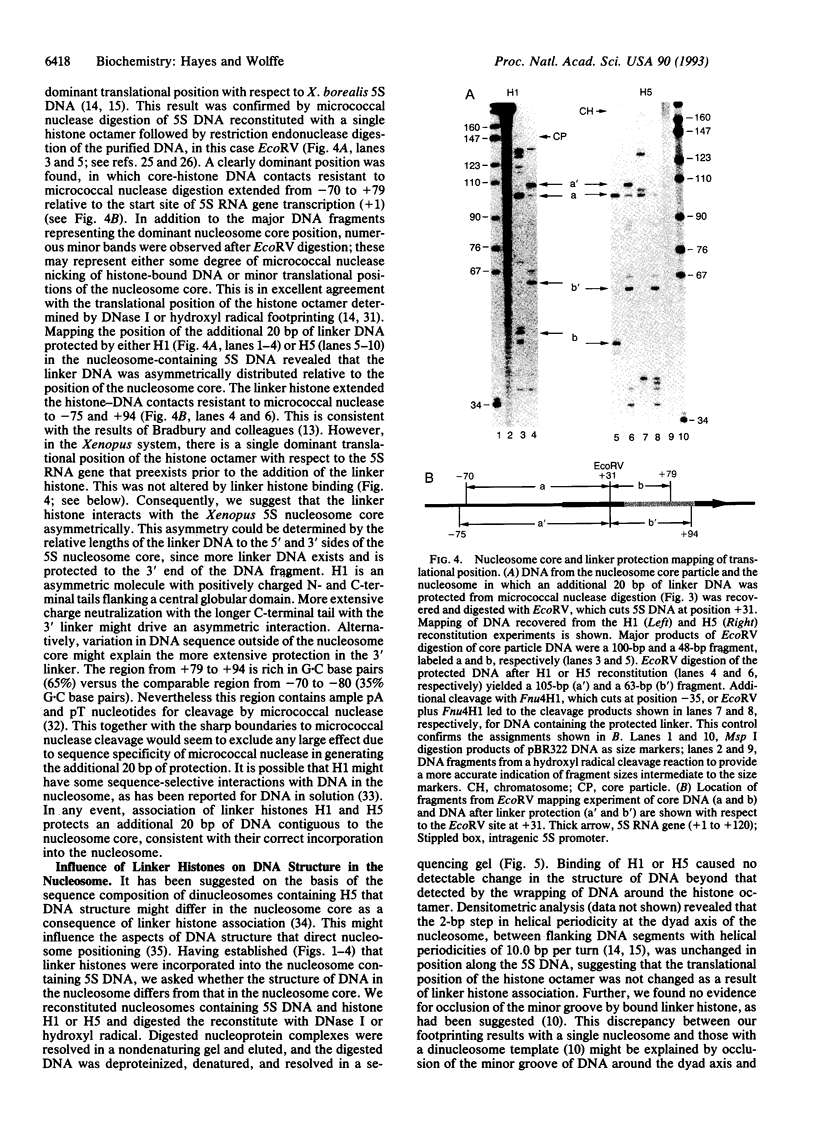
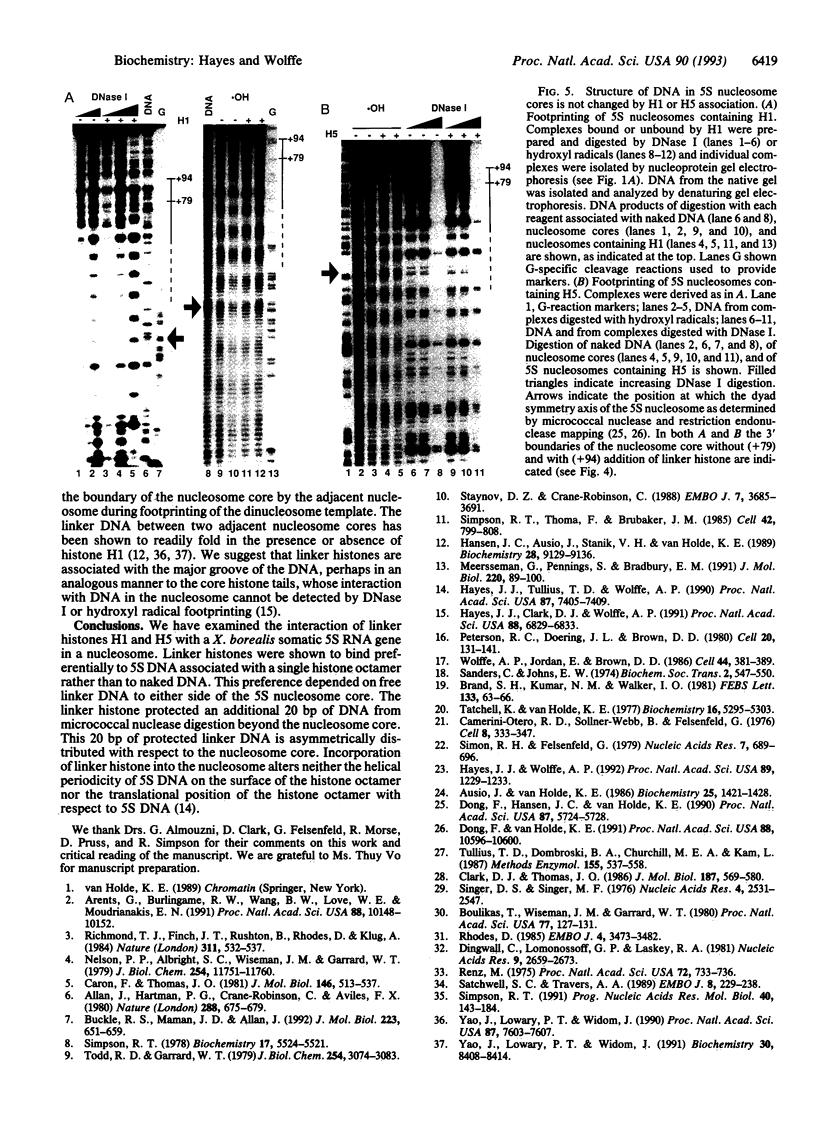
We establish that linker histones H1 and H5 bind preferentially to a Xenopus borealis somatic 5S RNA gene associated with an octamer of core histones rather than to naked 5S DNA. This preferential binding requires free linker DNA to either side of the nucleosome core. Incorporation of a single linker histone molecule into the nucleosome protects an additional 20 bp of linker DNA from micrococcal nuclease digestion. This additional DNA is asymmetrically distributed with respect to the nucleosome core. Incorporation of linker histones causes no change to the cleavage of DNA in the nucleosome by hydroxyl radical or DNase I.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S. H., Kumar N. M., Walker I. O. The properties of salt-extracted histone H1. FEBS Lett. 1981 Oct 12;133(1):63–66. doi: 10.1016/0014-5793(81)80471-1. [DOI] [PubMed] [Google Scholar]
- Buckle R. S., Maman J. D., Allan J. Site-directed mutagenesis studies on the binding of the globular domain of linker histone H5 to the nucleosome. J Mol Biol. 1992 Feb 5;223(3):651–659. doi: 10.1016/0022-2836(92)90981-o. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Hansen J. C., van Holde K. E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., van Holde K. E. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C., Ausio J., Stanik V. H., van Holde K. E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989 Nov 14;28(23):9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Clark D. J., Wolffe A. P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D., Wolffe A. P. The structure of DNA in a nucleosome. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1229–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5 S rDNA. J Mol Biol. 1991 Jul 5;220(1):89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Renz M. Preferential and cooperative binding of histone I to chromosomal mammalian DNA. Proc Natl Acad Sci U S A. 1975 Feb;72(2):733–736. doi: 10.1073/pnas.72.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Travers A. A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989 Jan;8(1):229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Singer D. S., Singer M. F. Studies on the interaction of H1 histone with superhelical DNA: characterization of the recognition and binding regions of H1 histones. Nucleic Acids Res. 1976 Oct;3(10):2531–2547. doi: 10.1093/nar/3.10.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staynov D. Z., Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988 Dec 1;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Reconstitution of chromatin core particles. Biochemistry. 1977 Nov 29;16(24):5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Overall pathway of mononucleosome production. J Biol Chem. 1979 Apr 25;254(8):3074–3083. [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Linker DNA bending induced by the core histones of chromatin. Biochemistry. 1991 Aug 27;30(34):8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]