Abstract
The possible association between CD28 IVS3 +17T>C (rs3116496) polymorphism and cancer susceptibility has been widely investigated. However, the results are conflicting. To verify the association more precisely, we performed a meta-analysis of 11 publications involving a total of 8,843 subjects. In this meta-analysis, 11 publications were included by searching PubMed and EMBASE databases up to May 23, 2014. The cancer susceptibility associated with the CD28 IVS3 +17T>C polymorphism was evaluated by odds ratios (ORs) with 95% confidence intervals (95% CIs). Heterogeneity, sensitivity and publication bias analyses were also assessed. The result suggested that the CD28 IVS3 +17T>C polymorphism is not associated with cancer susceptibility in overall cancer. In a stratified analysis by ethnicity, the association of CD28 IVS3 +17T>C polymorphism with cancer susceptibility was significant in Asians. In a stratified analysis by the origin of cancer cells and system of cancer, CD28 IVS3 +17T>C polymorphism was not associated with cancer susceptibility. In summary, this meta-analysis demonstrated that the CD28 IVS3 +17T>C polymorphism may be a cancer susceptibility factor in Asians.
Keywords: Cancer, polymorphism, CD28, cancer susceptibility, meta-analysis
Introduction
Cancer is a critical public health problem and one of the leading causes of death worldwide [1]. Accumulating evidence suggests that cancer results from complex mutual effect between genetic and environmental factors [2-4]. The immune reaction acts as an important natural barrier to cancer development and progression. All the principal antitumor responses are cell-mediated, such as by natural killer (NK) cells and T lymphocytes. Thus, genetic mutations of important immunological genes that regulate the function of T lymphocytes and NK cells may alter cancer susceptibility [5].
Effective activation of T cell results from the interaction between multiple costimulatory receptors and their ligands on an antigen presenting cell [6]. CD28, one of the best characterized costimulatory molecules, is expressed by the most T cells. CD28 competes with CTLA-4 for B7 binding, thus enhancing T-cell proliferation, which is inhibited by the CTLA-4-B7 interaction. In the last decade, several molecular epidemiological studies demonstrated an association between CD28 IVS3 +17T>C (rs3116496) polymorphism and cancer susceptibility. In the previous studies, it was reported that the CD28 IVS3 +17 TT genotype was associated with a low penetrance risk of cervical cancer and breast cancer in a Chinese Han population [7,8]. However, an individual investigation may have limited power to achieve a conclusive and reliable result. To further explore the role of the CD28 IVS3 +17T>C polymorphism in tumorigenesis, we conducted a comprehensive meta-analysis of all eligible publications. To the best of our knowledge, this study is the first meta-analysis considering the CD28 IVS3 +17T>C polymorphism and its association with cancer susceptibility.
Materials and methods
Search strategy
PubMed and EMBASE databases (the last search was updated in May 23, 2014) were searched simultaneously with combination of the following terms: ‘CD28’, ‘polymorphism’ or ‘SNP’ or ‘variant’, and ‘cancer’ or ‘malignance’ or ‘carcinoma’ or ‘Neoplasm’ or ‘tumor’. The search was limited to human studies and no language restrictions. All bibliographies in reviews and the retrieved articles were checked to identify additional publications.
Inclusion and exclusion criteria
For inclusion, recruited publications had to meet the major selection criteria: (1) evaluating the CD28 IVS3 +17T>C polymorphism and cancer susceptibility, (2) using a case-control study design, (3) containing complete data on genotype or allele frequency in case groups and control groups. Accordingly, reports without usable data, not case-control study, reviews and duplicated data were excluded.
Data extraction
For each recruited publications the following data was collected independently by two authors (S. Zhang and Y. Wang): (1) the name of first author, (2) cancer type, (3) published year, (4) country of origin, (5) ethnicity, (6) case number and control number, (7) allele and genotype frequency, (8) genotyping method and (9) evidence of Hardy-Weinberg equilibrium (HWE) in controls. When come to conflicting evaluations, disagreements were discussed until reaching conformity on items among all reviewers.
Statistical analysis
The HWE in controls was determined using an internet-based HWE calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The crude odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated to estimate the strength of association between CD28 IVS3 +17T>C polymorphism and cancer susceptibility. A P < 0.05 (two-tailed) was considered as statistically significant. A Chi-square-based I2 test was used to detect for heterogeneity and an I2 < 25% indicates low heterogeneity, 25% ≤ I2 ≤ 50% indicates moderate heterogeneity, and I2 > 50% indicates large heterogeneity [9]. When I2 > 50% or P < 0.10 (two-sided), the random-effects model (the DerSimonian-Laird method) [10] was utilized to analyze the data, otherwise the fixed-effects model (the Mantel-Haenszel method) was used [11]. Sub-group analyses were carried out according to ethnicity, system of cancer, the origin of cancer cells, sample size, and publication year to explore the source of heterogeneity among variables. Galbraith radial plot was used to detect the major source of heterogeneity. Publication bias of the literature was assessed by Begg’s funnel plot and Egger’s test. Nonparametric “trim-and-fill” method and one-way sensitivity analysis were both used to confirm the stability of our findings. In addition, for the results of publication bias test, statistical significance was defined as P < 0.1 (two-sided). All statistical analyses in meta-analysis were carried out using STATA version 12.0 software (Stata Corporation, College Station, TX).
Results
Characteristics
The detailed selecting and excluding process was shown in Figure 1. In total, there were 11 eligible studies [7,8,12-20] recruited in this meta-analysis, involving 4099 cancer cases and 4744 controls. Among them, four investigated cervical cancer [7,13,14,20], one investigated lung cancer [12], one investigated colorectal cancer [16], one investigated breast cancer [8], one investigated melanoma [15], one investigated myeloma [19], one investigated leukemia [17] and one study investigated lymphoma [18]. As for subjects, eight were Caucasians [12-17,19,20] and three were Asians [7,8,18]. Characteristics of these studies are presented in Table 1. The distribution of the CD28 IVS3 +17T>C polymorphism and allele among cases and controls is showed in Table 2.
Figure 1.
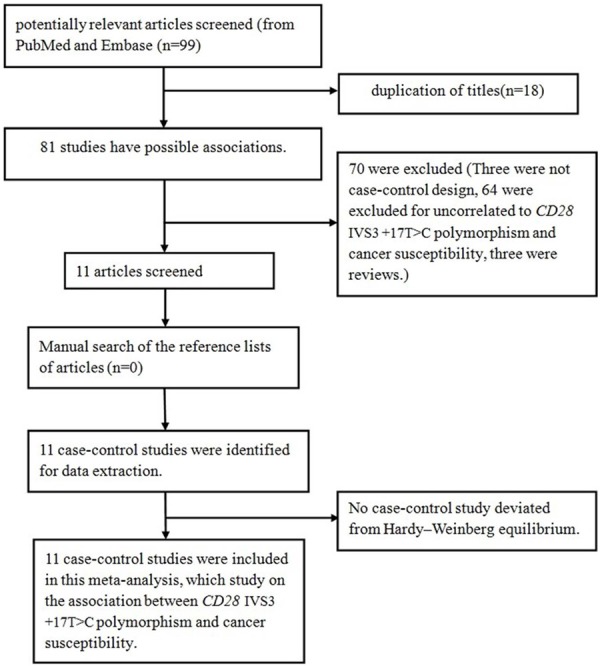
Flow diagram of articles selection process for CD28 IVS3 +17T>C polymorphism and cancer risk meta-analysis.
Table 1.
Characteristics of the individual studies included in the meta-analysis
| study | year | country | ethnicity | cancer type | No. of cases/controls | Genotype Method |
|---|---|---|---|---|---|---|
| Chen et al. | 2012 | China | Asians | breast cancer | 565/605 | PCR-RFLP |
| Karabon et al. | 2011 | Poland | Caucasians | lung cancer | 208/326 | single-nucleotide primer-extension methods |
| Chen et al. | 2011 | China | Asians | cervical cancer | 619/985 | PCR-RFLP |
| Ivansson et al. | 2010 | Sweden | Caucasians | cervical cancer | 1306/811 | Taqman |
| Pawlak et al. | 2010 | Poland | Caucasians | cervical cancer | 147/225 | single-nucleotide primer-extension methods |
| Bouwhuis et al. | 2010 | German | Caucasians | melanoma | 763/734 | Taqman |
| Karabon et al. | 2009 | Poland | Caucasians | myeloma | 150/238 | SNapShot |
| Dilmec et al. | 2008 | Turkey | Caucasians | colorectal cancer | 56/162 | PCR-RFLP |
| Suwalska et al. | 2008 | Poland | Caucasians | leukemia | 173/336 | single-nucleotide primer-extension methods |
| Cheng et al. | 2006 | China | Asians | lymphoma | 62/250 | PCR-RFLP |
| Wlodarska-Polinska et al. | 2006 | Poland | Caucasians | cervical cancer | 50/72 | SNapShot |
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
Table 2.
Distribution of CD28 IVS3 +17T>C polymorphisms genotype and allele among multiple cancer patients and controls
| Study | Year | Case | Control | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| TT | TC | CC | TT | TC | CC | C | T | C | T | |||
| Chen et al. | 2012 | 450 | 109 | 6 | 520 | 81 | 4 | 121 | 1009 | 89 | 1121 | 0.664536 |
| Karabon et al. | 2011 | 153 | 51 | 4 | 230 | 89 | 5 | 59 | 357 | 99 | 549 | 0.271397 |
| Chen et al. | 2011 | 492 | 120 | 7 | 853 | 123 | 9 | 134 | 1104 | 141 | 1829 | 0.057968 |
| Ivansson et al. | 2010 | 916 | 343 | 42 | 538 | 253 | 19 | 427 | 2175 | 291 | 1329 | 0.088850 |
| Pawlak et al. | 2010 | 100 | 31 | 1 | 172 | 49 | 2 | 33 | 231 | 53 | 393 | 0.462421 |
| Bouwhuis et al. | 2010 | 487 | 254 | 22 | 475 | 231 | 24 | 298 | 1228 | 279 | 1181 | 0.524521 |
| Karabon et al. | 2009 | 75 | 21 | 2 | 179 | 55 | 4 | 25 | 171 | 63 | 413 | 0.923949 |
| Dilmec et al. | 2008 | 32 | 19 | 5 | 106 | 50 | 6 | 29 | 83 | 62 | 262 | 0.972498 |
| Suwalska et al. | 2008 | 112 | 56 | 4 | 256 | 74 | 5 | 64 | 280 | 84 | 586 | 0.894690 |
| Cheng et al. | 2006 | 52 | 9 | 1 | 192 | 57 | 1 | 11 | 113 | 59 | 441 | 0.131639 |
| Wlodarska-Polinska et al. | 2006 | 39 | 9 | 2 | 52 | 18 | 2 | 13 | 87 | 22 | 122 | 0.771165 |
HWE: Hardy-Weinberg equilibrium.
Quantitative synthesis
A total of 8,843 subjects (4099 cancer cases and 4744 controls) from 11 studies were included to analyze the association of CD28 IVS3 +17T>C polymorphism with cancer susceptibility. After combining these studies, there was null association of CD28 IVS3 +17T>C polymorphism with overall cancer susceptibility (Table 3; Figures 2 and 3). In a stratified analysis by the origin of cancer cells and system of cancer, the association of CD28 IVS3 +17T>C polymorphism was also non-significant. While in a stratified analysis by ethnicity, a significant increase in cancer risk was detected among Asians in allele genetic models: C vs. T (OR, 1.38; 95% CI, 1.02-1.86; P = 0.039), but not Caucasians (Table 3).
Table 3.
Summary of results of the meta-analysis from different comparative genetic models in the subgroup analysis
| No. (cases/controls) | C vs. T | CC vs. TT | CC+TC vs. TT | CC vs. TC+TT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | ||
| Total | 4099/4744 | 1.14 (0.96-1.35) | 0.127 | 0.002 | 1.27 (0.93-1.73) | 0.134 | 0.945 | 1.13 (0.92-1.39) | 0.245 | <0.001 | 1.26 (0.93-1.72) | 0.135 | 0.950 |
| Ethnicity | |||||||||||||
| Asians | 1246/1840 | 1.38 (1.02-1.86) | 0.039 | 0.106 | 1.58 (0.74-3.34) | 0.236 | 0.789 | 1.37 (0.94-2.00) | 0.101 | 0.054 | 1.47 (0.69-3.11) | 0.315 | 0.723 |
| Caucasians | 2853/2904 | 1.01 (0.91-1.12) | 0.866 | 0.128 | 1.21 (0.86-1.70) | 0.265 | 0.867 | 1.04 (0.86-1.25) | 0.718 | 0.062 | 1.23 (0.88-1.72) | 0.234 | 0.876 |
| The origin of cancer cells | |||||||||||||
| epithelial tumor | 2951/3186 | 1.16 (0.92-1.48) | 0.208 | 0.001 | 1.41 (0.95-2.09) | 0.086 | 0.958 | 1.15 (0.86-1.53) | 0.356 | <0.001 | 1.43 (0.97-2.11) | 0.070 | 0.977 |
| non-epithelial tumor | 1148/1558 | 1.09 (0.83-1.45) | 0.525 | 0.092 | 1.06 (0.63-1.75) | 0.836 | 0.626 | 1.10 (0.78-1.54) | 0.599 | 0.057 | 1.02 (0.62-1.69) | 0.940 | 0.644 |
| System of cancer | |||||||||||||
| Reproductive cancer | 2122/2093 | 1.09 (0.77-1.56) | 0.624 | 0.003 | 1.29 (0.81-2.04) | 0.277 | 0.990 | 1.07 (0.69-1.66) | 0.769 | <0.001 | 1.34 (0.85-2.11) | 0.214 | 0.980 |
| Hematopoietic malignancy | 385/824 | 1.10 (0.69-1.76) | 0.684 | 0.069 | 1.71 (0.64-4.54) | 0.282 | 0.790 | 1.07 (0.59-1.93) | 0.823 | 0.032 | 1.60 (0.60-4.24) | 0.344 | 0.767 |
| Other system cancer | 1592/1827 | 1.17 (0.92-1.49) | 0.195 | 0.059 | 1.17 (0.74-1.85) | 0.512 | 0.396 | 1.18 (0.91-1.52) | 0.220 | 0.082 | 1.14 (0.72-1.79) | 0.587 | 0.433 |
| Sample sizes | |||||||||||||
| ≥ 1000 | 3253/3135 | 1.20 (0.91-1.57) | 0.192 | <0.001 | 1.17 (0.82-1.67) | 0.399 | 0.712 | 1.21 (0.87-1.68) | 0.249 | <0.001 | 1.17 (0.82-1.67) | 0.381 | 0.663 |
| < 1000 | 846/1069 | 1.11 (0.93-1.32) | 0.254 | 0.181 | 1.64 (0.89-3.01) | 0.114 | 0.947 | 1.08 (0.89-1.32) | 0.421 | 0.116 | 1.60 (0.87-2.93) | 0.129 | 0.960 |
| Publication year | |||||||||||||
| > 2009 | 3608/3686 | 1.13 (0.92-1.40) | 0.241 | 0.001 | 1.16 (0.83-1.63) | 0.390 | 0.920 | 1.14 (0.88-1.47) | 0.315 | <0.001 | 1.17 (0.83-1.64) | 0.367 | 0.893 |
| ≤ 2009 | 491/1058 | 1.21 (0.97-1.51) | 0.094 | 0.139 | 1.92 (0.94-3.94) | 0.075 | 0.916 | 1.09 (0.74-1.62) | 0.658 | 0.075 | 1.84 (0.90-3.74) | 0.095 | 0.923 |
Figure 2.
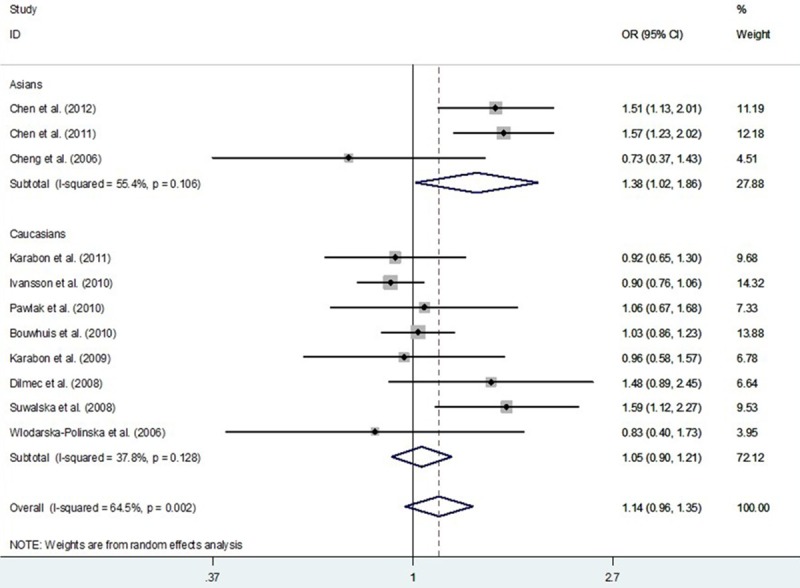
Meta-analysis with a random-effects model for the association between CD28 IVS3 +17T>C polymorphism and cancer risk (C vs. T compare genetic model).
Figure 3.
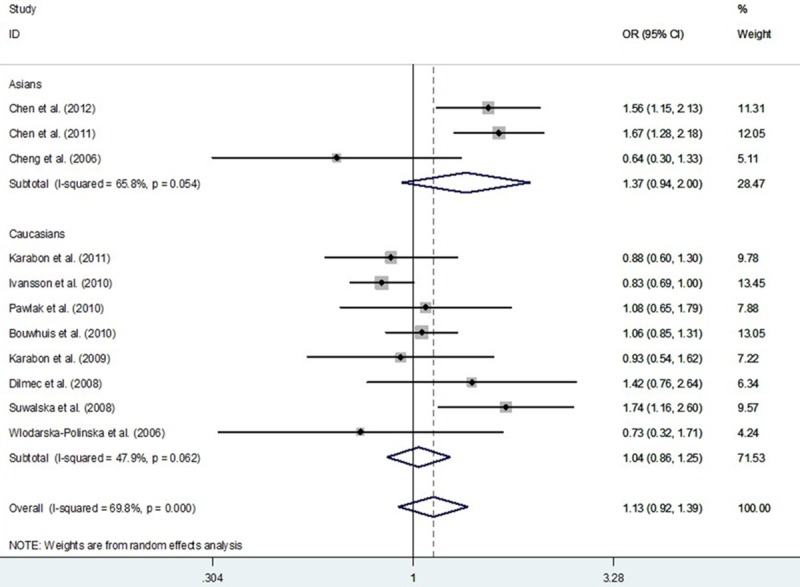
Meta-analysis with a random-effects model for the association between CD28 IVS3 +17T>C polymorphism and cancer risk (CC+TC vs. TT compare genetic model).
Tests for publication bias, sensitivity analyses, and heterogeneity
In our study, Begg’s funnel plot and Egger’s test were used to estimate the publication bias. The results showed that there was no evidence of publication bias (C vs. T: Begg’s test P = 0.755, Egger’s test P = 0.676; CC vs. TT: Begg’s test P = 0.876, Egger’s test P = 0.138; CC+TC vs. TT: Begg’s test P = 0.533, Egger’s test P = 0.852; CC vs. TC+TT: Begg’s test P = 1.000, Egger’s test P = 0.179) (Figure 4).
Figure 4.
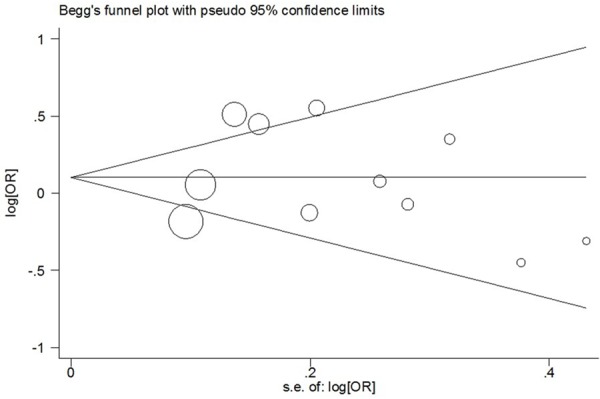
Begg’s funnel plot of meta-analysis of between the CD28 IVS3 +17T>C polymorphism and the risk of cancer in the dominant model.
We performed one-way sensitivity analysis to assess the influence of an individual study on the pooled OR by omitting one study in turn and the results suggested that our findings were stable (Figure 5) (data not shown). We also performed nonparametric “trim-and-fill” method as the other sensitivity analysis method. The adjusted ORs and CIs were not materially altered, suggesting that our findings were robust (CC+TC vs. TT: adjusted pooled OR = 1.13, 95% CI: 0.92-1.39, P = 0.245; CC vs. TC+TT: adjusted pooled OR = 1.17, 95% CI: 0.88-1.56, P = 0.288; CC vs. TT: adjusted pooled OR = 1.13, 95% CI: 0.85-1.50, P = 0.393; C vs. T: adjusted pooled OR = 1.14, 95% CI: 0.96-1.35, P = 0.127) (Figure 6).
Figure 5.
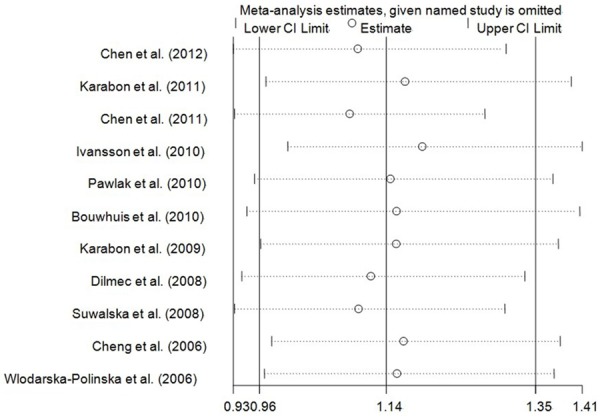
Sensitivity analysis of the influence of C vs. T compare genetic model in overall cancer meta-analysis (random-effects estimates).
Figure 6.
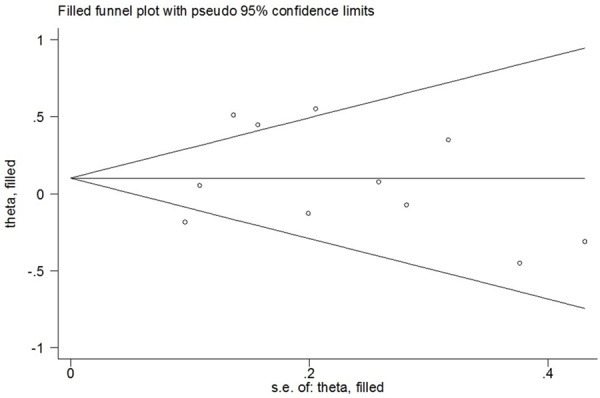
Filled funnel plot of meta-analysis of between the CD28 IVS3 +17T>C polymorphism and the risk of cancer in the dominant model.
As shown in Table 3, the significant heterogeneity was detected in current meta-analysis. Thus, we evaluated the sources of heterogeneity by the origin of cancer cells, system of cancer and ethnicity (Table 3). The results suggested that epithelial cancer and reproductive cancer subgroups might contribute to the major sources of heterogeneity. As shown in Table 3, heterogeneity was significant in the dominant model. We performed Galbraith radial plot to analyze the heterogeneity (Figure 7) and the result showed four outliers, which might contribute to the major source of heterogeneity. We conducted further stratified meta-analyses and the results suggested an association of studies designed in large sample size (≥ 1000 subjects) and publication year after 2009 with more prominent heterogeneity (Table 3).
Figure 7.
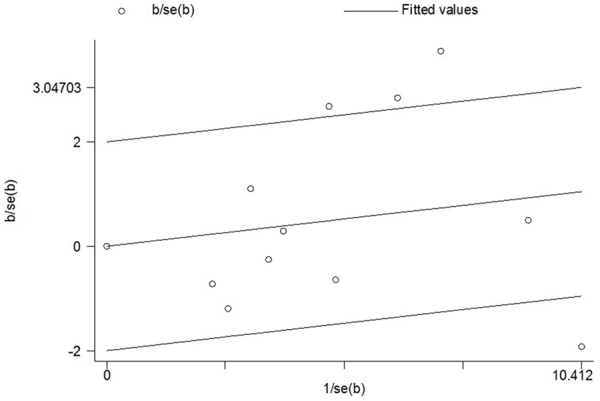
Galbraith radial plot of meta-analysis (CC+TC vs. TT compare genetic model).
Discussion
The possible association of CD28 IVS3 +17T>C polymorphism with cancer susceptibility has been widely studied; however, the results are conflicting. To more precisely determine this relationship, a meta-analysis was carried out. The results demonstrated that the CD28 IVS3 +17T>C polymorphism was not associated with overall cancer susceptibility. In a stratified analysis by the origin of the cancer cells and the cancer system, the association was also non-significant. Meanwhile, in a stratified analysis by ethnicity, a significant increase in cancer susceptibility was detected in the allele model among Asians, but not Caucasians (Table 3).
Recently, with the growing interest in the association between mutations of important immunological genes and cancer susceptibility, studies have examined the hypothesis whether CD28 IVS3 +17T>C polymorphism is relevant to cancer susceptibility; however, their findings were inconclusive and ambiguous. An individual study might be underpowered; therefore, the present study performed a meta-analysis to consider the association of the variant with cancer susceptibility in several cancer systems, the origin of the cancer cells and different ethnicities. One individual study has reported a borderline negative signal of CD28 IVS3 +17T>C polymorphism with cervical cancer [13]; another three studies reported a positive signal with breast cancer, cervical cancer and leukemia [7,8,17]. However, as presented in Table 3, the results among 8,843 subjects showed non-significance in overall cancer susceptibility, even in different cancer systems and the origin of the cancer cells. In the stratified analysis by ethnicity, increased susceptibility conferred by the allele model was observed for Asian populations. We also observed borderline evidence of an association between CD28 IVS3 +17T>C polymorphism and an increased risk of epithelial cancer in the recessive genetic model and homozygote comparison. Considering only 11 publications were included and some of them were designed as small sample sizes (< 1000), our results should be interpreted with caution. In the future, more extensive studies with large sample sizes, more types of cancer systems and origins of cancer cells are needed to confirm or refute our findings.
In the current meta-analysis, significant heterogeneity among recruited publications was detected (Table 3 and Figure 3). In general, the sources of heterogeneity included ethnicity, the cancer system, the origin of the cancer cells, sample size and publication year. We performed stratified analyses according to ethnicity, the cancer system and the origin of the cancer cells. In some subgroups, heterogeneity was significantly reduced, suggesting the different influences of these factors, even in the same polymorphism. Further subgroup analyses were conducted based on other factors, such as sample size and publication year (Table 3). The pooled subgroup analysis of a subset of large sample size (≥ 1000 subjects) design and publication year after 2009 suggested an association with more noteworthy heterogeneity. According to the Galbraith radial plot (Figure 7) and the forest plot (Figure 3), four major outliers were detected [7,8,13,17]. Reviewing these publications, they involved certain deficiencies, for example, one was a small sample size design [17] and the cervical cancer cases of the other study were selected from families with at least two affected women [13]. Begg’s funnel plots and Egger’s tests were used to explore publication bias and no significant publication bias was observed in the meta-analysis. Nonparametric “trim-and-fill” method and one-way sensitivity analysis were both used to conduct sensitivity analyses (Figures 5 and 6) and the results suggested that our findings were robust.
Although considerable efforts were made to detect the possible association between CD28 IVS3 +17T>C polymorphism and cancer susceptibility, there are certain limitations inherited from this meta-analysis that should be acknowledged. Large heterogeneity was detected in the allele genetic model and dominant genetic model, which means these results should be interpreted with caution. Additionally, only 11 published investigations were included in our work; therefore, unpublished studies, if any, might inevitably be missed and lead to bias. Finally, we only focused on IVS3 +17T>C polymorphism in CD28, and did not explore other susceptibility genes or polymorphisms.
In summary, despite its limitations, this meta-analysis suggests the CD28 IVS3 +17T>C polymorphism represents a low risk factor for Asian populations. In the future, further extensive studies with larger sample sizes and more types of cancer should be performed to confirm the influence of CD28 IVS3 +17T>C polymorphism on cancer susceptibility.
Acknowledgements
This study was supported in part by Jiangsu University Clinical Medicine Science and Technology Development Fund (JLY20140012), National Natural Science Foundation of China (81472332, 81341006), Fujian Province Natural Science Foundation (2013J01126, 2013J05116), Fujian Medical University professor fund (JS12008) and Fujian Province science and technology programmed fund (2012Y0030).
Disclosure of conflict of interest
None.
Abbreviations
- CI
confidence interval
- OR
odds ratio
- HWE
Hardy-Weinberg equilibrium
- NK
natural killer
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Ihsan R, Chauhan PS, Mishra AK, Yadav DS, Kaushal M, Sharma JD, Zomawia E, Verma Y, Kapur S, Saxena S. Multiple analytical approaches reveal distinct gene-environment interactions in smokers and non smokers in lung cancer. PLoS One. 2011;6:e29431. doi: 10.1371/journal.pone.0029431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Wu C, Wang Y, Zhong R, Wang F, Zhang X, Duan S, Lou J, Yu D, Tan W, Yuan J, Wu T, Nie S, Miao X, Lin D. Association of candidate genetic variations with gastric cardia adenocarcinoma in Chinese population: a multiple interaction analysis. Carcinogenesis. 2011;32:336–342. doi: 10.1093/carcin/bgq264. [DOI] [PubMed] [Google Scholar]
- 4.Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 5.Qi P, Ruan CP, Wang H, Zhou FG, Xu XY, Gu X, Zhao YP, Dou TH, Gao CF. CTLA-4 +49A>G polymorphism is associated with the risk but not with the progression of colorectal cancer in Chinese. Int J Colorectal Dis. 2010;25:39–45. doi: 10.1007/s00384-009-0806-z. [DOI] [PubMed] [Google Scholar]
- 6.Allison JP, Hurwitz AA, Leach DR. Manipulation of costimulatory signals to enhance antitumor T-cell responses. Curr Opin Immunol. 1995;7:682–686. doi: 10.1016/0952-7915(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Li H, Qiao Y, Yu D, Guo H, Tan W, Lin D. Association of CD28 gene polymorphism with cervical cancer risk in a Chinese population. Int J Immunogenet. 2011;38:51–54. doi: 10.1111/j.1744-313X.2010.00969.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Zhang Q, Shen L, Liu Y, Xu F, Li D, Fu Z, Yuan W, Pang D, Li D. Investigation of CD28 gene polymorphisms in patients with sporadic breast cancer in a Chinese Han population in Northeast China. PLoS One. 2012;7:e48031. doi: 10.1371/journal.pone.0048031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 11.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 12.Karabon L, Pawlak E, Tomkiewicz A, Jedynak A, Passowicz-Muszynska E, Zajda K, Jonkisz A, Jankowska R, Krzakowski M, Frydecka I. CTLA-4, CD28, and ICOS gene polymorphism associations with non-small-cell lung cancer. Hum Immunol. 2011;72:947–954. doi: 10.1016/j.humimm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecol Oncol. 2010;116:544–548. doi: 10.1016/j.ygyno.2009.10.084. [DOI] [PubMed] [Google Scholar]
- 14.Pawlak E, Karabon L, Wlodarska-Polinska I, Jedynak A, Jonkisz A, Tomkiewicz A, Kornafel J, Stepien M, Ignatowicz A, Lebioda A, Dobosz T, Frydecka I. Influence of CTLA-4/CD28/ICOS gene polymorphisms on the susceptibility to cervical squamous cell carcinoma and stage of differentiation in the Polish population. Hum Immunol. 2010;71:195–200. doi: 10.1016/j.humimm.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Bouwhuis MG, Gast A, Figl A, Eggermont AM, Hemminki K, Schadendorf D, Kumar R. Polymorphisms in the CD28/CTLA4/ICOS genes: role in malignant melanoma susceptibility and prognosis? Cancer Immunol Immunother. 2010;59:303–312. doi: 10.1007/s00262-009-0751-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilmec F, Ozgonul A, Uzunkoy A, Akkafa F. Investigation of CTLA-4 and CD28 gene polymorphisms in a group of Turkish patients with colorectal cancer. Int J Immunogenet. 2008;35:317–321. doi: 10.1111/j.1744-313X.2008.00782.x. [DOI] [PubMed] [Google Scholar]
- 17.Suwalska K, Pawlak E, Karabon L, Tomkiewicz A, Dobosz T, Urbaniak-Kujda D, Kuliczkowski K, Wolowiec D, Jedynak A, Frydecka I. Association studies of CTLA-4, CD28, and ICOS gene polymorphisms with B-cell chronic lymphocytic leukemia in the Polish population. Hum Immunol. 2008;69:193–201. doi: 10.1016/j.humimm.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Cheng TY, Lin JT, Chen LT, Shun CT, Wang HP, Lin MT, Wang TE, Cheng AL, Wu MS. Association of T-cell regulatory gene polymorphisms with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. J. Clin. Oncol. 2006;24:3483–3489. doi: 10.1200/JCO.2005.05.5434. [DOI] [PubMed] [Google Scholar]
- 19.Karabon L PE, Tomkiewicz A, Kielbinski M, Potoczek S, Woszczyk D, Jonkisz A, Kuliczkowski K, Frydecka I. Lack of association between CD28 gene polymorphism and multiple myeloma in a Polish population. Adv Clin Exp Med. 2009;18:129–133. [Google Scholar]
- 20.Wlodarska-Polinska I PE, Suwalska K, Dobosz T, Potoczek S, Kornafel J, Frydecka I. Lack of association between CD28 gene polymorphism and cervical cancer in lower silesian population. Adv Clin Exp Med. 2006;15:595–598. [Google Scholar]


