Abstract
The aim of this study was to evaluate the association between CTLA-4 +49 G>A polymorphism and esophageal cancer (EC) susceptibility in a hospital based case-control study and a subsequent meta-analysis. We implemented genotyping analyses for CTLA-4 +49 G>A polymorphism with 629 esophageal squamous cell carcinoma cases and 686 controls in a Chinese Han population. Polymerase chain reaction ligase detection reaction (PCR-LDR) method was used to identify genotypes of CTLA-4 +49 G>A polymorphism. We first assessed the association between CTLA-4 +49 G>A polymorphism and EC risk in a hospital based case-control study, and then performed a comprehensive meta-analysis to derive a more precise estimation. Our results demonstrated that CTLA-4 +49 G>A polymorphism was not associated with EC risk. This case-control study and further meta-analysis, failed to identify the association between CTLA-4 +49 G>A polymorphism and EC risk. And additional, further well designed studies with large sample sizes and detailed gene-environment data are required.
Keywords: CTLA4, polymorphism, esophageal cancer, susceptibility, meta-analysis
Introduction
Esophageal cancer (EC) is the sixth most common cancer with an estimated 482,300 new cases and more than eighty percent death rate occurred worldwide in 2008 [1]. In 2009, the incidence rate of EC was 22.14 per 10,000 in China [1,2]. Every year, there are about 250,000 new EC cases diagnosed in China, accounting for half of the global cases [3]. The death rate for EC patients is very high and the 5 years survival rate accounts only 12.3% [4]. In the highest risk area, such as Iran and China, the most frequent subtype of EC is esophageal squamous cell carcinoma (ESCC) and counts more than 90% [5]. However, the etiology of EC is still indistinct. Accumulating evidence suggests genetic components, environmental factors, and gene-environment interactions play vital roles in EC development and progression [6-10]. Recently, several studies have focused on the role of the immune system to explore the etiology of EC [11,12].
Cytotoxic T-lymphocyte antigen 4 (CTLA-4) is one of the most important members of the immunoglobulin superfamily. CTLA-4, a vital restraining regulator of T-cell proliferation and activation, induces Fas-independent apoptosis of activated T cells [11,13]. It suggests that CTLA-4 plays an important role in carcinogenesis. CTLA-4 gene is located on chromosome 2q33 and is composed of four exons that possess several vital single nucleotide polymorphisms (SNPs), such as the +49 G>A, -318 C>T, +6230 G>A (CT60), and -1722 T>C, etc. [11,14]. A meta-analysis conducted by Zhang et al. demonstrated that CTLA-4 +49 G>A polymorphism may be a risk factor for cancer, whereas in that analysis, only one case-control study conducted on EC [15]. However, up to now, three investigations [11,12,16] focused on the association of the CTLA-4 +49 G>A polymorphism with EC risk, and a definitive conclusion remained elusive. To further investigate this potential relationship, we first assessed the association between CTLA-4 +49 G>A polymorphism and ESCC risk in a hospital based case-control study, and then conducted a comprehensive meta-analysis to derive a more precise estimation.
Materials and methods
Subjects
A total of 629 esophageal squamous cell carcinoma (ESCC) patients and 686 cancer-free controls were consecutively recruited from the Affiliated People’s Hospital of Jiangsu University and Affiliated Hospital of Jiangsu University (Jiangsu Province, China) between October 2008 and December 2010. All patients were confirmed by postoperative pathologic means. In this study, the patients who had a history of cancer or autoimmune diseases before, or had undergone radiotherapy or chemotherapy were excluded. Ethnicity (Chinese), frequency of sex, and average age (±5 years) of the controls were well matched to patients. At recruitment, this investigation was approved by the Institutional Review Board of Jiangsu University (Zhenjiang, China) and each subject signed the written informed consent. Experienced doctors were assigned to administer a structured questionnaire and information of all subjects, such as demographic data (e.g. age, gender) and risk factors (including tobacco use and alcohol consumption), were collected. After completed the in-person interview, each individual donated 2-ml peripheral venous blood. The “smokers” criterion was subjects who smoked more than one cigarette per day over one year, and the “alcohol drinkers” criterion was those who consumed ≥3 alcoholic drinks a week for >6 months.
DNA extraction, SNP selection, and genotyping
Ethylenediamine tetra-acetic acid (EDTA)-anticoagulated peripheral venous blood samples were collected by using Vacutainers (BD Franklin Lakes NJ, USA). The QIAamp DNA Blood Mini Kit (Qiagen, Berlin, Germany) was used to isolate genomic DNA from peripheral blood lymphocytes and DNA samples were frozen at -80°C. Genotypes of CTLA-4 +49 G>A site were analyzed by using polymerase chain reaction ligase detection reaction (PCR-LDR) method [17,18]. Shanghai Biowing Applied Biotechnology Company provided the technical support. For quality control, one hundred and sixty samples were randomly selected and reciprocally tested with high DNA quality, and the reproducibility rate of was 100%.
Statistical analysis
Chi-square test (x2) was performed to test the differences in the distributions of demographic characteristics, selected variables and genotypes between patients and controls. The association of CTLA-4 +49 G>A genotypes with the risk of ESCC was evaluated by odds ratios (ORs) and corresponding 95% confidence intervals (CIs) using logistic regression analyses for crude ORs and adjusted ORs when it was appropriate. An internet-based Hardy-Weinberg equilibrium (HWE) calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) was used to measure the deviation from the HWE among the controls. Statistical analysis was implemented by SAS 9.1.3 software (SAS Institute, Cary, NC). Statistical significance was defined as P<0.05 with two-tailed for all statistical analyses.
Meta-analysis
The meta-analysis is reported on the basis of the Preferred Reporting Items for Meta-analyses (PRISMA) guideline (Table S1. PRISMA checklist) [19].
Embase, PubMed and CBM (Chinese BioMedical Disc), as well as CNKI (China National Knowledge Infrastructure) database were searched up to September 23th, 2014 for publications investigating the association of CTLA-4 +49 G>A polymorphism with EC. The combination terms were ‘esophageal’ or ‘esophagus’ and ‘cancer’ or ‘tumor’ or ‘carcinoma’ or ‘neoplasm’ or ‘malignance’ and ‘Cytotoxic T-lymphocyte antigen 4’ or ’CTLA-4’ or ’CD152’, annexed with ‘SNP’ or ’mutation’, ‘variant’ or ’polymorphism’. Additionally, the publication language was restricted to English and Chinese, and all studies carried out in human subjects were identified. The results of electronic retrieval were supplemented by manual search of all bibliographies listed in these studies or published reviews. The major included criteria were: (a) designed as case-control study, (b) evaluated the CTLA-4 +49 G>A polymorphism and EC risk, (c) provided genotype counts of CTLA-4 +49 G>A polymorphism between cases and controls. The major excluded criteria included the following: (a) not case-control study, (b) review publications and (c) overlapping data.
In this meta-analysis, the crude OR with the corresponding 95% CI was used to evaluate the strength of association between CTLA-4 +49 G>A polymorphism and EC risk. The Z-test and P-value (two-tailed) was used to measure the significance of pooled OR, and statistical significance was defined as P<0.05 (two-tailed). Heterogeneity among studies was evaluated by a Chi-square-based I2 test. If I2>50% or P<0.10, the pooled ORs were calculated by the random-effects model (the DerSimonian-Laird method) [20], otherwise the fixed-effects model was performed (the Mantel-Haenszel method) [21]. The funnel plot and Egger’s test were implemented to evaluate publication bias, which was measured by visual inspection of an asymmetric plot [22]. For the funnel plot, Egger’s test and the I2, statistical significance was considered at P<0.1. Sensitivity analysis was performed to determine whether any excluded studies affected the stability of our results. Galbraith radial plot was used to analyze the heterogeneity [23]. In current meta-analysis, all statistical analyses were performed by STATA software (version 12.0).
Results
Baseline characteristics
Characteristic of all subjects, such as the demographics and risk factors, are presented in Table 1. The terms of age and sex distributions were no significant differences between cases and controls (P=0.155 and P=0.185, respectively), which indicated that these factors were well matched. However, the results indicated that significant difference was found on drinking status and smoking rate between cases and controls (P<0.001). The primary information of CTLA-4 +49 G>A polymorphism is included in Table 2. For this SNP, the genotyping success rate was 96.43% in all samples. The minor allele frequency (MAF) of controls was similar to data for Han populations in Chinese database (Table 2). The genotypic frequencies for CTLA-4 +49 G>A polymorphism among controls were in HWE (P=0.284) (Table 2).
Table 1.
Distribution of selected demographic variables and risk factors in ESCC cases and controls
| Variable | Cases (n=629) | Controls (n=686) | P a | ||
|---|---|---|---|---|---|
|
|
|
||||
| n | % | n | % | ||
| Age (years) mean ± SD | 62.85 (±8.13) | 62.58 (±7.89) | 0.541 | ||
| Age (years) | 0.155 | ||||
| <63 | 310 | 49.28 | 365 | 53.21 | |
| ≥63 | 319 | 50.72 | 321 | 46.79 | |
| Sex | 0.185 | ||||
| Male | 444 | 70.59 | 461 | 67.20 | |
| Female | 185 | 29.41 | 225 | 32.80 | |
| Tobacco use | <0.001 | ||||
| Never | 355 | 56.44 | 499 | 72.74 | |
| Ever | 274 | 43.56 | 187 | 27.26 | |
| Alcohol use | <0.001 | ||||
| Never | 428 | 68.04 | 526 | 76.68 | |
| Ever | 201 | 31.96 | 160 | 23.32 | |
Two-sided χ2 test and student t test;
Bold values are statistically significant (P<0.05).
Table 2.
Primary information for CTLA4 rs231775 G>A polymorphism
| Genotyped SNPs | CTLA4 rs231775 G>A |
|---|---|
| Chromosome | 2 |
| Function | missense |
| Chr Pos (Genome Build 36.3) | 204440959 |
| Regulome DB Scorea | No Data |
| Splicing (ESE or ESS) | Y |
| nsSNP | Y |
| MAFb for Chinese in database | 0.314 |
| MAF in our controls (n=686) | 0.310 |
| P value for HWEc test in our controls | 0.284 |
| Genotyping methodd | LDR |
| % Genotyping value | 96.43% |
http://www.regulomedb.org/;
MAF: minor allele frequency;
HWE: Hardy-Weinberg equilibrium;
LDR: ligation detection reaction.
CTLA4 +49 G>A polymorphism and risk of ESCC
The genotype distributions of CTLA-4 +49 G>A were presented in Table 3. In the single locus analysis, the genotype frequencies of CTLA-4 +49 G>A were 50.83% (GG), 42.05% (GA) and 7.12% (AA) in cases, and 46.69% (GG), 44.58% (GA) and 8.73% (AA) in controls, and the difference was not statistically significant (P=0.270). When the CTLA-4 +49 GG homozygote genotype was used as the reference group, the GA genotype was not associated with the risk of ESCC (GA vs. GG: OR 0.87, 95% CI 0.69-1.09, P=0.223). When the CTLA-4 +49 GG homozygote genotype was used as the reference group, the AA genotype was not associated with the risk of ESCC (AA vs. GG: OR 0.75, 95% CI 0.49-1.15, P=0.182). In the recessive model, when the CTLA-4 +49 GG/GA genotypes were used as the reference group, the AA homozygote genotype was not associated with the risk of ESCC (OR 0.80, 95% CI 0.53-1.21, P=0.289). In the dominant model, the CTLA-4 +49 AA/GA variants were not associated with the risk of ESCC, compared with the CTLA-4 +49 GG genotype (AA/GA vs. GG: OR 0.85, 95% CI 0.68-1.06, P=0.141). When the CTLA-4 +49 G allele was used as the reference group, the A allele was not associated with the risk of ESCC (A vs. G: OR 0.87, 95% CI 0.73-1.03, P=0.114) (Table 3). After adjusting for age, gender, smoking and drinking status, no statistically increased or decreased risk of ESCC was observed in all genetic models (Table 3).
Table 3.
Logistic regression analyses of associations between CTLA4 rs231775 G>A, polymorphism and risk of ESCC
| Genotype | Cases (n=629) | Controls (n=686) | Crude OR (95% CI) | P | Adjusted ORa (95% CI) | P | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| n | % | n | % | |||||
| CTLA4 rs231775 G>A | ||||||||
| GG | 307 | 50.83 | 310 | 46.69 | 1.00 | 1.00 | ||
| GA | 254 | 42.05 | 296 | 44.58 | 0.87 (0.69-1.09) | 0.223 | 0.85 (0.68-1.08) | 0.189 |
| AA | 43 | 7.12 | 58 | 8.73 | 0.75 (0.49-1.15) | 0.182 | 0.70 (0.45-1.07) | 0.100 |
| GA+AA | 297 | 49.17 | 354 | 53.31 | 0.85 (0.68-1.06) | 0.141 | 0.83 (0.66-1.04) | 0.100 |
| GG+GA | 561 | 92.88 | 606 | 91.27 | 1.00 | 1.00 | ||
| AA | 43 | 7.12 | 58 | 8.73 | 0.80 (0.53-1.21) | 0.289 | 0.75 (0.49-1.14) | 0.177 |
| G allele | 868 | 71.85 | 916 | 68.98 | 1.00 | |||
| A allele | 340 | 28.15 | 412 | 31.02 | 0.87 (0.73-1.03) | 0.114 | ||
Adjusted for age, sex, smoking and drinking status.
Eligible articles for meta-analysis
The initial search yielded a total of 30 potentially relevant articles. After applying additional filters, three case-control studies and our study were eligible for inclusion. The detailed process is presented in Figure 1.
Figure 1.
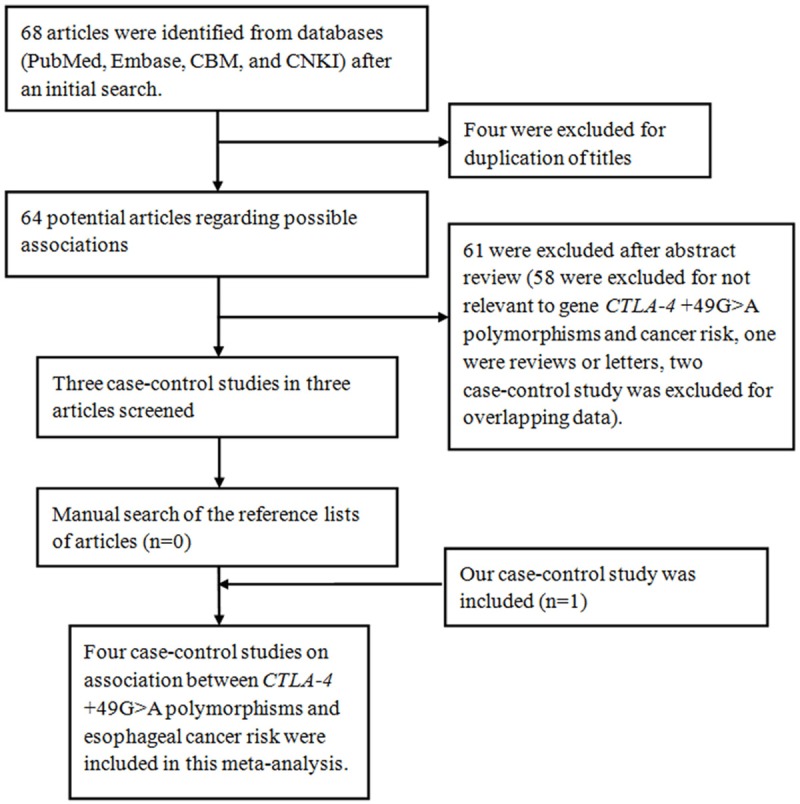
Flow diagram of articles selection process for CTLA-4 +49G/A (rs231775 G>A) polymorphism and esophageal cancer risk meta-analysis.
Study characteristics
In total, three previous studies plus our case-control study involving a total of 1969 EC cases and 2149 controls were recruited in this meta-analysis. As for subjects in these studies, all were Asians. Characteristics of all included study are presented in Table 4. The detailed distribution of the CTLA-4 +49 G>A polymorphism and allele among cases and controls is presented in Table 4.
Table 4.
Characteristics of populations and cancer types of the individual studies included in the meta-analysis
| Study | Year | Ethnicity | Country | Sample size (case/control) | Histologic subtype | Genotype method | Case | Control | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||
| GG | GA | AA | GG | GA | AA | G | A | G | A | ||||||||
| Cheng et al. | 2011 | Asians | China | 205/205 | ESCC | PCR-RFLP | 54 | 105 | 46 | 90 | 79 | 36 | 213 | 197 | 259 | 151 | no |
| Cai et al. | 2011 | Asians | China | 125/250 | ESCC | PCR-RFLP | 30 | 68 | 27 | 70 | 133 | 47 | 128 | 122 | 273 | 227 | Yes |
| Sun et al. | 2008 | Asians | China | 1010/1008 | ESCC | PCR-RFLP | 448 | 434 | 128 | 529 | 406 | 73 | 1330 | 690 | 1464 | 552 | Yes |
| Our study | 2013 | Asians | China | 629/686 | ESCC | PCR-LDR | 307 | 254 | 43 | 310 | 296 | 58 | 868 | 340 | 916 | 412 | Yes |
ESCC, esophageal squamous cell carcinoma. PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism. PCR-LDR, polymerase chain reaction-ligase detection reaction. HWE, Hardy-Weinberg equilibrium.
Meta-analysis results
After combining all recruited studies, a total of 1969 EC cases and 2149 controls from four investigations were included for meta-analysis of the association between CTLA-4 +49 G>A polymorphism and EC risk and the results indicated that there was null association (Table 5 and Figure 2). Among the four case-control studies, there was one case-control study deviated from HWE [12], after we excluded it and then obtained another result. However, this result was in accordance with the previous one (data not shown).
Table 5.
Summary of results of the meta-analysis from different comparative genetic models
| Polymorphism | Genetic comparison | OR (95%CI) | P | Test of heterogeneity | Model | |
|---|---|---|---|---|---|---|
|
| ||||||
| p-Value | I2 | |||||
| AA+GA vs. GG | 1.31 (0.90-1.89) | 0.160 | 0.000 | 85.1% | R | |
| AA vs. GA+GG | 1.26 (0.85-1.88) | 0.251 | 0.013 | 72.1% | R | |
| CTLA4 rs231775 G>A | AA vs. GG | 1.45 (0.85-2.47) | 0.171 | 0.001 | 81.2% | R |
| GA vs. GG | 1.26 (0.90-1.77) | 0.181 | 0.002 | 80.1% | R | |
| AA vs. GA | 1.15 (0.82-1.60) | 0.419 | 0.082 | 55.2% | R | |
| A vs. G | 1.21 (0.92-1.59) | 0.179 | 0.000 | 86.0% | R | |
R indicates random-effects model.
Figure 2.
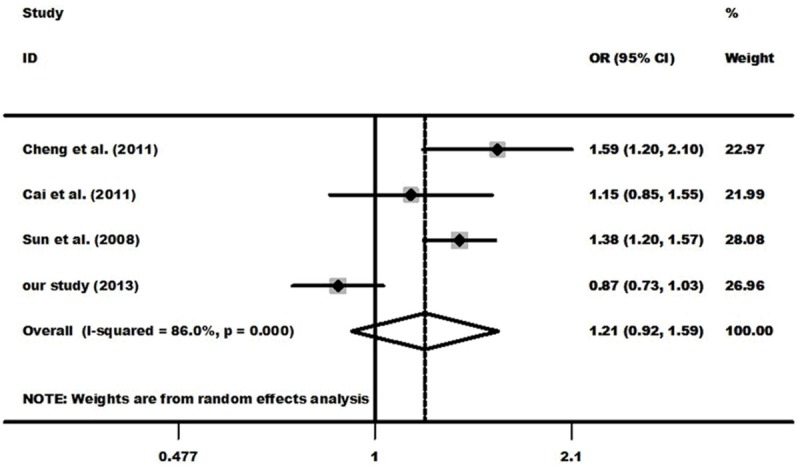
Meta-analysis with a random-effects model for the association between the risk of esophageal cancer and the CTLA-4 +49 G>A polymorphism (A vs. G).
In this meta-analysis, Begg’s Funnel plot and Egger’s test was created to detect potential publication bias (Figure 3). The shape of funnel was symmetry in all genetic models, suggesting that there were no publication bias in this meta-analysis (A vs. G: Begg’s test P=0.734, Egger’s test P=0.970; AA vs. GG: Begg’s test P=0.734, Egger’s test P=0.750; GA vs. GG: Begg’s test P=1.000, Egger’s test P=0.596; AA vs. GA: Begg’s test P=1.000, Egger’s test P=0.255; AA+GA vs. GG: Begg’s test P=1.000, Egger’s test P=0.725; AA vs. GG+GA: Begg’s test P=0.734, Egger’s test P=0.416).
Figure 3.

Begg’s funnel plot of meta-analysis of between the CTLA-4 +49 G>A polymorphism and the risk of cancer in the dominant model (random-effects model).
Sensitivity analyses were performed to evaluate the influence of each individual dataset on the pooled OR by omitting each dataset in turn. The results did not alter when any individual study was deleted, confirming the stability of our results (Figure 4).
Figure 4.
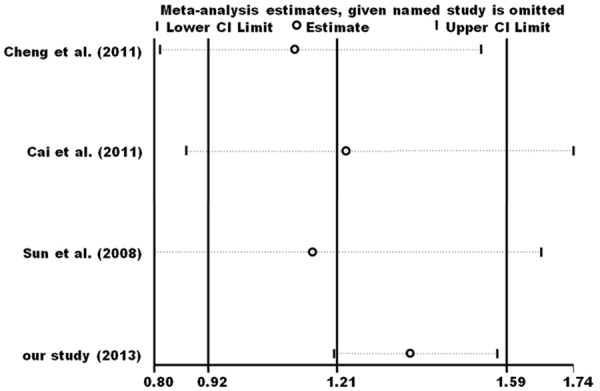
Sensitivity analysis of the influence of A vs. G in meta-analysis (random-effects estimates).
The results indicated there were large heterogeneities among four case-control studies enrolled. As shown in Table 5, heterogeneity was significant in allele genetic model. Galbraith radial plot was used to analyze the source of heterogeneity (Figure 5). The result identified one outlier which might contribute to the major source of heterogeneity. From the forest plot in allele genetic model (Figure 2), we can identify that one study which conducted by Cheng et al. [12] was the main source of heterogeneity.
Figure 5.
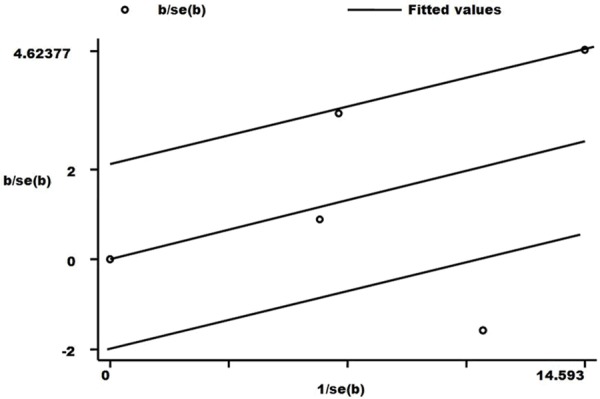
Galbraith radial plot of meta-analysis (A vs. G compare genetic model).
Discussion
Recently, the association between CTLA-4 +49 G>A polymorphism and EC risk have been investigated in several studies and a decisive answer is lacking. In this study, a hospital based case-control study in Chinese Han population, along with a meta-analysis on EC, attempted to derive a more precise evaluation and the results were remain non-significance. To the best of our knowledge, this is the first meta-analysis exploring the association of CTLA-4 +49 G>A polymorphism with EC risk.
With a growing interest in the associations of genetic polymorphisms and EC risk, several studies have examined the hypothesis that CTLA-4 +49 G>A polymorphism is relevant to the risk of EC; however, the results remain inconsistent and ambiguous. Considering the fact that most common SNPs usually make low cancer susceptibility, this meta-analysis recruits four case-control studies with relatively large sample sizes to get a more precise assessment. Two studies have reported positive signal of CTLA-4 +49 G>A polymorphism with EC risk [11,12]; the other individual study has reported negative signal [16]; however, as showed in the results of current meta-analysis among 4118 subjects, there were non-significance. Although there was one case-control study deviated from HWE [12], we excluded it or recruited it, this result was similar, suggesting our results were stable. The results should be interpreted with very caution. Considering only four case-control studies were conducted in EC and two studies were relatively small sample sizes, which might generate a fluctuated assessment or restrict the power to confirm a real influence. It was also possible that the real function of CTLA-4 +49 G>A polymorphism was covered or diluted by other genetic or environment factors, and these vital factors should not be ignored. In the future, well designed investigations with large sample sizes should be carried out to verify these results.
Some merit should be addressed in current study. First, this is a case-control study with large sample sizes in detecting the association of CTLA-4 +49 G>A polymorphism with EC risk and the meta-analysis is the first synthesis investigating the association Second, the results of our case-control study confirmed that of current meta-analysis. Third, in our case-control study, the genotype frequencies in the controls were in HWE, suggesting our results were less prone to selection bias, publication bias tests indicated that there was no bias in this meta-analysis.
Several limitations in this study should be acknowledged. First, in case-control study, all subjects were recruited from two hospitals and might not fully represent the general Chinese populations. Second, only published studies in four databases were recruited in this meta-analysis, publication bias might have occurred. Third, large heterogeneity was observed in our meta-analysis, which means the results should be interpreted with very caution. Fourth, due to lack of uniform background information for recruited investigations, data were not further stratified by other factors (such as, age, gender, smoking, alcohol consumption, ethnicity and other lifestyle factors). Fifth, in this study, we only focused on CTLA-4 +49 G>A polymorphism, and did not consider other polymorphisms in CTLA-4 or other susceptibility genes.
In summary, this case-control study and subsequent meta-analysis failed to confirm the association between CTLA-4 +49 G>A polymorphism and EC risk. Nevertheless, for practical reasons, further well designed studies with large sample sizes and detailed gene-environment data, should be performed to confirm or refute these results.
Acknowledgements
This study was supported in part by Jiangsu University Clinical Medicine Science and Technology Development Fund (JLY20140012), National Natural Science Foundation of China (81472332, 81341006), Fujian Province Natural Science Foundation (2013J01126, 2013J05116), Fujian Medical University professor fund (JS12008) and Fujian Province science and technology programmed fund (2012Y0030).
Disclosure of conflict of interest
None.
Abbreviations
- CI
confidence interval
- OR
odds ratio
- CTLA4
cytotoxic T-lymphocyte antigen 4
- HWE
Hardy-Weinberg equilibrium
- ESCC
esophageal squamous cell carcinoma
- PCR-LDR
polymerase chain reaction ligase detection reaction
- SNP
single nucleotide polymorphism
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res. 2013;25:10–21. doi: 10.3978/j.issn.1000-9604.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, He Y, Zheng R, Zhang S, Zeng H, Zou X, He J. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis. 2013;5:19–26. doi: 10.3978/j.issn.2072-1439.2013.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrino F, De Angelis R, Sant M, Rosso S, Bielska-Lasota M, Coebergh JW, Santaquilani M EUROCARE Working group. Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol. 2007;8:773–783. doi: 10.1016/S1470-2045(07)70245-0. [DOI] [PubMed] [Google Scholar]
- 5.Gholipour C, Shalchi RA, Abbasi M. A histopathological study of esophageal cancer on the western side of the Caspian littoral from 1994 to 2003. Dis Esophagus. 2008;21:322–327. doi: 10.1111/j.1442-2050.2007.00776.x. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Luo GJ, Zhang L, Shi J, Zhang DQ, Chen JM, Chen XB, Li ZD, Zhao Q. Interaction between alcohol consumption and CYP 2C19 gene polymorphism in relation to oesophageal squamous cell carcinoma. PLoS One. 2012;7:e43412. doi: 10.1371/journal.pone.0043412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar J, Dominguez E, Li G, Kusewitt DF, Johnson DG. Modeling gene-environment interactions in oral cavity and esophageal cancers demonstrates a role for the p53 R72P polymorphism in modulating susceptibility. Mol Carcinog. 2014;53:648–58. doi: 10.1002/mc.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y, Hu Z, He Z, Jia W, Abnet CC, Liang L, Hu N, Miao X, Zhou Y, Liu Z, Zhan Q, Liu Y, Qiao Y, Zhou Y, Jin G, Guo C, Lu C, Yang H, Fu J, Yu D, Freedman ND, Ding T, Tan W, Goldstein AM, Wu T, Shen H, Ke Y, Zeng Y, Chanock SJ, Taylor PR, Lin D. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012;44:1090–1097. doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- 9.Jain M, Kumar S, Rastogi N, Lal P, Ghoshal UC, Tiwari A, Pant MC, Baiq MQ, Mittal B. GSTT1, GSTM1 and GSTP1 genetic polymorphisms and interaction with tobacco, alcohol and occupational exposure in esophageal cancer patients from North India. Cancer Lett. 2006;242:60–67. doi: 10.1016/j.canlet.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 10.Talukdar FR, Ghosh SK, Laskar RS, Mondal R. Epigenetic, genetic and environmental interactions in esophageal squamous cell carcinoma from northeast India. PLoS One. 2013;8:e60996. doi: 10.1371/journal.pone.0060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun T, Zhou Y, Yang M, Hu Z, Tan W, Han X, Shi Y, Yao J, Guo Y, Yu D, Tian T, Zhou X, Shen H, Lin D. Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res. 2008;68:7025–7034. doi: 10.1158/0008-5472.CAN-08-0806. [DOI] [PubMed] [Google Scholar]
- 12.Cheng XL, Ning T, Xu CQ, Li Y, Zhu BS, Hou FT, Zhang SY, Chen ZP. Haplotype analysis of CTLA4 gene and risk of esophageal squamous cell carcinoma in Anyang area of China. Hepatogastroenterology. 2011;58:432–437. [PubMed] [Google Scholar]
- 13.Scheipers P, Reiser H. Fas-independent death of activated CD4(+) T lymphocytes induced by CTLA-4 crosslinking. Proc Natl Acad Sci U S A. 1998;95:10083–10088. doi: 10.1073/pnas.95.17.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Zhang J, Deng Y, Tian C, Li X, Huang J, Fan H. Polymorphisms in the cytotoxic T-lymphocyte antigen 4 gene and cancer risk: a meta-analysis. Cancer. 2011;117:4312–4324. doi: 10.1002/cncr.25979. [DOI] [PubMed] [Google Scholar]
- 16.Cai J. The screening value of immune-related gene and metabolic-related gene detection for Kazakh’s esophageal cancer in xinjiang: Shihezhi University. 2011:53. [Google Scholar]
- 17.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Y, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Y, Ma J, Zhao Y. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 18.Yin J, Wang X, Zheng L, Shi Y, Wang L, Shao A, Tang W, Ding G, Liu C, Liu R, Chen S, Gu H. rs4938723 T>C and rs6505162 C>A Polymorphisms Are Associated with the Risk of Esophageal Cancer in a Chinese Population. PLoS One. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269. W264. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 20.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 21.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 22.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Mou ZY, Zhai JX, Zong HX, Zhao XD. [Application of Stata software to test heterogeneity in meta-analysis method] . Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29:726–729. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


