Abstract
Hepatocellular carcinoma (HCC) is one of the most common primary malignant tumors of the liver worldwide; however, despite its significance, there is a lack of treatment methods and clear prognoses. MicroRNA-9 (miR-9) is known to play an important role in tumor tumorigenesis and progression. Talin-1, which plays a significant role in regulating the transmutation of carcinoma, has been demonstrated to be downregulated by miR-9 in epithelial ovarian cancer. In the present study, we researched the potential role of miR-9 in the prognosis of HCC. The expression levels of miR-9 and Talin-1 mRNA in HCC tissues (n = 60), adjacent non-cancerous tissues (n = 60), and normal liver tissues (n = 20) were detected using a real-time quantitative assay; protein expression levels of Talin-1 were detected using western blot. The expression levels of miR-9 were significantly higher in HCC tissues (P < 0.001) than in normal liver and adjacent non-cancerous tissues. These levels were significantly associated with tumor grade, tumor size, portal vein tumor thrombus, integral capsule, and 2.0-year disease-free survival rate (P < 0.05). High levels of miR-9 were strongly associated with the malignant progression of HCC, and overexpression of miR-9 is a risk factor that has a statistically significant effect on survival rate. miR-9 could play a role as an HCC tumor activator by regulating the expression of Talin-1; therefore, miR-9 might be a potentially valuable biomarker for the prognosis in HCC patients.
Keywords: Hepatocellular carcinoma, HCC, microRNA-9, miR-9, Talin-1, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common primary malignant tumors of the liver, is the third largest cause of cancer-associated deaths after stomach and lung cancer, and is the sixth most common neoplasm worldwide [1,2]; however, HCC is associated with a poor prognosis and has finite treatment options [3]. Although a combination of chemotherapy and surgery would improve survival, HCC that is associated with malignant metastasis and migration, has an unusually poor prognosis [4]; therefore, several investigators have focused on the molecular and cellular mechanisms of HCC to promote the prognosis and diagnosis of this malignant tumor [5-8].
MicroRNAs (miRNAs) are small, non-coding RNAs that adjust posttranscriptional gene expression by matching to the 3’-untranslated region (3’-UTR) of target mRNAs, and regulate essential cellular processes, such as proliferation, diversification, and apoptosis [9,10]. The connection between alterative miRNA expression and cancerogenesis is well constructed as well as the progress of tumor [11-13]. In addition, the expression of miRNAs and special miRNAs have proved to be potential prognostic or diagnostic instruments in different types of tumors, such as non-small-cell lung cancer, breast tumors, and colorectal cancer [14-16]. Until now, it has been determined from study data that a series of miRNA-9 (miR-9) targets, such as nuclear factor (NF)-κB1, caudal type homeobox 2 (CDX2), chromobox protein homolog 7 (CBX7), and methenyltetrahydrofolate cyclohydrolase (MTHFD2), were associated with cancer [17]. Previous research has shown that miR-9 expression is downregulated in some types of cancers, including gastric, ovarian, and neuroblastoma [18-21]; however, the levels of miR-9 expression have proved to be upregulated in colorectal cancer, breast cancer, lung cancer, and laryngeal squamous cell carcinomas [22-26]. These data indicate that miR-9 might play a crucial role in tumor progression and tumorigenesis, and might also have different effects on multifarious carcinomas.
Talin-1, which has a molecular mass of 270 kDa, is a cytoskeletal protein that has been found to play a significant role in regulating the development and transmutation of carcinoma and the overexpression of Talin-1 are related to decreased migration and invasion in human liver cancer cell lines; the low expression of Talin-1 promotes migration and invasion [27]. In addition, Talin-1 is mainly expressed in the liver, spleen, stomach, lung, kidney, and vascular smooth muscle [28].
Some studies have demonstrated that miR-9 could downregulate the levels of Talin-1 as a tumor suppressor in epithelial ovarian cancer [17]. Nevertheless, until now, there is little usable research on the role of miR-9 expression and the relationship between miR-9 and Talin-1 in HCC tissues.
In the present study, the potential role of miR-9 in the prognosis of HCC was researched by detecting the levels of miR-9 expression in HCC tissues, adjacent non-cancerous tissues, and normal liver tissues. To observe the relationship between miR-9 and Talin-1, the mRNA and protein expression levels of Talin-1 in the above tissues were also examined. The results showed that the expression level of miR-9 was significantly elevated in HCC tissues when compared with that in normal liver and adjacent non-cancerous tissues. On the contrary, the mRNA and protein expression level of Talin-1 in HCC tissues was significantly lower than that in the normal liver and adjacent non-cancerous tissues. By researching the levels of miR-9 and Talin-1 mRNA, the correlation between miR-9 level and that of Talin-1 mRNA was observed to be inverse in HCC tissue. In addition, the levels of miR-9 expression were significantly associated with tumor grade, tumor size, integral capsule, portal vein tumor thrombus, and 2.0-year disease-free survival rate; therefore, miR-9 is potentially involved in the process of the infiltration, metastasis, and carcinogenesis of HCC by regulating the expression of Talin-1 and can become a valuable biomarker of tumors in a prognosis for HCC patients.
Materials and methods
Tissue samples
Our research was approved by the Experimental Ethics Committee of The First Affiliated Hospital of Anhui Medical University, China. Informed consent was received from all patients. From February 2012 to February 2013, 60 HCC tissues and corresponding adjacent non-cancerous tissues (> 2.0 cm from the tumor) were collected from the operating room of First Affiliated Hospital of Anhui Medical University. No patients had access to chemotherapy or radiotherapy before surgery. The nature of these HCC tissues and corresponding adjacent non-cancerous tissues were confirmed by pathological examination. The clinicopathological information from these HCC patients is shown in Table 1. There were 20 cases of normal liver specimens that were obtained from liver tissues with hemangiomas as controls. Part of the fresh specimens was used for western blot and the remaining specimen was frozen in liquid nitrogen and subsequently stored at -80°C for analysis using the quantitative real-time reverse-transcriptase-polymerase chain reaction (qRT-PCR). The clinical follow-up data from phone interviews with patients or their family members and outpatient medical records were matched. Two-year disease-free survival was defined as the period immediately following surgery to the date of recidivation or death. Patients were regarded as censored cases when they died of unrelated diseases.
Table 1.
Correlation of micorRNA-9 expression levels with clinicopathological features of patients with HCC
| Clinicopathological features | Number of cases | miR-9 expression | P value | |
|---|---|---|---|---|
|
|
||||
| High (n, %) | Low (n, %) | |||
| Age (y) | 1.000 | |||
| < 60 | 25 | 12 | 13 | |
| ≥ 60 | 35 | 18 | 17 | |
| Gender | 0.411 | |||
| Male | 40 | 22 | 18 | |
| Female | 20 | 8 | 12 | |
| Tumor size (cm) | 0.01 | |||
| < 5 | 28 | 9 | 19 | |
| ≥ 5 | 32 | 21 | 11 | |
| Number of tumor | 0.595 | |||
| Single | 37 | 17 | 20 | |
| Multiple | 23 | 13 | 10 | |
| Venous invasion | 0.001 | |||
| - | 34 | 10 | 24 | |
| + | 26 | 20 | 6 | |
| Tumor capsule | 0.034 | |||
| With integral | 23 | 7 | 16 | |
| Without integral | 37 | 23 | 14 | |
| HBsAg | 0.171 | |||
| - | 20 | 7 | 13 | |
| + | 40 | 23 | 17 | |
| AFP (μg/L) | 0.111 | |||
| < 400 | 37 | 22 | 15 | |
| ≥ 400 | 23 | 8 | 15 | |
| Admondson stage | < 0.001 | |||
| I-II | 35 | 8 | 27 | |
| III | 25 | 22 | 3 | |
Western blot analysis
Tissue samples were cut into pieces and subsequently ground into a corresponding slurry. The total protein content from the slurry was extracted using a cell lysis buffer for western blot and immunoprecipitation (Beyotime Institution of Biotechnology, Haimen, China) with 100 mM phenylmethanesulfonylfluoride. Total proteins were then quantified using the Enhanced BCA Protein Assay Kit (Beyotime Institution of Biotechnology, Haimen, China). The protein samples (30 µg) were separated in 6.0% SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes, which were blocked in 5.0% skim milk for 1.0 h at ambient temperatures, followed by incubation with mouse anti-Talin-1 monoclonal primary antibody (ab104913, 1:1000 dilution, Abcam, Cambridge, MA, USA) overnight at 4.0°C. The membranes were then incubated with the species-specific peroxidase-conjugated secondary antibodies, Goat anti-Mouse IgG (BA1050, 1:5000 dilution, BOSTER, Wuhan, China). The proteins of the immunoreaction were made visible using the Pro-light HRP Chemiluminescent Kit (TIANGEN Biotech, Beijing, China). In addition, the levels of objective protein were normalized to the expression levels of β-actin protein. The results of western blotting were analyzed using ImageJ (National Institutes of Health, Maryland, USA).
microRNA qRT-PCR assay
To determine the relationship between miR-9 and Talin-1 and the expression levels of miR-9 and Talin-1 mRNA in HCC tissues, corresponding adjacent non-cancerous tissues and normal liver tissues were detected using qRT-PCR assay. Briefly, total tissue RNA from samples was extracted with Total RNA Extractor (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. In addition, the total RNA samples were detected with NanoDrop2000 (Thermo Scientific, Waltham, MA, USA) to control the concentration and purification of the products. cDNA that was extracted from total RNA samples was reverse transcribed with miRNA-specific primers using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Subsequently, the products of the first-strand cDNA were amplified directly by PCR using SuperReal Premix plus (TIANGEN Biotech, Beijing, China). qRT-PCR was executed using specialized primers from the Genscript Custom Primers (Genscript Biotech, Nanjing, China) as follows:
Talin-1-F, 5’-TCTCCCAAAATGCCAAGAAC-3’; Talin-1-R, 5’-TGGCTATTGGGGTCAGAGAC-3’; miR-9-F, 5’-GTGCAGGGTCCGAGGT-3’; miR-9-R, 5’-GCGCTCTTTGGTTATCTAGC-3’; U6-F, 5’-CTCGCTTCGGCAGCACA-3’; U6-R, 5’-AACGCTTCACGAATTTGCGT-3’.
The relative expression levels of miR-9 and Talin-1 mRNA were normalized to small nucleolar RNA U6 and the cycle threshold (CT) was calculated. The relative amount of miR-9 and Talin-1 mRNA was quantified using the 2-ΔCT method (ΔCT = CT miR-9 or Talin-1 mRNA-CT U6 RNA). Each survey was performed in triplicate.
Statistical analyses
SPSS 19.0 (IBM Corporation, Armonk, NY, USA) was used to analyze all of the data. The mean ± standard deviation (SD) was applied to the expressing continuous variables. The analysis of variance and Student-Newman-Keul test were used to assess the differences in the expression levels of miR-9 and Talin-1 in HCC tissues, corresponding adjacent non-cancerous tissues, and normal liver tissues. The chi-square test was used for categorical variables to show their differences. Linear correlation analysis was used to show the inverse correlation between miR-9 and Talin-1 mRNA in HCC tissues. The Kaplan-Meier method and log-rank test were used to evaluate the differences in 2.0-year disease-free survival rate of HCC patients. The multivariate analyses of prognostic values were determined using Cox proportional hazards regression analysis, and the difference was considered statistically significant if P < 0.05.
Results
Elevated expression levels of miR-9 in hepatocellular carcinoma tissues
The expression levels of miR-9 in HCC tissues, adjacent non-cancerous tissues, and normal liver tissues were detected using qRT-PCR normalized to U6. The results showed that the levels of miR-9 expression were significantly elevated in HCC tissues compared with that of corresponding adjacent non-cancerous tissues (P < 0.001) and normal liver tissues (P < 0.001) (Figure 1). The correlative expression levels of miR-9 normalized to U6 in HCC tissues (6.955 ± 2.478) were observably higher than that in corresponding adjacent non-cancerous tissues (4.386 ± 1.784) and normal liver tissues (4.200 ± 1.848). In addition, there was no significant difference between the levels of miR-9 expression in normal liver tissue samples and that in adjacent non-cancerous tissue samples (P = 0.734). The median miR-9 expression levels in all 60 patients with HCC was 6.895. Subsequently, the patients with HCC were separated into two groups based on their levels of miR-9 expression using the median as a cutoff: high levels of miR-9 expression (n = 30, 8.816 ± 1.640) and low levels of miR-9 expression (n = 30, 5.093 ± 1.622).
Figure 1.
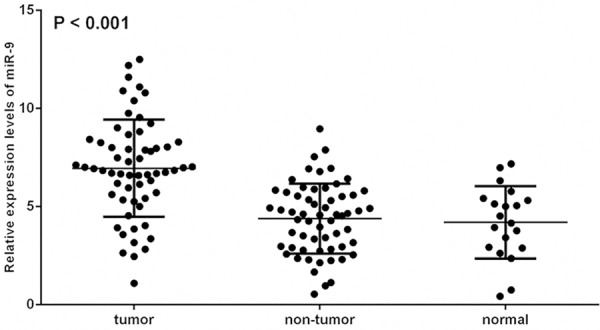
The relative expression of miR-9 in 60 pair of HCC tissues, corresponding adjacent non-cancerous tissues and 20 pairs of normal liver tissues. Compared to adjacent non-cancerous tissues and normal liver tissues, the levels of miR-9 expression in HHC tissues were significantly elevated (P < 0.001).
Depressed expression levels of Talin-1 in hepatocellular carcinoma tissues
The levels of Talin-1 protein expression in HCC tissues, corresponding adjacent non-cancerous tissues, and normal liver tissues were detected using western blot. The expression levels of Talin-1 mRNA in these tissues were also detected using qRT-PCR. The expression levels of Talin-1 protein in HCC tissues were significantly lower than that in the corresponding adjacent non-cancerous and normal liver tissues (P < 0.001). The results of the Talin-1 protein analysis are shown in Figure 3A and 3B. In addition, Figure 2 shows that the levels of Talin-1 mRNA in HCC tissues were also significantly lower than that in the adjacent non-cancerous tissues (P < 0.001) and normal control tissues (P = 0.002), and there was no significant difference between the expression levels of Talin-1 protein or Talin-1 mRNA (P = 0.798 and P = 0.702, respectively) in normal liver tissue samples and that in adjacent non-cancerous tissue samples.
Figure 3.
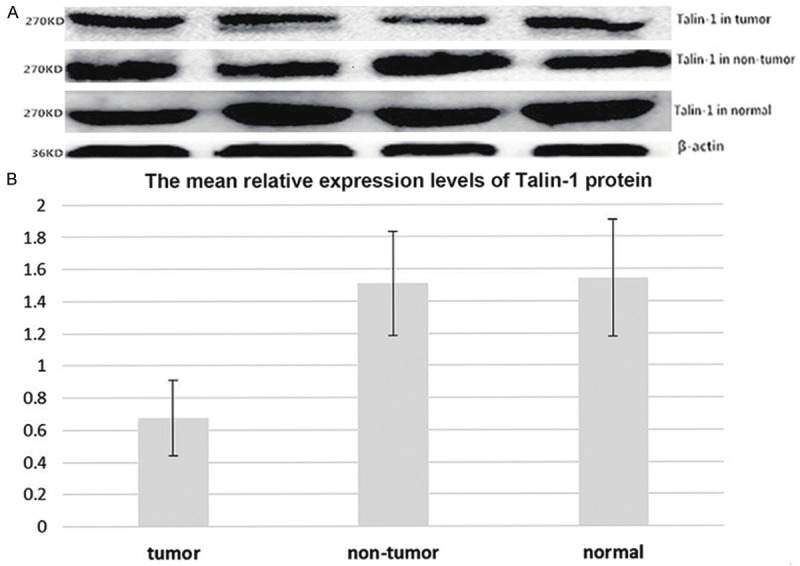
The expression levels of Talin-1 protein in HCC tissues, corresponding adjacent non-cancerous tissues and normal liver tissues.
Figure 2.
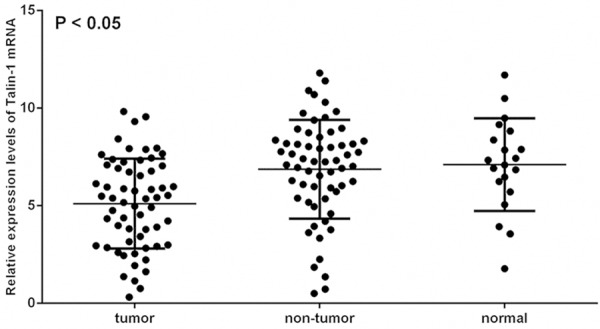
The relative expression levels of Talin-1 mRNA in 60 pair of HCC tissues, corresponding adjacent non-cancerous tissues and 20 pairs of normal liver tissues. Compared to adjacent non-cancerous tissues and normal liver tissues, the levels of Talin-1 mRNA expression in HHC tissues were significantly depressed (P < 0.05).
The relationship between miR-9 and Talin-1 in hepatocellular carcinoma tissues
The relationship between miR-9 and Talin-1 mRNA in HCC tissues was also investigated using qRT-PCR. Figure 1 shows that miR-9 expression levels in HCC tissues were elevated; however, the levels of Talin-1 mRNA expression in HCC tissues were reduced (Figure 2). After the above data were analyzed, it was found that the miR-9 expression levels and Talin-1 mRNA expression levels were observed by statistical analysis to be inversely related in HCC tissues (r = -0.337, P = 0.003) (Figure 4).
Figure 4.
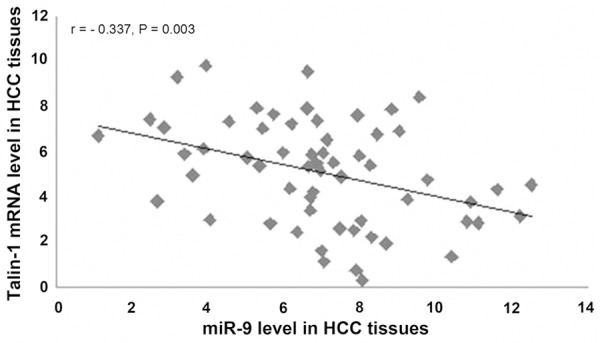
Linear regression analysis indicated a significant negative correlation between miR-9 and Talin-1 mRNA (r = -0.337, P = 0.003).
Correlation between clinicopathological features and miR-9 expression levels in hepatocellular carcinoma tissues
To further delineate the possible role of miR-9 in HCC, an investigation was designed to analyze the association between miR-9 expression and the clinicopathological features of patients with HCC. Table 1 summarizes the connections between miR-9 expression and the multifarious clinicopathological index of these patients with HCC; there was no significant statistical difference between the levels of miR-9 and patient age (P = 1.000), sex (P = 0.411), number of tumors (P = 0.595), the expression of alpha-fetoprotein (P = 0.111), and the hepatitis B antigen (P = 0.171); however, the expression levels of miR-9 were significantly associated with tumor size (P = 0.01), tumor grade (P < 0.001), integral capsule (P = 0.034), and portal vein tumor thrombus (P = 0.001).
The expression levels of miR-9 is a prognostic biomarker in patients with hepatocellular carcinoma correlated with 2.0-year disease-free survival rate
Statistical analysis was used to further investigate the correlation between the expression levels of miR-9 and 2.0-year disease-free survival rate to assess the potential role of miR-9 in the prognosis of HCC. Intact follow-up data were from 60 (60/60) HCC patients; the follow up time was > 2.0 years. 23 cases showed died or disease recurrence in the 60 cases. The variant that was connected with the 2.0-year disease-free survival rate was evaluated using the Kaplan-Meier survival analysis and compared by log-rank test. In addition, the expression levels of miR-9 in HCC tissues were divided into two groups as follows using the median as a cutoff: high levels of miR-9 expression (n = 30, 8.816 ± 1.640) and low levels of miR-9 expression (n = 30, 5.093 ± 1.622). The Kaplan-Meier curve indicated that for HCC patients with high levels of miR-9, the 2.0-year disease-free survival rate was 20.8%. By contrast, the 2.0-year disease-free survival rate of patients with low levels of miR-9 was 50.8%, which indicated that the HCC patients with low levels of miR-9 in their HCC tissues survived significantly longer than those with high levels of miR-9 (P < 0.001, by log-rank test) (Figure 5). In addition, multivariate analysis showed that the levels of miR-9 (P = 0.007) and clinical stage (P = 0.001) were crucial autocephalous prognostic elements for 2.0-year disease-free survival rate (Table 2).
Figure 5.
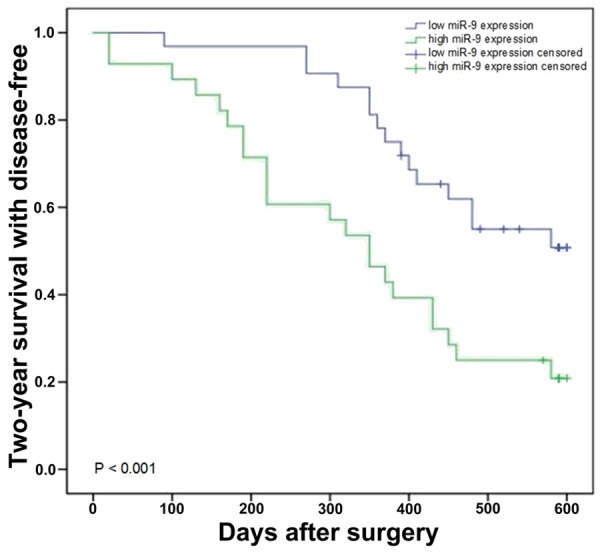
Kaplan-Meier survival curves for HCC patients with high or low expression of miR-9.
Table 2.
Multivariate analysis of two-year disease-free survival in patients with HCC
| Parameter | Hazard ratio | 95% confidence interval | P value |
|---|---|---|---|
| Age (y) | 1.299 | 0.833-2.006 | 0.256 |
| Gender | 0.875 | 0.561-2.098 | 0.784 |
| Tumor size (cm) | 1.526 | 0.996-2.550 | 0.059 |
| Number of tumor | 1.535 | 0.936-2.516 | 0.090 |
| Venous invasion | 1.315 | 0.886-2.036 | 0.315 |
| Edmondson stage | 3.279 | 1.654-6.502 | 0.001* |
| HBsAg | 1.239 | 0.784-1.989 | 0.394 |
| Tumor capsule | 1.337 | 0.847-2.110 | 0.205 |
| AFP (μg/L) | 1.500 | 0.987-2.469 | 0.078 |
| microRNA-9 | 2.681 | 1.306-5.504 | 0.007* |
Statistically significant difference.
Discussion
HCC is one of the most widespread and malignant human tumors worldwide and its prognosis is not optimistic, even with a series of advanced treatment [29]; therefore, an HCC biomarker must be identified to enable clinicians to better determine a prognosis in HCC patients. The results of this study showed that miR-9 plays an important role in the formation, progress, and distant metastasis of HCC and could become a valuable tumor marker in its prognosis.
It is well known that miRNAs are connected to a variety of cellular mechanism and are involved in multifarious tumors, and that they could participate in the regulation of cell immunity, development, differentiation proliferation, and apoptosis by regulating genetic expression at the post-transcriptional level [30]. A previous study has shown different results of miR-9 expression levels in different types of carcinomas [18-26]. These results illustrate that miR-9 might have different effects on multifarious carcinomas.
Cell adhesion molecules are a type of membrane receptor that regulate cell-cell and cell-stroma coactions, and are necessary for the signaling pathway, such as invasion, migration, metastasis, angiogenesis, and cell adhesion [31]. Cell adhesion is a critical factor in cellular motility in embryogenesis, inflammatory response, wound healing, and neoplasm metastasis [28]. Cell adhesion leads to a generation of focal adhesions that are regulatory procedures involved in cell movement. There are multitudinous proteins included with focal adhesions, such as tensin, vinculin, and talin. Talin-1 is a type of large adaptor protein that plays a significant role in coupling the integrin family of cell adhesion molecules to the actin cytoskeleton [32]. A preceding study demonstrated that miR-9 suppresses the expression levels of Talin-1 by targeting its 3’-UTR, indicating that miR-9 directly affects Talin-1 in ovarian serous carcinoma [17]; Therefore, it is hypothesized here that miR-9 might be involved in metastasis and adhesion by regulating Talin-1 expression levels in HCC and could become a significant biomarker for the prognosis of HCC.
There are a limited number of studies on the role of miR-9 expression levels in HCC tissues. In this research, our data reveal that the expression levels of miR-9 in HCC tissues is significantly higher than that in normal liver and adjacent non-cancerous tissues. On the other hand, the expression levels of Talin-1 protein and mRNA was observably decreased in HCC tissues compared with those in normal liver and corresponding adjacent non-cancerous tissues. In addition, the miR-9 and Talin-1 mRNA expression levels were observed to be an inverse relationship in HCC tissues, and the up-regulation of miR-9 in HCC tissues was significantly associated with invasive clinicopathological features, including integral capsule, tumor size, tumor grade and portal vein tumor thrombus. These results suggest that miR-9 might play a crucial role in HCC invasiveness, carcinogenesis, and infiltration. One of the most characteristic features of HCC is its distant metastasis. Our research further reveals that the expression levels of miR-9 is significantly lower in tissue specimens without portal vein cancer emboli than in tissue specimens with portal vein cancer emboli. The data indicate that high levels of miR-9 might increase the risk of tumor breaking away from the carcinoma in situ for lack of cell adhesion, eventually leading to tumor metastasis and invasion.
In previous studies, Shihong Xu et al. [33] found that the levels of miR-9 were increased in osteosarcoma. Comparatively, our results likewise proved that miR-9 expression in HCC tissues is higher than that in the normal liver and adjacent non-cancerous tissues. In addition, our study demonstrates that miR-9 levels were directly correlated with the 2.0-year disease-free survival rate. The data were further supported using the Cox proportional hazards regression model, indicating that the expression levels of miR-9 could be used as an independent risk factor and serve as a biomarker to estimate a prognosis in HCC patients. This conclusion has been proved in previous studies of osteosarcoma, glioma, lung cancer, cervical cancer, and laryngeal squamous cell carcinoma [24,30,33-35].
The results of our research suggest that high levels of miR-9 expression is strongly associated with malignant progression of HCC and that the overexpression of miR-9 is a statistically significant risk factor that affects survival rate. Our study also indicates that miR-9 might play a critical role as a tumor activator in HCC by regulating the expression of Talin-1 and might serve as a potential tumor marker for HCC prognosis; however, further studies are needed to illuminate the detailed molecular mechanism by which miR-9 plays a role in HCC.
Study limitations
Because of the relatively small number of biological tissue samples in the current research, additional research is necessary using a larger sample of HCC patients over a longer timeframe to confirm the prognostic significance of miR-9 expression.
Acknowledgements
This study was supported by the Natural Science Foundation of Anhui Province, No. 1508085MH173. We thank every teacher and doctor in the Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, for their assistance.
Disclosure of conflict of interest
None.
References
- 1.Faloppi L, Scartozzi M, Maccaroni E, Di Pietro Paolo M, Berardi R, Del Prete M, Cascinu S. Evolving strategies for the treatment of hepatocellular carcinoma: from clinical-guided to molecularly-tailored therapeutic options. Cancer Treat Rev. 2011;37:169–177. doi: 10.1016/j.ctrv.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Sapisochin G, de Sevilla EF, Echeverri J, Charco R. Management of “very early” hepatocellular carcinoma on cirrhotic patients. World J Hepatol. 2014;6:766–775. doi: 10.4254/wjh.v6.i11.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashworth RE, Wu J. Mammalian target of rapamycin inhibition in hepatocellular carcinoma. World J Hepatol. 2014;6:776–782. doi: 10.4254/wjh.v6.i11.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai X, Wang J, Guo Y, Pan J, Yang Q, Zhang M, Li H, Zhang L, Ma J, Shi F, Shu W, Wang Y, Leng J. Prostaglandin E2 stimulates beta1-integrin expression in hepatocellular carcinoma through the EP1 receptor/PKC/NF-kappaB pathway. Sci Rep. 2014;4:6538. doi: 10.1038/srep06538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ismail S, Mayah W, Battia HE, Gaballah H, Jiman-Fatani A, Hamouda H, Afifi MA, Elmashad N, Saadany SE. Plasma nuclear factor kappa B and serum peroxiredoxin 3 in early diagnosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:1657–1663. doi: 10.7314/apjcp.2015.16.4.1657. [DOI] [PubMed] [Google Scholar]
- 6.Schutte K, Schulz C, Link A, Malfertheiner P. Current biomarkers for hepatocellular carcinoma: Surveillance, diagnosis and prediction of prognosis. World J Hepatol. 2015;7:139–149. doi: 10.4254/wjh.v7.i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaheen KY, Abdel-Mageed AI, Safwat E, AlBreedy AM. The value of serum midkine level in diagnosis of hepatocellular carcinoma. Int J Hepatol. 2015;2015:146389. doi: 10.1155/2015/146389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu JP, Xu XG, Ma RJ, Qin SN, Wang CR, Wang XB, Li M, Li MS, Ma Q, Xu WW. Development of a clinical chemiluminescent immunoassay for serum GPC3 and simultaneous measurements alone with AFP and CK19 in diagnosis of hepatocellular carcinoma. J Clin Lab Anal. 2015;29:85–93. doi: 10.1002/jcla.21733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Li J, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettmer M, Vogetseder A, Durso MB, Moch H, Komminoth P, Perren A, Nikiforov YE, Nikiforova MN. MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J Clin Endocrinol Metab. 2013;98:E1–7. doi: 10.1210/jc.2012-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazeh H, Mizrahi I, Ilyayev N, Halle D, Brucher B, Bilchik A, Protic M, Daumer M, Stojadinovic A, Itzhak A, Nissan A. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer-a Comprehensive review. J Cancer. 2013;4:281–295. doi: 10.7150/jca.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H, Okubo K, Onozaki T, Zama T. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One. 2013;8:e71480. doi: 10.1371/journal.pone.0071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui EH, Li HJ, Hua F, Wang B, Mao W, Feng XR, Li JY, Wang X. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin. 2013;34:309–313. doi: 10.1038/aps.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perell K, Vincent M, Vainer B, Petersen BL, Federspiel B, Moller AK, Madsen M, Hansen NR, Friis-Hansen L, Nielsen FC, Daugaard G. Development and validation of a microRNA based diagnostic assay for primary tumor site classification of liver core biopsies. Mol Oncol. 2015;9:68–77. doi: 10.1016/j.molonc.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin H, Li X, Yang B, Zhang L, Han Z, Han C. Blood-based multiple-microRNA assay displays a better diagnostic performance than single-microRNA assay in the diagnosis of breast tumor. Tumour Biol. 2014;35:12635–12643. doi: 10.1007/s13277-014-2587-4. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Yao L, Tao X, Yu Y, Chen M, Zhang R, Xu C. miR-9 functions as a tumor suppressor in ovarian serous carcinoma by targeting TLN1. Int J Mol Med. 2013;32:381–388. doi: 10.3892/ijmm.2013.1400. [DOI] [PubMed] [Google Scholar]
- 18.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, Li X, Tang H. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, Huang KH, Lin WC. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Qi M, Li S, Qi T, Mei H, Huang K, Zheng L, Tong Q. microRNA-9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol Cancer Ther. 2012;11:1454–1466. doi: 10.1158/1535-7163.MCT-12-0001. [DOI] [PubMed] [Google Scholar]
- 21.Zheng L, Qi T, Yang D, Qi M, Li D, Xiang X, Huang K, Tong Q. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8:e55719. doi: 10.1371/journal.pone.0055719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muraoka T, Soh J, Toyooka S, Maki Y, Shien K, Furukawa M, Ueno T, Tanaka N, Yamamoto H, Asano H, Tsukuda K, Miyoshi S. Impact of aberrant methylation of microRNA-9 family members on non-small cell lung cancers. Mol Clin Oncol. 2013;1:185–189. doi: 10.3892/mco.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Zhao H, Tang D, Wu J, Yao G, Zhang Q. Overexpressions of microRNA-9 and microRNA-200c in human breast cancers are associated with lymph node metastasis. Cancer Biother Radiopharm. 2013;28:283–288. doi: 10.1089/cbr.2012.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Jia S, Xu P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal sq- uamous cell carcinoma. Int J Clin Exp Med. 2014;7:5523–5528. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Marian C, Makambi KH, Kosti O, Kallakury BV, Loffredo CA, Zheng YL. MicroRNA-9 as potential biomarker for breast cancer local recurrence and tumor estrogen receptor status. PLoS One. 2012;7:e39011. doi: 10.1371/journal.pone.0039011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S, Ge W. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037–1043. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
- 27.Fang KP, Zhang JL, Ren YH, Qian YB. Talin-1 correlates with reduced invasion and migration in human hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15:2655–2661. doi: 10.7314/apjcp.2014.15.6.2655. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JL, Qian YB, Zhu LX, Xiong QR. Talin1, a valuable marker for diagnosis and prognostic assessment of human hepatocelluar carcinomas. Asian Pac J Cancer Prev. 2011;12:3265–3269. [PubMed] [Google Scholar]
- 29.Liang L, Li Q, Huang LY, Li da W, Wang YW, Li XX, Cai SJ. Loss of ARHGDIA expression is associated with poor prognosis in HCC and promotes invasion and metastasis of HCC cells. Int J Oncol. 2014;45:659–666. doi: 10.3892/ijo.2014.2451. [DOI] [PubMed] [Google Scholar]
- 30.Xu T, Liu X, Han L, Shen H, Liu L, Shu Y. Up-regulation of miR-9 expression as a poor prognostic biomarker in patients with non-small cell lung cancer. Clin Transl Oncol. 2014;16:469–475. doi: 10.1007/s12094-013-1106-1. [DOI] [PubMed] [Google Scholar]
- 31.Xin M, Dong XW, Guo XL. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomed Pharmacother. 2015;69:179–185. doi: 10.1016/j.biopha.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, Hanein D. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J Struct Biol. 2013;184:21–32. doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu SH, Yang YL, Han SM, Wu ZH. MicroRNA-9 expression is a prognostic biomarker in patients with osteosarcoma. World J Surg Oncol. 2014;12:195. doi: 10.1186/1477-7819-12-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Wang L, Li G, Liu H, Fan F, Li Z, Li Y, Gao G. Increased expression of microRNA-9 predicts an unfavorable prognosis in human glioma. Mol Cell Biochem. 2013;384:263–268. doi: 10.1007/s11010-013-1805-5. [DOI] [PubMed] [Google Scholar]


