Abstract
Background: MALAT1, a newly discovered long noncoding RNA (lncRNA), has been reported to be highly expressed in many types of cancers. This meta-analysis summarizes its potential prognostic value in digestive system malignancies. Methods: A quantitative meta-analysis was performed through a systematic search in PubMed, Cochrane Library, Web of Science and Chinese National Knowledge Infrastructure (CNKI) for eligible papers on the prognostic impact of MALAT1 in digestive system malignancies from inception to Apr. 25, 2015. Pooled hazard ratios (HRs) with 95% confidence interval (95% CI) were calculated to summarize the effect. Results: Five studies were included in the study, with a total of 527 patients. A significant association was observed between MALAT1 abundance and poor overall survival (OS) of patients with digestive system malignancies, with pooled hazard ratio (HR) of 7.68 (95% confidence interval [CI]: 4.32-13.66, P<0.001). Meta sensitivity analysis suggested the reliability of our findings. No publication bias was observed. Conclusions: MALAT1 abundance may serve as a novel predictive factor for poor prognosis in patients with digestive system malignancies.
Keywords: MALAT1, LncRNA, digestive system malignancies, prognosis, meta-analysis
Introduction
It has reported that 3.4 million new diagnosed cases and 1.5 million deaths of digestive system malignancies occurs each year [1]. Despite recent advances in treatment of surgery, chemotherapy and radiotherapy, the prognosis of tumor is still extremely poor. Although the WHO classification can act as a criterion to predict the patient clinical outcomes, recent studies have suggested that the criteria alone may not be sufficient to estimate patient prognosis [2,3]. Therefore, it is vital to identify new potential biomarker for early diagnosis, more accurate prognosis prediction, and novel therapeutic target.
LncRNAs are non-protein coding RNA molecules greater than 200 nucleotides in length. More and more evidences have been presented to suggest that lncRNAs play an important role in cell proliferation and differentiation, chromosome inactivation, and organization of nuclear domains [4]. It has been found and could be used as novel biomarkers and therapeutic targets for cancers [5,6].
LncRNA MALAT1, also known as nuclear-enriched abundant transcript 2 (NEAT2), is a highly abundant and ubiquitously expressed long non coding RNA with a length of ~8000 nt [7]. MALAT1 is aberrantly expressed in tumor tissues and correlates with clinical parameters and prognosis in several types of human cancer. It has been suggested that MALAT1 expression may play a useful prognostic role in many digestive system malignancies, such as pancreatic cancer [8,9], gastric cancer [10], colorectal cancer [11], and hepatocellular carcinoma [12]. However, most studies examining the implications of MALAT1 expression are limited by small sample size. Therefore, we conducted a systematic review and quantitative meta-analysis to clarify the prognostic value of MALAT1 expression in digestive system malignancies.
Materials and methods
Study strategy
The present review was performed in accordance with the standard guidelines for meta-analyses and systematic reviews of tumor marker prognostic studies [13,14]. To obtain relevant articles for this review, two authors (H Zhai and XM Li) independently used the following research tools: PubMed, Cochrane Library, Chinese National Knowledge Infrastructure (CNKI) and Web of Science to identify all relevant articles about MALAT1 as a prognostic factor for survival of patients with digestive system malignancies. The literature search ended on Apr. 25, 2015. The search strategy used both MeSH terms and free-text words to increase the sensitivity of the search. The following search terms were used: “MALAT1” or “long noncoding RNA”, or “noncoding RNA”, or “lncRNA”, or “lincRNA”, “cancer” or “tumor” or “carcinoma” or “neoplasm”, “prognosis” or “survival” or “clinical outcome” or “mortality”.
Study selection
The same two investigators independently assessed all the eligible studies and extracted the data. Studies were considered eligible if they met the following criteria: digestive system malignancies was studied; MALAT1 expression was determined in human tissue using quantitative PCR or microarray expression analysis; the relationship between MALAT1 expression and survival was examined; sufficient data was provided to estimate hazard ratios (HRs) for survival rates and their 95% confidence intervals. If data subsets were published in more than one article, only the most recent one was included. Animal studies and single case reports were excluded. If the data could not be extracted or calculated from the original article, the study was excluded. Disagreements were resolved through discussion with a third investigator (Yi-Ning Yang).
Data extraction
The two investigators (A Maimaiti and W Liao) extracted data independently and reached a consensus on all items. For each study, the following characteristics of the individual research articles were collected: author, year of publication, region, tumor type, number of patients, clinical stage of tumor, cut-off values, elevated MALAT1 (%), study design, follow-up, overall survival (OS), methods, treatment data, disease-free survival (DFS), and disease-specific survival (DSS).
Quality assessment of primary studies
Quality assessment was performed independently by three investigators (QJ Chen, F Liu and HM Lai). The final scores are expressed as percentages, with a higher percentage denoting better methodological quality.
Statistical analysis
We extracted HRs according to the following three methods [15]. The first and most accurate method was to obtain the reported HRs directly from the publication, or to estimate the HRs from O-E statistic and variance. If that was not possible, we calculated the HRs from the published data including the number of patients at risk in each group, the number of events and the log rank statistic or its p value. All statistical analyses used Stata SE12.0 (Stata Corporation). To determine the heterogeneity among the included studies, chi-square-based Q test and I2 statistics were used. For the Q test, a P value less than 0.05 indicated significant heterogeneity; for the I2 statistics, an I2 value greater than 50% was considered severe heterogeneity. We also conducted sensitivity analyses to test the effect of each study on the overall pooled results. The presence of publication bias was evaluated by using funnel plots, Begg’s test. Because there was no significant statistical heterogeneity among the studies, the fixed effects model was applied for the analysis. By analyzing the HR of digestive system malignancies and high MALAT1 expression, we tried to make a thorough inquiry on the relationship between MALAT1 expression levels and prognosis of digestive system malig-nancies.
Results
Included studies and characteristics
As shown in the flow diagram (Figure 1), our search terms revealed 151 articles. After the titles and abstracts were reviewed, 142 irrelevant or duplicate articles were excluded. After a more careful inspection of the abstracts, a total of 9 articles were reviewed in detail. 4 papers were excluded because of insufficient data to estimate HR for further analysis. As a result, 5 published articles were included in the current meta-analysis [8-12]. Among these 5 studies, a total of 527 patients were included, with a maximum sample size of 150 and a minimum sample size of 45 patients (Mean 105.4). Three studies enrolled more than 100 participants. The accrual period of these studies ranged from 2012 to 2014. Four studies came from China and one study from America. All the research methods were qRT-PCR. A total of 4 different types of cancer were evaluated in studies in this meta-analysis (2 pancreatic cancer, 1 gastric cancer, 1 colorectal cancer, 1 hepatocellular carcinoma). Treatment information was not available in 3 studies, the participants in 2 didn’t receive preoperative treatment.
Figure 1.
Flowchart presenting the steps of literature search and selection.
Table 1 summarizes the main characteristics of the included studies. A total of 6 HRs were analyzed. HRs could be obtained directly in 5 studies. All of the studies were comprised of a high MALAT1 expression arm and a low MALAT1 expression arm. The average percentage of digestive system malignancies with increased MALAT1 expression was 54.3%, with a maximum of 58.7% in gastric cancer and a minimum of 50% in pancreatic cancer and colorectal cancer. OS, DFS, and DSS were estimated as survival outcome measures in 80% (4/5), 20% (1/5) and 20% (1/5) of the studies, respectively. Multivariable analyses were performed in 40% (2/5) of studies and univariate analyses were performed in 80% (4/5) of studies.
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Region | Tumor type | Sample size (n) | Clinical stage of tumor | Cut-off value | Elevated MALAT1 (%) | Preoperative treatment | Outcome measures | Survival analysis | HR (95% CI) | Quality score (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pang et al. | 2014 | China | PC | 126 | ≤II a or >II a | Median value (6.23) | 50 | No | OS | a, b | 2.439 (1.577-3.772) | 78.1 |
| 1.761 (1.101-2.815) | ||||||||||||
| Okugawa et al. | 2014 | USA | GC | 150 | III-IV | 0.985 | 58.7 | NA | OS | a | 1.54 (0.92-2.58) | 83.5 |
| Zheng et al. | 2014 | China | CC | 146 | II, III | 2.26 times higher than noncancerous tissues | 50 | No | OS, DFS | b | 2.863 (1.659-4.943) | 79.3 |
| Liu et al. | 2014 | China | PC | 45 | I-IV | Mean value | 57.8 | NA | DSS | a | 1.798 (1.177-7.747) | 80.5 |
| Lai et al. | 2012 | China | HC | 60 | I-IV | Mean value | 55 | NA | OS | a | 2.435 (1.182–5.019) | 75.0 |
PC: pancreatic cancer; GC: gastric cancer; CC: colorectal cancer; HC: hepatocellular carcinoma. NA: not available; overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS); a: Univariate analysis; b: Multivariate analysis.
Association between MALAT1 and survival in four types of digestive system malignancies
There was no significant heterogeneity among the studies (I2=0%, P=0.549), and then the fixed-effects model was adopted (Figure 2). Three studies reported the overall survival (OS), one study reported the disease-specific survival (DSS) and one study reported the overall survival (OS) and disease-free survival (DFS) of four types of digestive system malignancies based on different MALAT1 expression levels in a total of 527 patients. A significant association was observed between MALAT1 and OS/DSS/DFS in cancer patients (pooled HR 7.68, 95% CI: 4.32-13.66) (Figure 2). MALAT1 was significantly associated with OS/DSS/DFS. Therefore, it showed that patients with high MALAT1 expression were more likely to have significantly shorter OS or DSS or DFS. This analysis showed that MALAT1 was an independent prognostic factor for digestive system malignancies.
Figure 2.
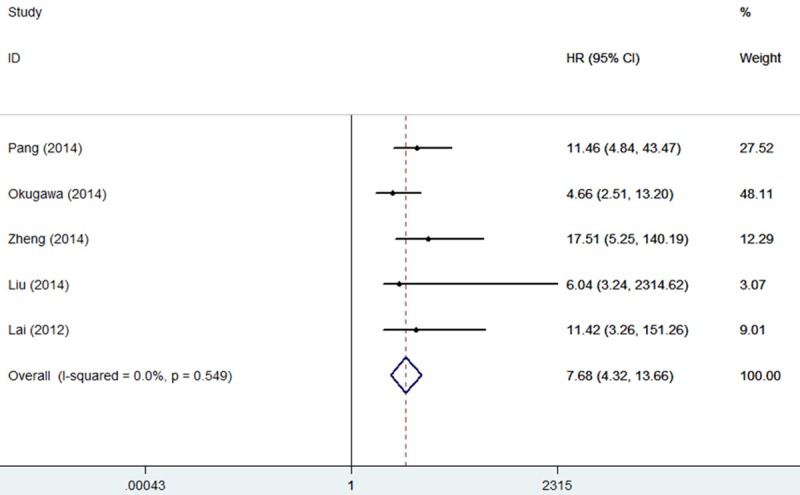
Forest plot for the association between MALAT1 expression levels and the overall survival of patients with digestive system malignancies (HR=7.68, 95% CI=4.32-13.66).
Sensitivity analysis
Sensitivity analysis was performed to examine the effect of a single study on the overall meta-analysis results by omitting one study at a time in total population. When each study was excluded sequentially, none of the results were significantly altered each time (Figure 3).
Figure 3.
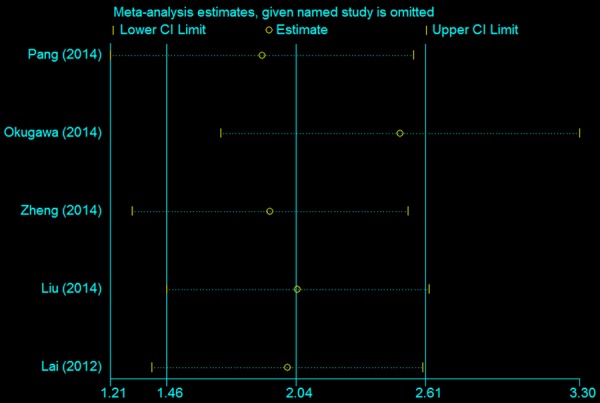
Results of sensitivity analysis.
Publication bias
We used a funnel plot to test for publication bias (Figure 4). No significant publication bias was observed. Bias was assessed statistically using the Begg’s test; Still, the results of Begg’s test (Pr>|z|=0.806) revealed no publication bias (P>0.05).
Figure 4.
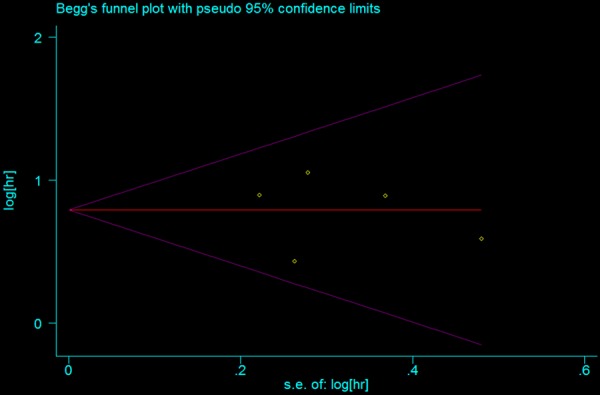
Begg’s funnel plot analysis for publication bias.
Discussion
In recent years, numerous studies have demonstrated that lncRNAs are involved in various biological processes, including cancer progression and metastasis, via chromosome remodeling, transcription and post-transcriptional processing [16]. LncRNAs are also involved in the progression of diverse diseases, especially cancers [17]. Many studies have proved that lncRNAs are associated with the development and progression of cancer through diverse pathways, including regulation of the cell cycle [18], chemotherapy resistance, apoptosis, metastasis, autophagy, and epigenetic regulation [19-23] in tumor tissues or malignant cell lines. Emerging lncRNAs can be commonly detected in various tumors, such as SRHC, HOTAIR, CCAT1, ANRIL, and MALAT1 [24-28]. Thus, lncRNAs have opened a new field of cancer genomics.
The lncRNA MALAT1 is a ~8000-nt long non coding RNA, which is located on chromosome 11q13.1. It regulates gene expression and post transcriptionally modifies primary transcripts [29]. Recently, more and more studies showed that dysregulation in lncRNAs is proved to contribute in tumor development in many cancer types and can be used to develop as biomarkers and prognosis factors [30]. MALAT1 is aberrantly expressed in different types of cancer, especially in digestive system cancer. A number of studies have reported that expression levels of MALAT1 in paired primary cancerous tissues exhibited higher than that in adjacent noncancerous tissues. Overexpression of MALAT1 was associated with high-risk grade, metastasis, and poor overall survival of cancer patients [31,32].
In this meta-analysis, we have examined the prognostic role of MALAT1 in digestive system malignancies. We believe that this meta-analysis is the first to investigate the relationship between MALAT1 and digestive system malignancies prognosis. And sensitivity analysis was all performed in the current study, enhancing the statistical power to detect a role of MALAT1 in different types of digestive system malignancies. A total of 5 papers comprising 527 patients were included into this meta-analysis. We found that MALAT1 expression was associated with a poorer prognosis in patients with different types of digestive system malignancies. Since significant heterogeneity did not exist across these studies, subgroup analyses were not performed. The above findings suggest that MALAT1 expression might be more meaningful in predicting OS or DFS or DSS of patients with digestive system carcinoma than those with non-digestive system cancer.
Nevertheless, it should be emphasized that there are several limitations in our study. First, it is still necessary to conduct larger-size and better design studies to confirm our results. Second, the major limitation of this meta-analysis was that patients included in our study were most of Asians, only one study was from USA. Because of this, our finding may just represent patients from Asia. Third, the cut-off value of high and low MALAT1 expression varied in different studies. It was difficult to reach a consensus value. Fourth, the treatment protocols after surgery differed in the various studies, and these differences might have a great impact on survival and thus result in some heterogeneity. Fifth, most of the included studies reported positive results because those with negative results are generally less likely to be published. Thus, our results might overestimate the predictive significance of MALAT1 in prognosis of digestive system malignancies to some extent.
In conclusion, our study found that MALAT1 might be a novel predictive factor for assessing poor prognosis in different types of digestive system malignancies. This is the first example of lncRNA MALAT1 being shown to be a biomarker in predicting digestive system malignancies prognosis.
Acknowledgements
This work was supported by the science and technology supporting project of Xinjiang Autonomous region in China (201491176).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Dellaretti M, Reyns N, Touzet G, Dubois F, Gusmao S, Pereira JL, Blond S. Diffuse brainstem glioma: prognostic factors. J Neurosurg. 2012;117:810–4. doi: 10.3171/2012.7.JNS111992. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DR, Galanis E. Incorporation of prognostic and predictive factors into glioma clinical trials. Curr Oncol Rep. 2013;15:56–63. doi: 10.1007/s11912-012-0279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Wang Z, Zhou D. Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med Hypotheses. 2013;81:319–21. doi: 10.1016/j.mehy.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–41. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA malat1 regulates alternative splicing by modulating sr splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang EJ, Yang R, Fu XB, Liu YF. Over expression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 9.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–7. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 10.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–81. [PMC free article] [PubMed] [Google Scholar]
- 12.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–6. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 13.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM Statistics Subcommittee of NCI-EORTC Working Group on Cancer Diagnostics. Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 14.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 23.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long Noncoding RNA ANRIL Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol Cancer Ther. 2015;14:268–77. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 27.Zheng H, Yang S, Yang Y, Yuan SX, Wu FQ, Wang LL, Yan HL, Sun SH, Zhou WP. Epigenetically silenced long noncoding-SRHC promotes proliferation of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2015;141:1195–203. doi: 10.1007/s00432-014-1871-4. [DOI] [PubMed] [Google Scholar]
- 28.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–92. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 29.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–6. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 31.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in earlystage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 32.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: A long noncoding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]


