Abstract
Deregulated microRNAs and their roles in tumorigenesis have attracted much attention in recent years. Although miR-503 has been reported to be aberrant expression in several cancers, its role in glioma remains unknown. In this study, we focused on the expression and mechanisms of miR-503 in glioma development. We found that miR-503 was downregulated in glioma cell lines and tumor tissues, and the restoration of miR-503 reduced cell proliferation invasion. Furthermore, bioinformatics analysis indicated that L1CAM was a putative target of miR-503. In a Luciferase reporter system, we confirmed that L1CAM was a direct target gene of miR-503. These findings indicate that miR-503 suppresses glioma cell growth by negatively regulating the expression of L1CAM. Collectively, our data identify the important roles of miR-503 in glioma pathogenesis, indicating its potential application in cancer therapy.
Keywords: miR-503, glioma, L1CAM, proliferation, invasion
Introduction
Gliomas are the most common malignant primary brain tumors in adults and exhibit a spectrum of aberrantly aggressive phenotype. Although there has been great improvement in chemotherapy and molecular-targeted therapy, the outcome of lung cancer remains poor. The prognosis of the disease remains poor with a median survival in the range of 15-17 months [1-3]. Therefore, it is urgent to investigate the mechanism involved in the development and progression of glioma and to find new therapeutic targets.
MicroRNAs (miRNAs), an abundant class of ~22 nucleotide small noncoding RNAs, post-transcriptionally regulate gene expression through binding to multiple target mRNAs (mRNAs) [4-6]. miRNAs, as important regulators, are significantly involved in the development of human diseases. Recent studies showed that the dysregulated miRNAs is closely associated with carcinogenesis and cancer progression [7,8]. For example, expression levels of microRNA miR-25, miR-584, miR-663 and miR-155 are associated with progression and prognosis of glioma [9-12]. MiRNAs contribute to tumorigenesis and can function as oncogenes or tumor suppressors by regulating the expressions of their target genes [13]. Thus, investigation of aberrant miRNA expression in glioma might lead to the discovery of novel miRNA biomarkers for glioma [14].
In the present study, we confirmed that miR-503 was down-regulated in glioma. Also, overexpression of miR-503 suppressed glioma cell proliferation and invasion in vitro. Furthermore, miR-503 targets LICAM and inhibited the expression of L1CAM both at the mRNA and protein levels. In conclusion, we found that miR-503 functions as a tumor suppressor by directly targeting L1 cell adhesion molecule (L1CAM). Thus, our findings provide significant clues regarding the role of miR-503 as a tumor suppressor in glioma.
Materials and methods
Human tissue specimens
Glioma tissues and normal brain tissues were obtained from patients undergoing surgery at the third Xiangya hospital, China. Collected tissues were immediately snap-frozen and stored at -80°C. Informed consents were obtained from each patient to approve the use of their tissues for research purposes. The study protocol was approved by the Institute Research Ethics Committee at Shandong University.
Cell lines
The human glioma cell lines U87, T98G, U373 and U251 were obtained from the American Type Culture Collection (ATCC, Rockville, MD). These cell lines were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin/streptomycin (Invitrogen). Normal human astrocytes (NHA) cell line was obtained from the Lonza group (Lonza, Basel, Switzerland) and cultured according to the manufacturer’s instructions.
miRNAs and siRNA transfection
Negative control (NC), miR-503 mimics (mimics) were purchased from GenePharma (Shanghai, China). The cells were transfected with NC and mimics using Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s protocol. Transfection efficiency was monitored by qRT-PCR.
Luciferase reporter gene assays
The 3’-UTR of L1CAM containing the putative binding site of miR-503 was amplified and subcloned into pGL3 luciferase promoter vector (Promega, Madison, WI, USA). The vector was co-transfected with miR-503 mimics into HEK293T cells for 48 h. The cells were harvested and relative luciferase activity was detected using a dual-luciferase reporter assay kit (Promega) according to the manufacturer’s instructions. All experiments were performed at least three times.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissues and cell lines using the miRNeasy Mini Kit (Qiagen). The miRNA Q-PCR Detection Kit (GeneCopoeia) was used for quantification of miRNA levels according to the manufacturer’s protocol. For quantification of PRMT1 mRNA levels, the RT reactions were conducted with the RevertAid TM H Minus First Strand cDNA Synthesis Kit (Fermentas). qRT-PCR was performed using an ABI 7900 System (Bio-Rad). RNU6B and β-actin were used as normalizing controls for miRNA and mRNA quantification, respectively. The 2-ΔΔCt method was employed to calculate the relative expression levels. The primers were as follows: miR-503, forward primer: 5’-CCTATTTCCCATGATTCCTTCATA-3’ and reverse primer: 5’-GTAATACGGTTATCCACGCG-3’; L1CAM, forward primer: 5’-GGCATCTCCTGTGACTGCAG-3’ and reverse primer: 5’-GGCATCTCCTGTGACTGCAG-3’.
Western blotting analysis
Whole cell extracts were prepared with a cell lysis reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manual, and then, the protein was quantified by a BCA assay (Pierce, Rockford, IL, USA). Then, the protein samples were separated by SDS-PAGE (10%) and detected by Western blot using polyclonal (rabbit) anti-L1CAM (Santa Cruz Bio-technology, Santa Cruz, CA, USA). Goat anti-rabbit IgG (Pierce, Rockford, IL, USA) secondary antibody conjugated to horseradish peroxidase and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
MTT assay
The 3-(4,5-dimethylthiazal-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was used to estimate cell viability [15]. Briefly, cells were plated at a density of 1×104 cells per well in 96-well plates. After exposure to specific treatment, the cells were incubated with MTT at a final concentration of 0.5 mg/ml for 4 h at 37°C. After the removal of the medium, 150 mM DMSO solutions were added to dissolve the formazan crystals. The absorbance was read at 570 nm using a multi-well scanning spectrophotometer reader. Cells in the control group were considered 100% viable.
Invasion assay
The capability of cell invasion was examined by transwell invasion assay. Cells were cultivated to 80% confluence on the 12-well plates. Then, we observed the procedures of cellular growth at 24 h. All the experiments were repeated in triplicate. The transwell migration chambers were used to evaluate cell invasion. The invading cells across the membrane were counted under a light microscope.
Statistical analysis
Data are expressed as mean ± SD and analyzed by Student’s t-test. Compared with respective controls, P values of <0.05 were considered statistically significant.
Results
miR-503 expression in glioma tissues and cells
We first employed qRT-PCR to detect miR-503 levels in glioma tissues. As shown in Figure 1A, the expression level of miR-503 was markedly downregulated in glioma tissues compared with normal brain tissues. Moreover, real-time PCR analysis showed that the expression level of miR-503 was markedly downregulated in four of the glioma cell lines (U87, T98G, U373 and U251), in comparison with the expression levels in Normal human astrocytes (NHA) cell line (Figure 1B). Taken together these results indicate that miR-503 may be a tumor inhibitor a in the progression of glioma.
Figure 1.
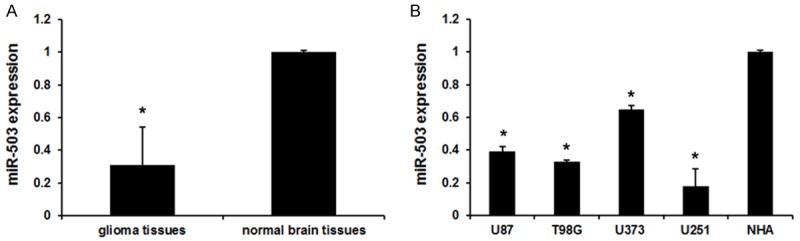
miR-503 expression in glioma tissues and cells. A. miR-503 expression in glioma tissues and normal brain tissues. Error bars represent ± S.E. and *P<0.01 versus normal brain tissues. B. miR-503 expression in glioma cells (U87, T98G, U373 and U251) and Normal human astrocytes (NHA) cell. Error bars represent ± S.E. and *P<0.01 versus NHA cell.
miR-503 inhibited invasion and migration of glioma cells
To further verify the role of miR-503 as an antitumor properties in U251 cells, we then performed rescue experiments. The transient transfection of miR-503 mimics was used to restore miR-503 expression in glioma cells. As shown in Figure 2A, expression level of miR-503 was greatly increased by miR-503 mimics. To further characterize the functional importance of miR-503 in glioma progression, we examined its effect on the proliferation and invasion of glioma cells. The MTT assay and transwell invasion were employed. The results showed that miR-503 mimics decreased the proliferation of glioma cells (Figure 2B). Similar results were observed in invasion assays of glioma cells (Figure 2C). Together, these findings demonstrate that miR-503 inhibits glioma cell proliferation and invasion in vitro.
Figure 2.
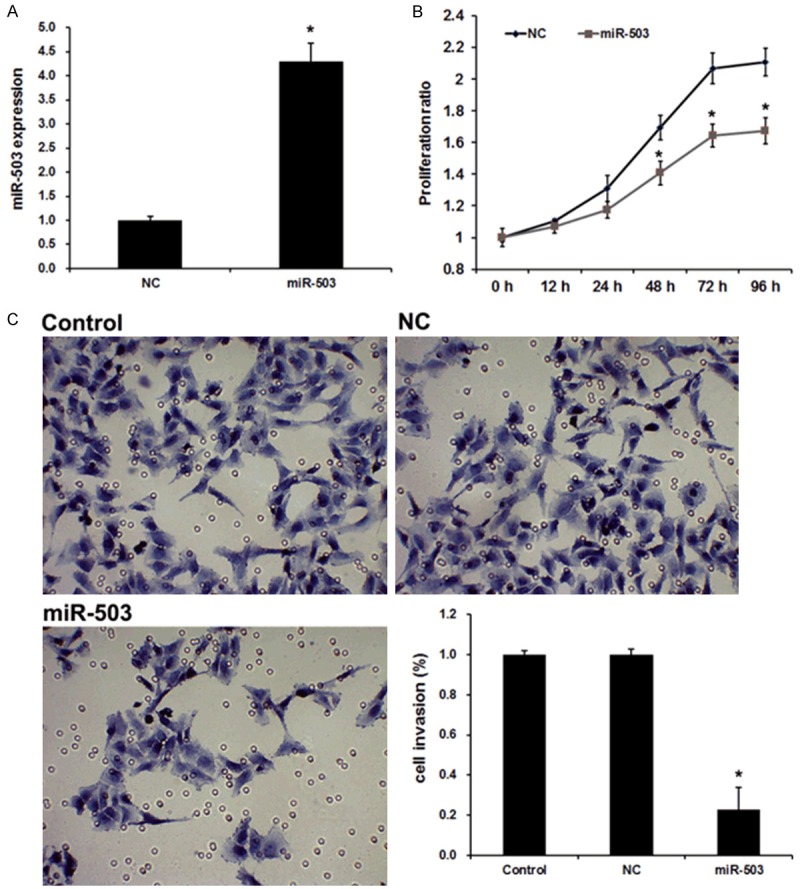
miR-503 affects the proliferation and invasion of glioma cells. A. qRT-PCR analysis revealed the effects of miR-503 mimics on the expression level of miR-503. B. MTT assays revealed the invasion ability of U251 cell transfected with miR-NC and miR-503. C. Transwell assays revealed the invasion ability of U251 cell transfected with miR-NC and miR-503. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments. *P<0.01 vs. NC group.
miR-503 directly targets L1CAM in glioma
The miRNA target prediction websites www.microRNA.org and TargetScan were used and demonstrated L1CAM is a potential downstream target gene of miR-503 in glioma with a conserved miR-503-binding site in the 3’-UTR of L1CAM mRNA. To confirm this prediction and verify whether L1CAM is direct targets of miR-503, a dual-luciferase reporter system was employed by co-transfection of miR-503 and luciferase reporter plasmids containing 3’UTR of L1CAM, or mutated L1CAM (bearing deletions of the putative miR-503 target sites). As shown in Figure 3A, co-transfection of miR-503 mimics suppressed the luciferase activity of the reporter containing wild-type L1CAM 3’UTR sequence by dual-luciferase reporter assay. However, miR-503 mimics did not have any effect on luciferase activity when target cells were transfected with mutated L1CAM. These data suggest that L1CAM may be a direct functional target of miR-503 in glioma.
Figure 3.
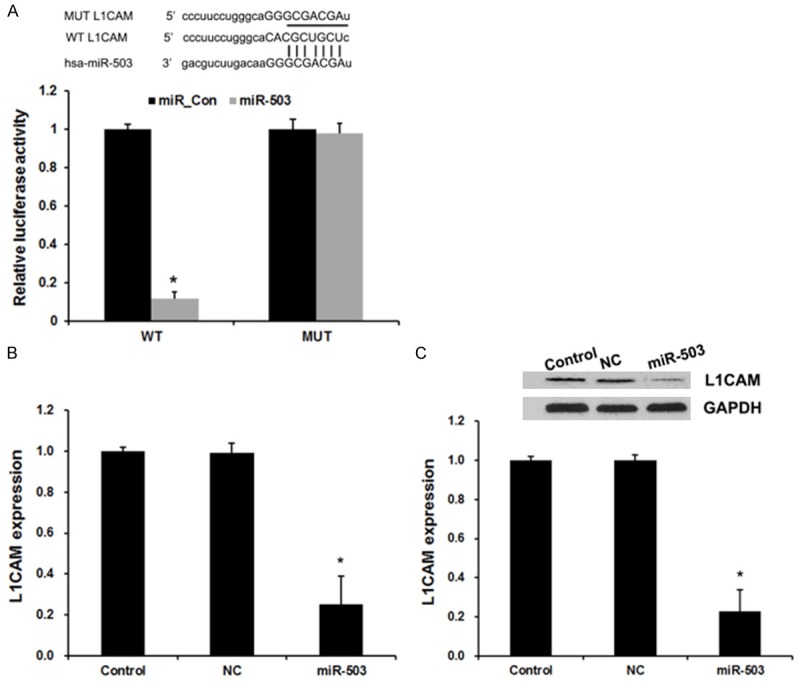
miR-503 directly targeted L1CAM. A. Sequence alignment of miR-503 and 3’UTR of L1CAM using mirco-RNA. org. Luciferase reporter assay with co-transfection of wild-type or mutant L1CAM and miR-503 mimics or miR-control in U251 cells. Error bars represent ± S.E. and *P<0.01 versus negative control (NC). B. qRT-PCR analysis revealed the effects of miR-503 mimics on the expression level of L1CAM. C. Western blot analysis revealed the effects of miR-503 mimics on the expression level of L1CAM. Error bars represent ± S.E. and *P<0.01 versus negative control (NC).
In additional, to confirm the regulatory effect of miR-503 on L1CAM, we performed qRT-PCR and western blot assay to detect the expression of L1CAM responses to the changes of miR-503 expression in glioma cell lines. As shown in Figure 2B and 2C, the assay showed a negative regulatory effect of miR-503 on L1CAM. Up-regulated miR-503 could decrease the expression of L1CAM.
Discussion
Accumulated studies revealed deregulated miRNAs in various human cancers including glioma. Identifying the miRNAs and their targets that are essential for glioma progression may provide promising therapeutic opportunities [11,16-18]. In this study, we demonstrated miR-503 as tumor suppressor and revealed that miR-503 inhibits proliferation and invasion of glioma via targeting L1CAM.
miR-503 is an intragenic miRNA clustered with miR-424 on chromosomal location Xq26.3 [19]. Several studies identified miR-503 to be involved in malignant tumors. miR-503 expression was found up-regulated in human parathyroid carcinomas [20] and in adrenocortical carcinomas [21]. miR-503 promotes tumor progression and acts as a novel biomarker for prognosis in oesophageal cancer [22]. Moreover, miR-503 acts as a tumor suppressor in various tumors. For example, microRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition [23]. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1 [24]. MiR-503 targets PI3K p85 and IKK-beta and suppresses progression of non-small cell lung cancer [25]. miR-503 was frequently downregulated in HCC cell lines and tissues, and it inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma [26]. However, the expression and role of miR-503 in glioma remains unknown. Here, we confirmed that miR-503 expression was significantly downregulated in glioma tissues and cells, and the restoration of miR-503 reduced cell proliferation invasion. These results suggested that miR-503 acted as a tumor-suppressor whose downregulation may contribute to the progression and metastasis of glioma.
L1CAM is the prototype member of the L1-family of closely related neural adhesion molecules [27]. Recent studies in tumor biology have showed that L1CAM is overexpressed in many human cancers, such as melanoma, pancreatic ductal adenocarcinoma and ovarian, endometrial carcinoma and glioblastoma [28-31]. L1CAM expression is generally associated with poor prognosis, an aggressive phenotype, and advanced tumor stages [32]. Investigations in a variety of tumor types demonstrated that increased expression of L1CAM significantly increased the migration and proliferation capacity of cancer cells in vitro [33]. In addition, multiple clinical pathology studies have indicated that L1CAM might promote cancer cell invasion and metastasis [33]. In this study, a dual-luciferase reporter assay we showed that miR-503 directly bound to the 3’-UTR of L1CAM, which contains a miR-503-binding site. Overexpression of miR-503 significantly repressed L1CAM expression at the both mRNA and protein levels. Collectively, these results confirmed that miR-503 might function as a tumor suppressor in part by repressing L1CAM expression during the development of glioma.
In conclusion, our results demonstrated that miR-503 was downregulated in glioma and played critical roles in the growth and invasion of glioma cells. L1CAM is a direct target of miR-503 and overexpression of miR-503 downregulated the expression of L1CAM protein and mRNA simultaneously, suggesting that miR-503 function as a tumor suppressor probably through down-regulating L1CAM in glioma. Thus, these results indicate that miR-503 functions as a tumor suppressor miRNA and can be used as a potential target in the gene therapy of glioma.
Disclosure of conflict of interest
None.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Hottinger AF, Stupp R, Homicsko K. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin J Cancer. 2014;33:32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posti JP, Bori M, Kauko T, Sankinen M, Nordberg J, Rahi M, Frantzen J, Vuorinen V, Sipila JO. Presenting symptoms of glioma in adults. Acta Neurol Scand. 2015;131:88–93. doi: 10.1111/ane.12285. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Tutar L, Tutar E, Tutar Y. MicroRNAs and Cancer; an Overview. Curr Pharm Biotechnol. 2014;15:430–7. doi: 10.2174/1389201015666140519095304. [DOI] [PubMed] [Google Scholar]
- 7.Xu C, Zheng Y, Lian D, Ye S, Yang J, Zeng Z. Analysis of MicroRNA expression profile identifies novel biomarkers for non-small cell lung cancer. Tumori. 2015;101:104–10. doi: 10.5301/tj.5000224. [DOI] [PubMed] [Google Scholar]
- 8.Del Vescovo V, Grasso M, Barbareschi M, Denti MA. MicroRNAs as lung cancer biomarkers. World J Clin Oncol. 2014;5:604–620. doi: 10.5306/wjco.v5.i4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo JW, Wang X, Yang Y, Mao Q. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med Pharmacol Sci. 2015;19:1630–1639. [PubMed] [Google Scholar]
- 10.Zhang J, Gong X, Tian K, Chen D, Sun J, Wang G, Guo M. miR-25 promotes glioma cell proliferation by targeting CDKN1C. Biomed Pharmacother. 2015;71:7–14. doi: 10.1016/j.biopha.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Chen C, Yu S, Liu Q, Rao J, Zhang HR, Xiao H, Fu TW, Long H, He ZC, Zhou K, Yao XH, Cui YH, Zhang X, Ping YF, Bian XW. miR-663 suppresses oncogenic function of CXCR4 in glioblastoma. Clin Cancer Res. 2015;21:4004–13. doi: 10.1158/1078-0432.CCR-14-2807. [DOI] [PubMed] [Google Scholar]
- 12.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q. miR-155 contributes to the progression of glioma by enhancing Wnt/beta-catenin pathway. Tumour Biol. 2015;36:5323–31. doi: 10.1007/s13277-015-3193-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang BC, Ma J. Role of MicroRNAs in Malignant Glioma. Chin Med J (Engl) 2015;128:1238–1244. doi: 10.4103/0366-6999.156141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Che W, Xue J, Zheng C, Tang K, Zhang J, Wen J, Xu Y. SIRT4 prevents hypoxia-induced apoptosis in H9c2 cardiomyoblast cells. Cell Physiol Biochem. 2013;32:655–662. doi: 10.1159/000354469. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y, Wang Q, Yan XL, Zhang Y, Li W, Tang F, Li X, Yang P. miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition via EphA8. FEBS Lett. 2015;589:756–765. doi: 10.1016/j.febslet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, Zhang H, James CD, Sznajder JI, Chin L, Giljohann DA, Kessler JA, Peter ME, Mirkin CA, Stegh AH. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL, Zhang MH, Liang HQ. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, Vicentini L, Chiodini I, Bisceglia M, Beck-Peccoz P, Bosari S, Spada A. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer. 2010;17:135–146. doi: 10.1677/ERC-09-0134. [DOI] [PubMed] [Google Scholar]
- 21.Tombol Z, Szabo PM, Molnar V, Wiener Z, Tolgyesi G, Horanyi J, Riesz P, Reismann P, Patocs A, Liko I, Gaillard RC, Falus A, Racz K, Igaz P. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocr Relat Cancer. 2009;16:895–906. doi: 10.1677/ERC-09-0096. [DOI] [PubMed] [Google Scholar]
- 22.Ide S, Toiyama Y, Shimura T, Kawamura M, Yasuda H, Saigusa S, Ohi M, Tanaka K, Mohri Y, Kusunoki M. MicroRNA-503 promotes tumor progression and acts as a novel biomarker for prognosis in oesophageal cancer. Anticancer Res. 2015;35:1447–1451. [PubMed] [Google Scholar]
- 23.Peng Y, Liu YM, Li LC, Wang LL, Wu XL. microRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition. Oncol Lett. 2014;7:1233–1238. doi: 10.3892/ol.2014.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y, Yin LR. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J. 2013;280:3768–3779. doi: 10.1111/febs.12365. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Liu L, Zhang Y, Guan H, Wu J, Zhu X, Yuan J, Li M. MiR-503 targets PI3K p85 and IKK-beta and suppresses progression of non-small cell lung cancer. Int J Cancer. 2014;135:1531–1542. doi: 10.1002/ijc.28799. [DOI] [PubMed] [Google Scholar]
- 26.Xiao F, Zhang W, Chen L, Chen F, Xie H, Xing C, Yu X, Ding S, Chen K, Guo H, Cheng J, Zheng S, Zhou L. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma. J Transl Med. 2013;11:195. doi: 10.1186/1479-5876-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer MK, Frotscher M. Role of L1CAM for axon sprouting and branching. Cell Tissue Res. 2012;349:39–48. doi: 10.1007/s00441-012-1345-4. [DOI] [PubMed] [Google Scholar]
- 28.Schafer MK, Altevogt P. L1CAM malfunction in the nervous system and human carcinomas. Cell Mol Life Sci. 2010;67:2425–2437. doi: 10.1007/s00018-010-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grage-Griebenow E, Jerg E, Gorys A, Wicklein D, Wesch D, Freitag-Wolf S, Goebel L, Vogel I, Becker T, Ebsen M, Rocken C, Altevogt P, Schumacher U, Schafer H, Sebens S. L1CAM promotes enrichment of immunosuppressive T cells in human pancreatic cancer correlating with malignant progression. Mol Oncol. 2014;8:982–997. doi: 10.1016/j.molonc.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirmer U, Doberstein K, Rupp AK, Bretz NP, Wuttig D, Kiefel H, Breunig C, Fiegl H, Muller-Holzner E, Zeillinger R, Schuster E, Zeimet AG, Sultmann H, Altevogt P. Role of miR-34a as a suppressor of L1CAM in endometrial carcinoma. Oncotarget. 2014;5:462–472. doi: 10.18632/oncotarget.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Held-Feindt J, Schmelz S, Hattermann K, Mentlein R, Mehdorn HM, Sebens S. The neural adhesion molecule L1CAM confers chemoresistance in human glioblastomas. Neurochem Int. 2012;61:1183–1191. doi: 10.1016/j.neuint.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Yamada S, Tanaka C, Ito S, Murai T, Kobayashi D, Fujii T, Nakayama G, Sugimoto H, Koike M, Nomoto S, Fujiwara M, Kodera Y. Overexpression of L1CAM is associated with tumor progression and prognosis via ERK signaling in gastric cancer. Ann Surg Oncol. 2014;21:560–568. doi: 10.1245/s10434-013-3246-5. [DOI] [PubMed] [Google Scholar]
- 33.Chen DL, Zeng ZL, Yang J, Ren C, Wang DS, Wu WJ, Xu RH. L1cam promotes tumor progression and metastasis and is an independent unfavorable prognostic factor in gastric cancer. J Hematol Oncol. 2013;6:43. doi: 10.1186/1756-8722-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]


