Abstract
Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms are associated with many types of cancers. The purpose of our study was to evaluate the effect of MTHFR single nucleotide polymorphisms (SNPs) on gastric cardia adenocarcinoma (GCA). We conducted a hospital-based case-control study. Three hundred and thirty cases with GCA and 608 controls were recruited. The ligation detection reaction (LDR) method was used to determine genotypes. The genotype MTHFR rs1801133 TT was significantly more frequent in cases than in controls (adjusted odds ratio (OR) = 1.46, 95% confidence interval (CI) = 1.04-2.05, P = 0.029) in a recessive model, after adjusting for age, sex and smoking and alcohol use. The haplotype MTHFR Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 was more frequent in cases than in controls (crude OR = 5.32, 95% CI = 2.34-12.10, P < 0.001). No association between other genotypes and haplotypes was observed. Our results suggest that the genotype MTHFR rs1801133 TT and the MTHFR Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 haplotype may be associated with susceptibility to GCA. Further studies are needed to confirm these findings.
Keywords: Polymorphism, MTHFR, gastric cardia adenocarcinoma, susceptibility
Introduction
Gastric cardia adenocarcinoma (GCA) is a lethal malignancy common in Chinese population. Epidemiological studies have shown a steady decline in non-cardia gastric cancer but a continuous increase in morbidity and mortality of GCA. This underlines the importance of preventative strategies for GCA undertaken in the past twenty years [1]. Many studies have demonstrated the importance of various environmental factors [2]. Genetic factors, including single nucleotide polymorphisms (SNPs), may also be important. It has been suggested that SNPs might partly explain differences in individual susceptibility to GCA [3].
DNA methylation is one of the principal mechanisms leading to loss of gene function [4]. DNA methylation involves cytosine on carbon 5. It is one of the epigenetic mechanisms currently being researched most intensively in mammals. It regulates the transcriptional plasticity of the mammalian genome. It plays a vital role in diverse cellular processes, including gene expression and regulation, and in the control of cell differentiation. During normal cell aging and differentiation, variations in DNA methylation may contribute to tumorigenesis [5]. DNA methylation is facilitated and regulated by the level of the methyl donor, S-adenosylmethionine (SAM), and in return DNA methylation affects the level of SAM [6]. SAM is synthesized by SAM synthetase from ATP and methionine. As a precursor of SAM, methionine is regulated by several pathways, including the methionine salvage pathway, the folate pathway and the diet and transmethylation pathway. The folate pathway is an important factor in one-carbon (C1) metabolism [7].
Folate is an important nutrient. It has roles in a number of fundamental physiological processes, including cell division, DNA synthesis and methylation [8]. It is catalyzed by dihydrofolate reductase to form tetrahydrofolate (H4F). Once formed, H4F enters C1 metabolism and form methylene-H4F [9]. Catalyzed by methylenetetrahydrofolate reductase (MTHFR), methylene-H4F takes part in methionine synthesis.
MTHFR is a key enzyme that catalyzes the transformation of 5, 10-methylenetetrahydrofolate into 5-methyltetrahydrofolate. This is a crucial hydrolysis step in the re-methylation of homocysteine into methionine and folate [10]. The gene coding MTHFR is located on 1p36.3 and contains 11 exons. In humans, the MTHFR enzyme is made up of dimers, each of which has a C-terminal regulatory domain and an N-terminal catalytic domain [11]. Functional polymorphisms of MTHFR may result in reduced enzyme activity and decreased plasma levels of 5-methyl tetrahydrofolic acid. This leads to decreased transformation of homocysteine to methionine, and may play a role in carcinogenesis [12]. The gene coding for MTHFR contains more than 20 SNPs, some of which are non-synonymous. These are the most frequently studied.
Given the biological and pathologic importance of MTHFR, functional genetic variations in MTHFR may contribute to the development of GCA. To explore the association between MTHFR tagging SNPs rs9651118 T>C, rs4846048 A>G, rs1801133 C>T, rs4845882 G>A and rs3753584 A>G and susceptibility to GCA, we performed a hospital-based case-control study in the Chinese Han population.
Materials and methods
Subjects
The study was approved by the Institutional Review Board of Jiangsu University (Zhenjiang, China). All subjects in the study, including the controls, were of Chinese Han origin. Participants were recruited consecutively from the Affiliated People’s Hospital of Jiangsu University and the Affiliated Hospital of Jiangsu University (Zhenjiang City, China) between October 2008 and June 2013. In all of the cases, the diagnosis of GCA cases has been established by postoperative pathological studies. Potential participants were excluded if they had a past history of cancer, an autoimmune disorder, or had received chemotherapy or radiotherapy. Controls were recruited randomly. Most of them were hospitalized due to trauma. Those with a past history of any form of malignancy were ineligible to be controls. The controls were matched to the study participants for ethnicity, sex and age (±5 years).
A previously piloted questionnaire was administered to the participants and to the controls by one of two specially trained interviewers. This was used to obtain demographic data and data on known risk factors for GCA, including cigarette smoking and alcohol drinking, as has been previously described [13].
DNA extraction
Each subject donated 2 ml peripheral venous blood, which was stored in tubes containing ethylene diamine tetraacetic acid (EDTA) disodium salt at 4°C. Genomic DNA was extracted using a commercially available DNA Blood Mini Kit (Qiagen, Berlin, Germany), within a week of blood sampling.
Polymorphism genotyping
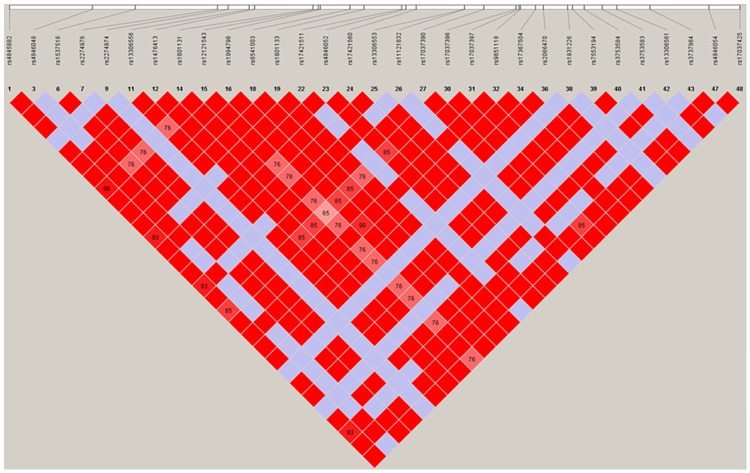
For our study we selected MTHFR tagging SNPs according to the HapMap Project and Haploview 4.2 software described previously (Figure 1) [13]. The MTHFR SNPs mentioned above genotyping was performed utilizing using the ligase detection reaction (LDR) method. Technical support was provided by Shanghai Biowing Applied Biotechnology Company [13-15]. For quality control purpose, we randomly selected 110 samples for repeat test. This confirmed an accuracy rate of 100%. We used the SHEsis Program (Bio-X Inc., Shanghai, China, available at http://202.120.7.14/analasis/myAnalasis.php) to construct haplotypes of the five SNPs [16].
Figure 1.
Linkage disequilibrium (LD) plot of MTHFR: the plot was drawn by Haploview 4.2 with D’Color Scheme. The cells are gradient color representing strength of LD between five polymorphisms. Light-colored cells indicate low LD and dark-colored indicate high LD.
Statistical analysis
The ages of the cases and controls were compared using the t-test. Deviations from the Hardy-Weinberg equilibrium (HWE) in controls were tested using an internet-based HWE calculator (available at http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) [15]. Differences in the genotype, haplotype and demographic characteristics between cases and controls were estimated using the χ2 test. Unconditional logistic regression analysis was used to evaluate associations between the MTHFR genotypes and susceptibility to GCA by computing odds ratios (Ors, crude or adjusted appropriate) and 95% confidence intervals (CIs). Statistical analysis was performed using the SAS 9.1.3 software (SAS Institute, Cary, NC). Differences were considered statistically significant when P < 0.05; with two-sided probabilities.
Results
Subject characteristics
A total of 330 cases and 608 controls were included in the study. Their demographic characteristics and risk factors for GCA are shown in Table 1. The cases and controls were matched for age and sex. There was no statistical difference between them in alcohol use, but GCA cases were significantly more likely to use tobacco.
Table 1.
Distribution of selected demographic variables and risk factors in GCA cases and controls
| Variable | Cases (n = 330) | Controls (n = 608) | P a | ||
|---|---|---|---|---|---|
|
| |||||
| n | % | n | % | ||
| Age (years) mean ± SD | 65.06 (±8.37) | 64.19 (±6.66) | 0.103 | ||
| Age (years) | 0.746 | ||||
| < 60 | 89 | 26.97 | 170 | 27.96 | |
| ≥ 60 | 241 | 73.03 | 438 | 72.04 | |
| Sex | 0.965 | ||||
| Male | 223 | 67.58 | 410 | 67.43 | |
| Female | 107 | 32.42 | 198 | 32.57 | |
| Tobacco use | 0.006 | ||||
| Never | 209 | 63.33 | 438 | 72.04 | |
| Ever | 121 | 36.67 | 170 | 27.96 | |
| Alcohol use | 0.072 | ||||
| Never | 233 | 70.61 | 462 | 75.99 | |
| Ever | 97 | 29.39 | 146 | 24.01 | |
Two-sided χ2 test and student t test;
Bold values are statistically significant (P < 0.05).
Associations between MTHFR polymorphisms and GCA risk
Our principle findings concerning MTHFR rs9651118 T>C, rs4846048 A>G, rs1801133 C>T, rs4845882 G>A and rs3753584 A>G polymorphisms are shown in Table S1. With the exception rs1801133 C>T (P = 0.033), the genotype distribution of these SNPs in the controls conformed to the HWE (P > 0.05).
With regard to rs1801133 C>T, in the recessive model, the TT homozygote genotype was associated with a borderline statistically increased risk of GCA (P = 0.054). In the same model, and after adjusted for age, sex and tobacco and alcohol use, the TT genotype increased the risk of GCA (adjusted OR = 1.46, P = 0.029; Table 2).
Table 2.
Logistic regression analyses of associations between MTHFR polymorphisms and the risk of GCA
| Genotype | Cases (n = 330) | Controls (n = 608) | Crude OR (95% CI) | P | Adjusted ORa (95% CI) | P | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| n | % | n | % | |||||
| MTHFR rs1801133 C>T | ||||||||
| CC | 102 | 31.48 | 170 | 28.72 | 1.00 | 1.00 | ||
| CT | 148 | 45.68 | 318 | 53.72 | 0.78 (0.57-1.06) | 0.112 | 0.76 (0.55-1.04) | 0.086 |
| TT | 74 | 22.84 | 104 | 17.57 | 1.19 (0.81-1.75) | 0.387 | 1.23 (0.83-1.82) | 0.300 |
| CT+TT | 222 | 68.52 | 422 | 71.28 | 0.88 (0.65-1.18) | 0.381 | 0.87 (0.65-1.17) | 0.362 |
| CC+CT | 250 | 77.16 | 488 | 82.43 | 1.00 | 1.00 | ||
| TT | 74 | 22.84 | 104 | 17.57 | 1.39 (0.99-1.94) | 0.054 | 1.46 (1.04-2.05) | 0.029 |
| T allele | 296 | 45.68 | 526 | 44.43 | ||||
| MTHFR rs3753584 A>G | ||||||||
| AA | 275 | 84.88 | 518 | 87.50 | 1.00 | 1.00 | ||
| AG | 48 | 14.81 | 72 | 12.16 | 1.26 (0.85-1.86) | 0.257 | 1.22 (0.82-1.82) | 0.319 |
| GG | 1 | 0.31 | 2 | 0.34 | 0.94 (0.09-10.43) | 0.961 | 1.03 (0.09-11.41) | 0.984 |
| AG+GG | 49 | 15.12 | 74 | 12.50 | 1.25 (0.85-1.84) | 0.266 | 1.22 (0.82-1.81) | 0.324 |
| AA+AG | 323 | 99.69 | 590 | 99.66 | 1.00 | 1.00 | ||
| GG | 1 | 0.31 | 2 | 0.34 | 0.91 (0.08-10.12) | 0.942 | 1.00 (0.09-11.12) | 1.000 |
| G allele | 50 | 7.72 | 79 | 6.67 | ||||
| MTHFR rs4845882 G>A | ||||||||
| GG | 216 | 66.26 | 416 | 69.10 | 1.00 | 1.00 | ||
| GA | 99 | 30.37 | 161 | 26.74 | 1.18 (0.88-1.60) | 0.268 | 1.18 (0.87-1.60) | 0.287 |
| AA | 11 | 3.37 | 25 | 4.15 | 0.85 (0.41-1.76) | 0.656 | 0.81 (0.39-1.68) | 0.562 |
| GA+AA | 110 | 33.74 | 186 | 30.90 | 1.14 (0.85-1.52) | 0.375 | 1.13 (0.84-1.51) | 0.421 |
| GG+GA | 315 | 96.63 | 577 | 95.85 | 1.00 | 1.00 | ||
| AA | 11 | 3.37 | 25 | 4.15 | 0.81 (0.39-1.66) | 0.558 | 0.77 (0.37-1.59) | 0.475 |
| A allele | 121 | 18.56 | 211 | 17.52 | ||||
| MTHFR rs4846048 A>G | ||||||||
| AA | 253 | 79.56 | 490 | 81.26 | 1.00 | 1.00 | ||
| AG | 63 | 19.81 | 103 | 17.08 | 1.19 (0.84-1.68) | 0.340 | 1.22 (0.86-1.74) | 0.262 |
| GG | 2 | 0.63 | 10 | 1.66 | 0.39 (0.08-1.78) | 0.223 | 0.33 (0.07-1.54) | 0.158 |
| AG+GG | 65 | 20.44 | 113 | 18.74 | 1.11 (0.79-1.57) | 0.534 | 1.14 (0.80-1.60) | 0.473 |
| AA+AG | 316 | 99.37 | 593 | 98.34 | 1.00 | 1.00 | ||
| GG | 2 | 0.63 | 10 | 1.66 | 0.38 (0.08-1.72) | 0.208 | 0.32 (0.07-1.48) | 0.145 |
| G allele | 67 | 10.53 | 123 | 10.20 | ||||
| MTHFR rs9651118 T>C | ||||||||
| TT | 129 | 40.31 | 241 | 41.13 | 1.00 | 1.00 | ||
| TC | 148 | 46.25 | 276 | 47.10 | 1.00 (0.75-1.34) | 0.990 | 0.98 (0.73-1.31) | 0.874 |
| CC | 43 | 13.44 | 69 | 11.77 | 1.16 (0.75-1.80) | 0.495 | 1.18 (0.76-1.83) | 0.469 |
| TC+CC | 191 | 59.69 | 345 | 58.87 | 1.03 (0.78-1.37) | 0.812 | 1.02 (0.77-1.34) | 0.912 |
| TT+TC | 277 | 86.56 | 517 | 88.23 | 1.00 | 1.00 | ||
| CC | 43 | 13.44 | 69 | 11.77 | 1.16 (0.77-1.75) | 0.468 | 1.19 (0.79-1.80) | 0.403 |
| C allele | 234 | 36.56 | 414 | 35.32 | ||||
Adjusted for age, sex, smoking and drinking status;
Bold values are statistically significant (P < 0.05).
The genotype frequencies of MTHFR rs9651118 T>C, rs4846048 A>G, rs4845882 G>A and rs3753584 A>G polymorphisms were not statistically different between the cases and the controls (P = 0.912, P = 0.473, P = 0.421 and P = 0.324, respectively).
Further analysis of the association between the haplotypes of these SNPs and the susceptibility to GCA was further performed. Compared with the Grs4845882Ars4846048Trs1801133Trs9651118Ars3753584 haplotype, the Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 haplotype was associated with an increased susceptibility to GCA (crude OR = 5.32, 95% CI = 2.34-12.10, P < 0.0001; Table 3). No significant associations were observed between other haplotypes and GCA risk.
Table 3.
MTHFR haplotype frequencies (%) in cases and controls and risk of GCA
| Haplotypes | Cases (n = 660) | Controls (n = 1216) | Crude OR (95% CI) | P | ||
|---|---|---|---|---|---|---|
|
| ||||||
| n | % | n | % | |||
| Grs4845882Ars4846048Trs1801133Trs9651118Ars3753584 | 273 | 41.36 | 528 | 43.42 | 1.00 | |
| Grs4845882Ars4846048Crs1801133Crs9651118Ars3753584 | 210 | 31.82 | 407 | 33.47 | 1.00 (0.80-1.25) | 0.985 |
| Ars4845882Grs4846048Crs1801133Trs9651118Ars3753584 | 64 | 9.70 | 118 | 9.70 | 1.05 (0.75-1.47) | 0.780 |
| Ars4845882Ars4846048Crs1801133Trs9651118Grs3753584 | 42 | 6.36 | 73 | 6.00 | 1.11 (0.74-1.67) | 0.607 |
| Grs4845882Ars4846048Crs1801133Trs9651118Ars3753584 | 27 | 4.09 | 48 | 3.95 | 1.09 (0.66-1.78) | 0.738 |
| Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 | 22 | 3.33 | 8 | 0.66 | 5.32 (2.34-12.10) | < 0.0001 |
| Ars4845882Ars4846048Crs1801133Crs9651118Ars3753584 | 7 | 1.06 | 7 | 0.58 | 1.93 (0.67-5.57) | 0.222 |
| Grs4845882Ars4846048Trs1801133Trs9651118Grs3753584 | 5 | 0.76 | 0 | 0.00 | — | 0.980 |
| Ars4845882Ars4846048Crs1801133Trs9651118Ars3753584 | 3 | 0.45 | 9 | 0.74 | 0.65 (0.17-2.40) | 0.513 |
| Ars4845882Grs4846048Crs1801133Trs9651118Grs3753584 | 2 | 0.30 | 0 | 0.00 | — | 0.981 |
| Grs4845882Ars4846048Crs1801133Trs9651118Grs3753584 | 0 | 0.00 | 4 | 0.33 | — | 0.983 |
| Ars4845882Ars4846048Trs1801133Trs9651118Ars3753584 | 2 | 0.30 | 3 | 0.25 | 1.29 (0.21-7.76) | 0.781 |
| Ars4845882Grs4846048Crs1801133Crs9651118Ars3753584 | 0 | 0.00 | 3 | 0.25 | — | 0.978 |
| Grs4845882Grs4846048Trs1801133Trs9651118Ars3753584 | 0 | 0.00 | 3 | 0.25 | — | 0.978 |
| others | 3 | 0.45 | 5 | 0.41 | 1.16 (0.28-4.89) | 0.839 |
With the order of MTHFR rs4845882 G>A, rs4846048 A>G, rs1801133 C>T, rs9651118 T>C and rs3753584 A>G in gene position.
Discussion
In this study, we performed a hospital-based case-control study to investigate whether functional SNPs in MTHFR affect the susceptibility of the Chinese Han population to GCA. We found evidence that MTHFR rs1801133 TT genotype and the MTHFR Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 haplotype increased the risk of GCA.
The MTHFR gene produces methylenetetrahydrofolate reductase, which is a rate-limiting enzyme in folate metabolism and DNA methylation. It is an active 77 kDa protein that catalysis the conversion of 5, 10-methylenetetrahydrofolate into 5-methyltetrahydrofolate [17]. The MTHFR gene is highly polymorphic in the general population. There is evidence that MTHFR gene mutations lead to increased thymidylate synthase (TS) activity in cancer cells, as a consequence of increased level of 5, 10-methylenetetrahydrofolate. The latter supplies methyl for the methylation of dUMP to dTMP [18]. TS is a critical and rate-limiting enzyme for maintaining an appropriate supply of DNA to ensure accurate DNA synthesis and repair [19]. It follows that SNPs in the MTHFR gene may contribute to the genetic susceptibility to cancer [20].
Several earlier studies have also suggested that MTHFR rs1801133 TT genotype may increase the risk of GCA in the Chinese Han population [21-23]. It is reported that a C→T mutation at nucleotide 677 loci (in exon 4 at the folate-binding site) led to valine substitution for alanine (677 C>T, rs1801133 C>T) and that this is functionally relevant, causing a reduction in the activity of methylenetetrahydrofolate reductase [24]. Studies have shown that individuals who are heterozygous for MTHFR rs1801133 polymorphism have 70% of normal enzyme activity, but those who are homozygous have only 30% of the normal enzyme activity [25]. With regard to relationship between MTHFR and folate, some studies have suggested that compound heterozygosity for the 677T allele is associated with decreased plasma folate levels [26]. A different study found that the functional polymorphism rs1801133 C>T is associated with low plasma folate content and significantly decrease MTHFR activity [27]. MTHFR plays a role in the formation of dimers, with flavin adenine dinucleotide (FAD) being a cofactor. However, mutant MTHFR (677T) dissociates into monomers leading to decreased enzymatic activity. Docking studies have established that mutant MTHFR (677T) has less affinity with FAD than the wild type enzyme (677C) [11]. When combined with our results, findings suggest that a C-to-T mutation in MTHFR results in lower enzyme activity and lower folate concentrations and that these may be associated with an increased risk of GCA.
The rs9651118 T>C SNP is situated in the intron region of the MTHFR gene. Several recent studies have shown that it has moderate protective effect against carcinoma of the lung and breast [28,29], but not against esophageal squamous cell carcinoma (ESCC) [13]. We did not find any association between it and GCA risk. Together, these findings indicate that MTHFR rs9651118 T>C has different effects depending on the type of cancers. MTHFR rs4845882 G>A is located on the intron region of the MTHFR gene, with almost complete linkage disequilibrium with rs1801131 A>C. A previous study showed that there was no significant association between the combined AC/CC variant genotypes and the risk of GCA [30]. Our results are consistent with this. MTHFR rs3753584 A>G is also situated in the intron region of the MTHFR gene. A previous study found that there was an increased risk of lung cancer in carriers of the variant allele of this SNP when compared with subjects who were homozygote for the wild type. The risk was more marked in those over 60 years [31]. No similar association was found with ESCC [13]. Our study failed to find an association between this SNP and GCA. MTHFR rs4846048 A>G is situated 463 base pairs (bp) up stream of a polyadenylation signal [32]. It has been associated with the decreased risk of ESCC [13], but no association has been found between it and the risk of breast cancer [33]. We found no evidence of an association between it and the risk of GCA. Further studies are required in order to better determine the biological significance of these SNPs in the pathogenesis of GCA.
We studies five potentially functional MTHFR SNPs in order to ascertain the association between MTHFR haplotypes and the susceptibility to GCA. Haplotype analysis suggested a significant association with susceptibility to GCA. In a previous reported study of the association between the haplotype of three SNPs (MTHFR rs1801133 C>T, rs1801131 A>C and rs2274976 G>A) and GCA risk, it was found that individuals with six mutant alleles had a significantly increased risk when compared to those with 0-2 mutant alleles [30]. However, further studies with larger sample sizes are needed to conform these findings.
Our study used a fine-mapping approach to obtain the MTHFR SNPs we used. It is the first reported study to investigate rs9651118 T>C, rs4846048 A>G, rs4845882 G>A and rs3753584 A>G MTHFR SNPs. Moreover, in comparison to previously reported study, its sample size was large.
However, the study did have several limitations. Given that the cases and controls were recruited from hospitals, the study population may not have been representative of the general Chinese Han population. Folate status may influence the association between MTHFR SNPs and GCA susceptibility. We did not have data on the folate intake of those we studied. Finally, an even larger sample size than we were able to recruit might be expected to lead to more definitive findings. Further studies are needed to better understand the role of interactions between genes and the environment in the causation of GCA.
In conclusion, our results suggest that the functional MTHFR rs1801133 C>T polymorphism and the MTHFR Grs4845882Ars4846048Trs1801133Crs9651118Ars3753584 haplotype may contribute to susceptibility to GCA in Chinese Han individuals.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (81472332, 81370001, 81300037 and 81341006), Jiangsu Province Natural Science Foundation (BK2010333 and BK2011481), Fujian Province Natural Science Foundation (2013J01126 and 2013J05116), Fujian Medical University professor fund (JS12008) and Fujian Province science and technology programmed fund (2012Y0030). We appreciate all subjects who participated in this study. We wish to thank Dr. Yiqun Chen (Biowing Applied Biotechnology Company, Shanghai, China) for technical support.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer. 2005;113:456–463. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 2.Zhang XM, Zhong R, Liu L, Wang Y, Yuan JX, Wang P, Sun C, Zhang Z, Song WG, Miao XP. Smoking and COX-2 functional polymorphisms interact to increase the risk of gastric cardia adenocarcinoma in Chinese population. PLoS One. 2011;6:e21894. doi: 10.1371/journal.pone.0021894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin J, Wang X, Wei J, Wang L, Shi Y, Zheng L, Tang W, Ding G, Liu C, Liu R, Chen S, Xu Z, Gu H. Interleukin 12B rs3212227 T>G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis Esophagus. 2014;28:291–8. doi: 10.1111/dote.12189. [DOI] [PubMed] [Google Scholar]
- 4.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013;27:e420–423. doi: 10.1111/jdv.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trivedi MS, Deth R. Redox-based epigenetic status in drug addiction: a potential contributor to gene priming and a mechanistic rationale for metabolic intervention. Front Neurosci. 2014;8:444. doi: 10.3389/fnins.2014.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou HY, Lin YH, Shiu GL, Tang HY, Cheng ML, Shiao MS, Pai LM. ADI1, a methionine salvage pathway enzyme, is required for Drosophila fecundity. J Biomed Sci. 2014;21:64. doi: 10.1186/s12929-014-0064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho Y, Kim JO, Lee JH, Park HM, Jeon YJ, Oh SH, Bae J, Park YS, Kim OJ, Kim NK. Association of Reduced Folate Carrier-1 (RFC-1) Polymorphisms with Ischemic Stroke and Silent Brain Infarction. PLoS One. 2015;10:e0115295. doi: 10.1371/journal.pone.0115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ly A, Hoyt L, Crowell J, Kim YI. Folate and DNA methylation. Antioxid Redox Signal. 2012;17:302–326. doi: 10.1089/ars.2012.4554. [DOI] [PubMed] [Google Scholar]
- 10.Wu YL, Hu CY, Lu SS, Gong FF, Feng F, Qian ZZ, Ding XX, Yang HY, Sun YH. Association between methylenetetrahydrofolate reductase (MTHFR) C677T/A1298C polymorphisms and essential hypertension: a systematic review and meta-analysis. Metabolism. 2014;63:1503–1511. doi: 10.1016/j.metabol.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Rai V. Folate pathway gene MTHFR C677T polymorphism and risk of lung cancer in Asian populations. Asian Pac J Cancer Prev. 2014;15:9259–9264. doi: 10.7314/apjcp.2014.15.21.9259. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Du C, Guo X, Yuan L, Niu W, Yu W, Er L, Wang S. Interleukin-8-251A/T polymorphism and Helicobacter pylori infection influence risk for the development of gastric cardiac adenocarcinoma in a high-incidence area of China. Mol Biol Rep. 2010;37:3983–3989. doi: 10.1007/s11033-010-0057-7. [DOI] [PubMed] [Google Scholar]
- 13.Tang W, Zhang S, Qiu H, Wang L, Sun B, Yin J, Gu H. Genetic variations in MTHFR and esophageal squamous cell carcinoma susceptibility in Chinese Han population. Med Oncol. 2014;31:915. doi: 10.1007/s12032-014-0915-6. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Y, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Y, Ma J, Zhao Y. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 15.Tang W, Qiu H, Jiang H, Sun B, Wang L, Yin J, Gu H. Lack of association between cytotoxic T-lymphocyte antigen 4 (CTLA-4) -1722T/C (rs733618) polymorphism and cancer risk: from a case-control study to a meta-analysis. PLoS One. 2014;9:e94039. doi: 10.1371/journal.pone.0094039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 17.Xuan C, Li H, Zhao JX, Wang HW, Wang Y, Ning CP, Liu Z, Zhang BB, He GW, Lun LM. Association between MTHFR polymorphisms and congenital heart disease: a meta-analysis based on 9,329 cases and 15,076 controls. Sci Rep. 2014;4:7311. doi: 10.1038/srep07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn KJ, Croxford R, Yates Z, Lucock M, Kim YI. Effect of the methylenetetrahydrofolate reductase C677T polymorphism on chemosensitivity of colon and breast cancer cells to 5-fluorouracil and methotrexate. J Natl Cancer Inst. 2004;96:134–144. doi: 10.1093/jnci/djh015. [DOI] [PubMed] [Google Scholar]
- 19.Tan W, Miao X, Wang L, Yu C, Xiong P, Liang G, Sun T, Zhou Y, Zhang X, Li H, Lin D. Significant increase in risk of gastroesophageal cancer is associated with interaction between promoter polymorphisms in thymidylate synthase and serum folate status. Carcinogenesis. 2005;26:1430–1435. doi: 10.1093/carcin/bgi090. [DOI] [PubMed] [Google Scholar]
- 20.Kim YI. Methylenetetrahydrofolate reductase polymorphisms, folate, and cancer risk: a paradigm of gene-nutrient interactions in carcinogenesis. Nutr Rev. 2000;58:205–209. doi: 10.1111/j.1753-4887.2000.tb01863.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang LD, Guo RF, Fan ZM, He X, Gao SS, Guo HQ, Matsuo K, Yin LM, Li JL. Association of methylenetetrahydrofolate reductase and thymidylate synthase promoter polymorphisms with genetic susceptibility to esophageal and cardia cancer in a Chinese high-risk population. Dis Esophagus. 2005;18:177–184. doi: 10.1111/j.1442-2050.2005.00492.x. [DOI] [PubMed] [Google Scholar]
- 22.Miao X, Xing D, Tan W, Lu W, Lin D. [Single nucleotide polymorphisms in methylenetetrahydrofolate reductase gene and susceptibility to cancer of the gastric cardia in Chinese population] . Zhonghua Yi Xue Za Zhi. 2002;82:669–672. [PubMed] [Google Scholar]
- 23.Kurokawa Y, Sasako M, Sano T, Yoshikawa T, Iwasaki Y, Nashimoto A, Ito S, Kurita A, Mizusawa J, Nakamura K Japan Clinical Oncology Group (JCOG9502) Ten-year follow-up results of a randomized clinical trial comparing left thoracoabdominal and abdominal transhiatal approaches to total gastrectomy for adenocarcinoma of the oesophagogastric junction or gastric cardia. Br J Surg. 2015;102:341–348. doi: 10.1002/bjs.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai W, Zhang Z, Ni M, Geng P, Lian Z, Zhang G, Shi LL, Chen J. Genetic Association between Methylenetetrahydrofolate Reductase Gene Polymorphism and Risk of Osteonecrosis of the Femoral Head. Biomed Res Int. 2015;2015:196495. doi: 10.1155/2015/196495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain M, Pandey P, Tiwary NK, Jain S. MTHFR C677T polymorphism is associated with hyperlipidemia in women with polycystic ovary syndrome. J Hum Reprod Sci. 2012;5:52–56. doi: 10.4103/0974-1208.97802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fodinger M, Wagner OF, Horl WH, Sunder-Plassmann G. Recent insights into the molecular genetics of the homocysteine metabolism. Kidney Int Suppl. 2001;78:S238–242. doi: 10.1046/j.1523-1755.2001.59780238.x. [DOI] [PubMed] [Google Scholar]
- 27.Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, Mendel M, Kidron M, Bar-On H. A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr. 1999;129:1656–1661. doi: 10.1093/jn/129.9.1656. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Hu C, Xiao SH, Wan B. Association of tagging SNPs in the MTHFR gene with risk of type 2 diabetes mellitus and serum homocysteine levels in a Chinese population. Dis Markers. 2014;2014:725731. doi: 10.1155/2014/725731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swartz MD, Peterson CB, Lupo PJ, Wu X, Forman MR, Spitz MR, Hernandez LM, Vannucci M, Shete S. Investigating multiple candidate genes and nutrients in the folate metabolism pathway to detect genetic and nutritional risk factors for lung cancer. PLoS One. 2013;8:e53475. doi: 10.1371/journal.pone.0053475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen H, Newmann AS, Hu Z, Zhang Z, Xu Y, Wang L, Hu X, Guo J, Wang X, Wei Q. Methylenetetrahydrofolate reductase polymorphisms/haplotypes and risk of gastric cancer: a case-control analysis in China. Oncol Rep. 2005;13:355–360. [PubMed] [Google Scholar]
- 31.Liu H, Jin G, Wang H, Wu W, Liu Y, Qian J, Fan W, Ma H, Miao R, Hu Z, Sun W, Wang Y, Jin L, Wei Q, Shen H, Huang W, Lu D. Association of polymorphisms in one-carbon metabolizing genes and lung cancer risk: a case-control study in Chinese population. Lung Cancer. 2008;61:21–29. doi: 10.1016/j.lungcan.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Gaughan DJ, Barbaux S, Kluijtmans LA, Whitehead AS. The human and mouse methylenetetrahydrofolate reductase (MTHFR) genes: genomic organization, mRNA structure and linkage to the CLCN6 gene. Gene. 2000;257:279–289. doi: 10.1016/s0378-1119(00)00392-9. [DOI] [PubMed] [Google Scholar]
- 33.Lu Q, Jiang K, Li Q, Ji YJ, Chen WL, Xue XH. Polymorphisms in the MTHFR gene are associated with breast cancer risk and prognosis in a Chinese population. Tumour Biol. 2015;36:3757–62. doi: 10.1007/s13277-014-3016-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.