Abstract
Objective: To study the effect of DNMT1 on CD4+ T cells in the peripheral blood of systemic lupus erythematosus (SLE) patients. Methods: To investigate the differential expression of DNMT1 in CD4+ T cells of SLE patients and healthy individuals, a DNMT1 lentiviral plasmid (pLenti6.3/V5-DNMT1) and a control plasmid (pLenti6.3/V5-GW/LacZ) were constructed and transfected into CD4+ T cells from the peripheral blood of SLE patients. The transcriptional and translational expression of DNMT1, global genomic DNA methylation, and the production of IgG antibody in the CD4+ T cells in the peripheral blood of SLE patients were assessed using qPCR analysis, western blotting, flow cytometry, and ELISA, respectively. Results: The expression level of DNMT1 in SLE patients was significantly lower than that in normal humans. The expression of DNMT1 was found to be positively correlated with the methylation level of genomic DNA and negatively correlated with the IgG titration level. DNA sequencing results confirmed that the DNMT1 lentiviral plasmid was successfully constructed. After the CD4+ T cells from the peripheral blood of SLE patients were transfected with the pLenti6.3/V5-DNMT1 plasmid, the transcription level of the DNMT1 gene was upregulated and abundance of DNMT1 protein significantly increased. Global genomic DNA methylation was enhanced, while the production of IgG antibody was reduced. Conclusion: DNMT1 can inhibit the autoimmune response in SLE patients by reversing the abnormally low DNA methylation level in the CD4+ T cells.
Keywords: Lentivirus, pLenti6.3/V5-DNMT1, SLE, CD4+ T cells, methylation
Introduction
Because of high prevalence, extremely harmful effects, and the unavailability of effective medical treatments, systemic lupus erythematosus (SLE) has become a serious threat to human health. Recent studies have found that demethylation of DNA in CD4+ T cells can increase the expression of autoimmune-related genes, such as CD11a (ITFAL) [1] and CD70 (TNFSF7) [2], in the T cells of SLE patients. CD11a can trigger the autoreactivity of T cells, while the overexpression of CD70 can stimulate B cells to increase the production of autoantibodies. A study by Lu et al. [3] also found that the high susceptibility of women to lupus erythematosus is related to DNA methylation levels in CD4+ T cells. DNA demethylation leads to the inactivation of regulatory genes in the X chromosome and the overexpression of autoimmune-related genes such as CD40L (TNFSF5), which enhances the effect of anti-chromatin antibodies in females. The association of the increasing susceptibility of female patients to lupus erythematosus with abnormal DNA methylation and its regulatory mechanism is highly complex. To date, the regulatory mechanism of DNA methylation in the T cells of SLE patients remains a highly debated subject of study within epigenetics. Sawalha AH et al. [4] claimed that DNA demethylation is caused by the downregulation of DNA methyltransferase (DNMTs) due to the disruption of ERK signaling pathways in T cells of SLE patients. Deng C et al. [5] suggested that DNMTs are directly inhibited by inhibitors such as 5-AZAC and procainamide. However, the research groups of Barreto G [6] and Li Y [7] proved that GADD45A erases methylation marks by promoting DNA repair, resulting in demethylation of CD11a and CD70 promoter sequences in the T cells, resulting in autoimmunity in SLE patients. Some transcriptional regulatory proteins, such as Sp and CTCF (isolated proteins, CCCTC binding factor), protect parts of regulatory genetic sequences from methylation by the formation of a specific demethylated region via inhibition of the enzymatic transfer of the methyl group to cytosine by DNMTs. This enhances chromatin-remodeling activators and maintains gene activation [8]. However, these studies did not fully investigate the relationship between gene demethylation in the CD4+ T cells of SLE patients and the pathological mechanism of the disease. In this study, the effect of the DNMT1 on CD4+ T cells was investigated in vitro by overexpressing the DNMT1 gene on the DNMT1 lentiviral plasmid transfected into the CD4+ T cells. This study proved that DNMT1 is key to the occurrence and development of SLE. This study has contributed to the development of an effective treatment for the disease by providing a more comprehensive description of the epigenetics of SLE pathogenesis.
Materials and methods
Subjects of the study
A total of 15 SLE outpatients and SLE inpatients who met the 1997 updates of the American College of Rheumatology (ARA) revised criteria for classification of SLE were selected from our hospital [9]. The disease activity assessment was performed in accordance with the SLE Disease Activity Index (SLEDAI-2000) [10]. The selected patients were sex- and age-matched and consisted of 8 males and 7 females aged 25-62 years, with an average age of 34 years. Sex- and age-matched control groups of 12 healthy subjects were also selected. These subjects included 6 males and 6 females aged 30-55 years with an average age of 43 years.
Cell lines and cell cultures
E. coli DH5α and the human embryonic kidney 293FT cell line were purchased from the Shanghai Institute of Biochemistry and Cell Biology (SIBCB). The 293FT cell line was cultured in complete medium: D-MEM (high glucose), 10% fetal bovine serum, 0.1 mmol/l of non-essential amino acids, 2 mmo1/l of L-glutamine thalidomide, 100 U/ml of penicillin/streptomycin, and 500 µg/ml of Geneticin® 293FT.
Isolation of peripheral blood CD4+ T cells
A total of 20 ml of blood was withdrawn from the peripheral veins of fasting subjects early in the morning (with the informed consent of the patients and healthy volunteers). The blood was injected into a sterile anticoagulant tube containing 3.8% sodium citrate. The mononuclear cells in the peripheral blood were isolated by Ficoll density gradient centrifugation, and the MACS technique was then used to separate the subtypes of CD4+ T cells.
Construction of the pLenti6.3/V5-DNMT1 expression plasmid
The amplification primers for the ORF of human DNMT1 (GenBank ID: NM001379) were purchased from Shanghai Invitrogen Co., Ltd. The forward primer was 5’ TCGCCCCTCCCCATCGGTTT 3’, and the reverse primer was 5’ TGGGGCTAGGTGAAGGTTCAGGC 3’. These primers were used for PCR amplification with the following protocol for 32 amplification cycles: 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s. Five microliters of the PCR products was loaded onto a 1.5% agarose gel for gel electrophoresis. The PCR-amplified DNMT1 product was recovered from the gel using the Gel Extraction Kit purchased from Shanghai Watson Biotechnologies, Inc. Fifteen microliters of the recovered PCR product and 0.5 µl of plasmid pMD18T were added to the ligation reaction mixture (3 µl sterile water, 2 µl of 5X ligation buffer, and 1 µl of T4 DNA ligase). The reaction mixture was incubated for 16 h, and the ligation product was transformed into competent DH5α cells. The transformed plasmid was extracted and sent to Shanghai Invitrogen Co., Ltd for sequencing using primers for the universal T7 promoter. The sequencing results were compared against the GenBank database by using BLAST in order to verify the topology of the constructed pMD18T-DNMT1 plasmid.
Lentiviral packaging and viral titer determination
The lentivirus was packaged using the lentivirus transfection kit and the 293FT packaging cell line according to the manufacturer’s instructions. The supernatant was collected and concentrated. The primers and probes were designed for targeting the common cDNA sequences on the lentiviral plasmids and control plasmid to determine viral titers using qPCR.
Lentivirus transfection of CD4+ T cells from the peripheral blood of SLE patients
The pLenti6.3/V5-DNMT1 plasmid and pLenti6.3/V5-GW/LacZ (control plasmid) were transfected into the CD4+ T cells from the peripheral blood of SLE patients. The cells were collected for detection of positive isolates after 72 h of transfection.
Determination of DNMT1 Gene expression by qPCR
TRIzol reagent was used to extract the total RNA from the 72-h treatment group (pLenti6.3/V5-DNMT1 plasmid) and the control group (pLenti6.3/V5-GW/LacZ plasmid). Total RNA was reverse transcribed into cDNA, and the target gene was detected using the primers F1: 5’-CCTTGGAGAACGGTGCTCAT-3’ and R1: 5’-TCTCCATCGGACTTGCTCCT-3’. The PCR reaction protocol consisted of denaturation at 95°C for 2 min, followed by 40 cycles of 10-s denaturation at 95°C, 30-s annealing at 60°C, and 45-s extension at 70°C.
Western blot detection of DNMT1 protein expression
Cultures of the treatment group (pLenti6.3/V5-DNMT1 plasmid) and the control group (pLenti6.3/V5-GW/LacZ plasmid) were harvested 2 weeks after the transfection. Cell lysis buffer was added to the cells to extract the proteins. The protein extracts were quantified using the BCA method. Twenty picograms of each sample were used for SDS-PAGE gel electrophoresis and blotting to the transfer membrane. After blocking, the transfer membrane was incubated overnight at 4°C with the primary antibody (DNMT1 antibody or the housekeeping GAPDH antibody). After washing, the membrane was hybridized with the secondary antibody at room temperature for l h, followed by a second washing. The membrane was then visualized using ECL light emission and imaged.
Genomic DNA extraction and determination of methylation levels
Cellular DNA was extracted using the Tiangen Genomic DNA kit (blood/tissue/cell genomic DNA extraction kit). DNA methylation levels were determined using the MethylFlash™ Methylated DNA Quantification Kit (Colorimetric) (Epigentek Inc., USA).
IgG antibody test
The CD4+ T cells were cocultured with the non-CD4+ T cells. The non-CD4+ T cells were isolated and quantified, and the cell suspension with a total of 8 × 105 cells was centrifuged at 400 × g for 10 min to discard the supernatant. The cells were resuspended in 100 µl of RPMI 1640 medium and transferred in duplicate into a 96-well plate. A total of 1 × 105 of the 72-h-transfected CD4+ T cells were collected from each group and were added to the non-CD4+ T cell wells with gentle mixing. The plate was incubated at 37°C in a 5% CO2 incubator. Fifty microliters of RPMI 1640 medium was added to each well on the fourth and eighth days of incubation. The plate was centrifuged at 400 × g for 10 min on the tenth day of incubation to collect the supernatant. The IgG antibody was determined using the Human Immunoglobulin G (IgG) ELISA kit according to the instructions provided by the manufacturer (Nanjing Bo Pai Biofactory Co. Ltd.).
Results
Differential expression of DNMT1 mRNA and protein in the CD4+ T cells of SLE patients and normal humans
As indicated in Figure 1A, the qPCR assay showed that the expression level of DNMT1 mRNA in CD4+ T cells of SLE patients was significantly lower than that in normal humans (P < 0.05). Figure 1B illustrates the western Blot experimental results, which showed that the expression level of DNMT1 protein in CD4+ T cells of SLE patients was significantly lower than that of the normal humans.
Figure 1.
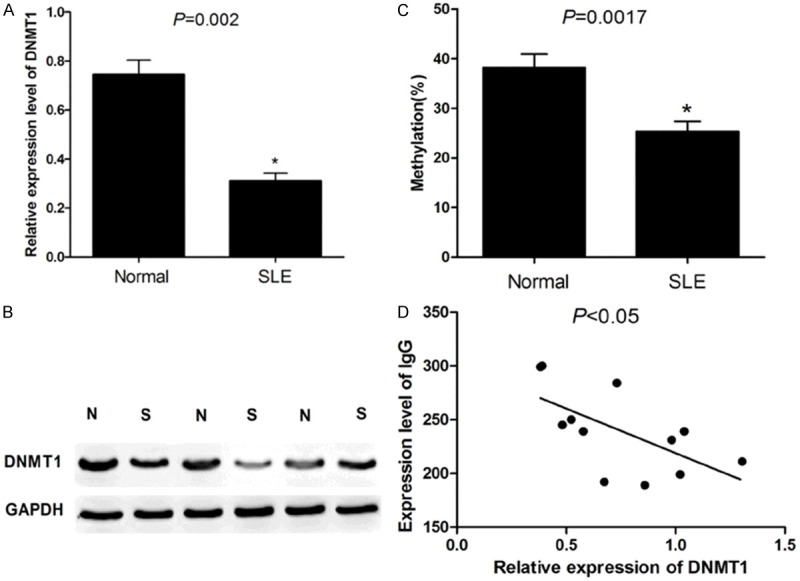
A: PCR results showed that in CD4+ T lymphocytes from SLE patients, DNMT1 is significantly lower than that in normal population (P < 0.05); B: Western Blot results showed that in CD4+ T lymphocytes from SLE patients, the protein expression of DNMT1 was significantly lower than that in normal population; C: The DNA total methylation level of CD4+ T lymphocyte in SLE patients were significantly lower than that in healthy people; D: In SLE patients, the expression of DNMT1 and IgG antibody titers were negatively correlated (P < 0.05).
Global genomic DNA methylation in the CD4+ T cells of the SLE patients and normal humans
Global genomic DNA methylation levels in the CD4+ T cells of SLE patients were significantly lower than those in the normal humans (P = 0.0017) (Figure 1C).
Correlation test of DNMT1 expression and IgG antibody titers
As indicated in Figure 1D, the DNMT1 expression and IgG antibody titers were negatively correlated in SLE patients (P < 0.05).
Construction and sequencing validation of plenti6.3/v5-DNMT1 viral vector
Refer to Figure 2.
Figure 2.
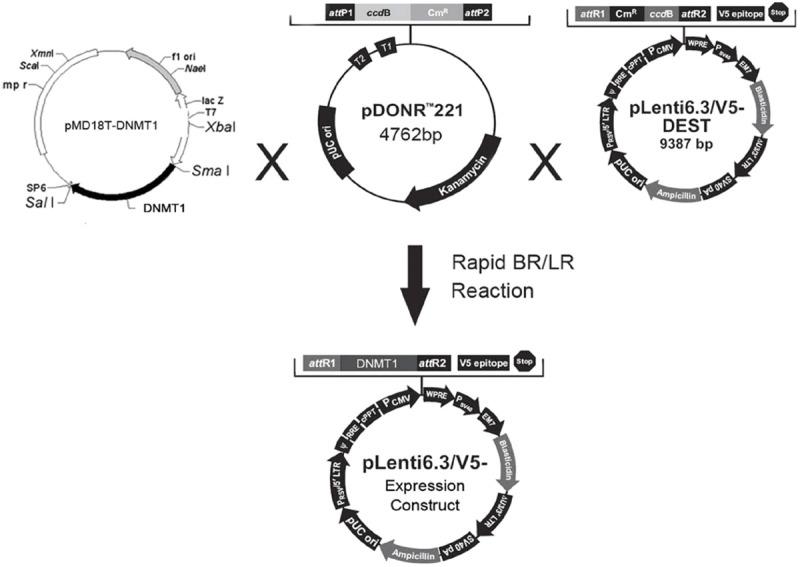
Synthesis process of plenti6.3/v5-DNMT1 viral vector.
Effect of plenti6.3/v5-DNMT1 plasmid transfection on the DNMT1 mRNA and protein expression levels in CD4+ T cells of SLE patients
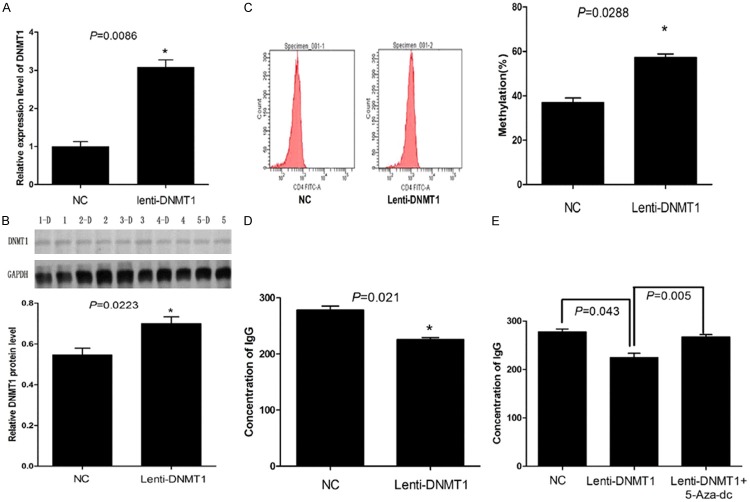
As shown in Figure 3A, after transfection with the plenti6.3/v5-DNMT1 plasmid, DNMT1 mRNA expression levels in the CD4+ T cells of SLE patients were significantly higher than those of the control group (P = 0.0086). After transfection with the plenti6.3/v5-DNMT1 plasmid, DNMT1 protein expression levels in the CD4+ T cells of SLE patients were significantly higher than those of the control group (P = 0.0223) (Figure 3B).
Figure 3.
A: PCR results showed that, after the transfection of plenti6.3/v5-DNMT1 viral vector into CD4+ T lymphocytes, the mRNA level of DNMT1 were markedly elevated; B: Western Blot results showed that, after the transfection of plenti6.3/v5-DNMT1 viral vector into CD4+ T lymphocytes, the protein level of DNMT1 were markedly elevated; C: Flow cytometry results showed that, after the transfection of plenti6.3/v5-DNMT1 viral vector into CD4+ T lymphocytes, the methylation level of total DNA raised; D: ELISA results show that, compared with the control group, the production of IgG antibody was reduced after the transfection of plenti6.3/v5-DNMT1 viral vector into CD4+ T lymphocytes (P = 0.021); E: 5-Aza-dc cell demethylation can significantly reverse the level of IgG antibody titers in CD4+ T lymphocytes.
Effect of the plenti6.3/v5-DNMT1 plasmid on global DNA methylation in CD4+ T cells of SLE patients
Flow cytometry analysis showed that the DNA methylation levels of the CD4+ T cells from the peripheral blood of SLE patients transfected with the pLenti6.3/V5-DNMT1 plasmid were significantly increased by 1.35-fold (1.35 ± 0.059) as compared to the control group (Figure 3C).
ELISA evaluation of the effect of plenti6.3/v5-DNMT1 plasmid on the production of IgG antibodies by the SLE patients’ peripheral blood CD4+ T cells
As indicated in Figure 3D, the amount of IgG antibody produced from the CD4+ T cells that were obtained from the peripheral blood of SLE patients and transfected with Lenti6.3/V5-DNMT1 plasmid was significantly lower than that from the control group, and the difference was statistically significant (P = 0.021).
Effect of 5-AZA on the production of IgG antibody in CD4+ T cells obtained from SLE patients and transfected with the plenti6.3/v5-DNMT1 plasmid
After the CD4+ T cells that were obtained from the peripheral blood of SLE patients and transfected with the plenti6.3/v5-DNMT1 plasmid were treated with 5-Aza-2-dc, the production of the IgG antibody increased significantly compared to that from the untreated cells (P = 0.005) (Figure 3E).
Discussion
DNA methylation is closely tied to gene silencing. It plays an important role in body development and gene marking in mammals. After the T-cells are stimulated, the methylation group of the IL-2 gene promoter is erased, thus leading to activation of the T cells. This indicates that methylation is a reversible and dynamic mechanism of DNA modification [11]. Recent studies on SLE patients and gene expression of DNA demethylation-associated genes have gradually revealed the epigenetics of the SLE pathological mechanism.
A study found that DNA demethylation can lead to the overexpression of LFA-1, which was detected in the SLE patients and in the T cells of DNA methylation inhibitor-induced lupus erythematosus-like autoimmune disease [12]. The level of overexpressed LFA-1 is correlated with SLE-DAI [13]. LFA-1 is expressed as a heterodimer (CD11α/CD18) on the surface of the lymphocytes and belongs to the β2-integrin family, which mediates cell-cell adhesion and plays an important role in inflammation and immune responses with co-stimulatory functions [14]. The mRNA expression levels of LFA-1/CD11α/CD18 have been found to increase by 10-fold in human T-cells after treatment with azacitidine or procaine. Flow cytometry also detected the overexpression of CD11α/CD18 on the cell surface. Transfected cells can exert an immune response in the presence of low levels of antigen, but equal levels of antigen do not trigger an immune response in non-transfected cells [15], suggesting that the overexpression of LFA-1 and the autoreactivity of T-cells play important roles in the pathological mechanism of autoimmune diseases.
Recent studies have shown that the perforin expression in the CD4+ T cells of lupus erythematosus patients was significantly higher than that of the control group, and the increase in expression levels was found to be positively correlated with those of SLE-DAI. Perforins, which are mainly expressed on the activated cytotoxic T cells and natural killer cells, serve as the main toxic proteins that kill the target cells. Perforins are usually stored in the cytoplasmic granules. T-cells and macrophages from an active lupus erythematosus patient were cocultured and divided into two groups; one group was treated with concanamycin A (a selective inhibitor of perforin). The results showed that concanamycin A completely inhibited the killing of macrophages, indicating that perforin might play an important role in the process of killing target cells [16]. The reduction in the functionality or the abundance of DNA methyltransferase enzyme can cause DNA demethylation, leading to onset of the disease. Therefore, any disruption in the gene or the expression of DNMT1 may play an important role in the onset of SLE.
Our previous study [17] used real time RT-PCR to determine mRNA expression levels of DNMT1 in CD4+ T cells from 30 SLE patients and 18 normal humans. In addition, global methylation of genomic DNA was analyzed in parallel to DNMT1 expression in order to determine the correlation between the two and their relevance with the disease activity score (SLEDAI). Our study found that expression levels of DNMT1 mRNA in the CD4+ T cells of SLE patients decreased and were associated with DNA demethylation, which plays an important role in the pathological mechanism of SLE.
In this study, a genetically modified lentivirus served as the vector. A lentivirus plasmid, pLenti6.3/V5-DNMT1, containing the human DNMT1 gene was constructed, and the sequence was validated by DNA sequencing; the lentiviral expression plasmids were packaged into the lentivirus in 293FT cells to produce the infectious viral stock. The construction of the human DNMT1 gene expression plasmid served as the foundation for the study of the physiological function of DNMT1 protein and the pathological mechanisms in SLE. Using our lentiviral vector, the pLenti6.3/V5-DNMT1 plasmid was transfected into the CD4+ T cells of the SLE patients and compared to the control group. qPCR analysis showed an increase in DNMT1 transcription levels, which was further confirmed by western blotting, which showed increase in protein levels in the cells. Flow cytometry verified that global genomic DNA methylation levels increased, and that production of the IgG antibodies decreased after the cocultivation. The capability of the pLenti6.3/V5-DNMT1 plasmid in promoting methylation in CD4+ T cells of SLE patients has provided a new tool and insights for future research on DNA methylation and its regulation factors.
Acknowledgements
This study was supported by Basic Research Creative Program of Science Technologe of Shanghai (Grant No. 08JC1403100).
Disclosure of conflict of interest
None.
References
- 1.Lu Q, Kaplan M, Ray D, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in lupus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 2.Kozlowska A, Hrycaj P, Lacki JK, Jagodzinki PP. Fyn and CD70 Expression in CD4+ T Cells from Patients with Systemic Lupus Erythematosus. J Rheumatol. 2010;37:53–59. doi: 10.3899/jrheum.090424. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the Inactive X in T Cells from Women with Lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 4.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 6.Barreto G, Scher A, Marhold J, Stach D, Swaminathan SK, Handa V, Derlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Zhao M, Yin H, Gao F, Wu X, Luo Y, Zhao S, Zhang X, Su Y, Hu N, Long H, Richardson B, Lu Q. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62:1438–1447. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 8.Hark AT, Schoenherr CJ, Katz DJ, Inqram RS, Levorse JM, Tilqhman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 9.Hochberg MC. For the diagnostic and therapeutic criteria committee of the american college of rheumatology: updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 10.Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29:288–291. [PubMed] [Google Scholar]
- 11.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–40. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 12.Richardson BC, Strahler JR, Pivirotto TS, Quddus J, Bayliss GE, Gross LA, O’Rourke KS, Powers D, Hanash SM, Johnson MA. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35:647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi T, Amano K, Sekine H, Koide J, Abe T. Upregulated expression and function of integrin adhesive receptors in systemic lupus erythematosus patients with vasculitis. J Clin Invest. 1993;92:3008–3016. doi: 10.1172/JCI116924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 15.Richardson B, Powers D, Hooper F, Yung RL, O’Rourke K. Lymphocyte function-associated antigen 1 overexpression and T cell autoreactivity. Arthritis Rheum. 1994;37:1363–1372. doi: 10.1002/art.1780370915. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 17.Qin HH, Zhu XH, Liang J, Yang YS, Wang SS, Shi WM, Xu JH. Associations between aberrant DNA methylation and transcript levels of DNMT1 and MBD2 in CD4+ T cells from patients with systemic lupus erythematosus. Australas J Dermatol. 2013;54:90–5. doi: 10.1111/j.1440-0960.2012.00968.x. [DOI] [PubMed] [Google Scholar]