Graphical abstract
Keywords: Bacterial hydrogen metabolism, Fermentation, Protein engineering, Molecular genetics, [FeFe]-hydrogenase, Electron-bifurcation
Highlights
-
•
Engineering microbial biohydrogen production may have biotechnological applications.
-
•
A synthetic operon encoding an NADH-linked [FeFe]-hydrogenase was designed.
-
•
The enzyme was heterologously produced, activated and characterised.
-
•
The addition of ferredoxin and pyruvate oxidoreductase was necessary for in vivo activity.
Abstract
Biohydrogen is a potentially useful product of microbial energy metabolism. One approach to engineering biohydrogen production in bacteria is the production of non-native hydrogenase activity in a host cell, for example Escherichia coli. In some microbes, hydrogenase enzymes are linked directly to central metabolism via diaphorase enzymes that utilise NAD+/NADH cofactors. In this work, it was hypothesised that heterologous production of an NAD+/NADH-linked hydrogenase could connect hydrogen production in an E. coli host directly to its central metabolism. To test this, a synthetic operon was designed and characterised encoding an apparently NADH-dependent, hydrogen-evolving [FeFe]-hydrogenase from Caldanaerobacter subterranus. The synthetic operon was stably integrated into the E. coli chromosome and shown to produce an active hydrogenase, however no H2 production was observed. Subsequently, it was found that heterologous co-production of a pyruvate::ferredoxin oxidoreductase and ferredoxin from Thermotoga maritima was found to be essential to drive H2 production by this system. This work provides genetic evidence that the Ca.subterranus [FeFe]-hydrogenase could be operating in vivo as an electron-confurcating enzyme.
1. Introduction
Biohydrogen (Bio-H2), which is hydrogen produced biologically from sustainable sources, is a possible future source of biofuel or industrial chemical feedstock [38]. While hydrogen metabolism is rare in higher eukaryotes, many microorganisms naturally produce H2 in order to dispose of excess reducing equivalents under some growth conditions. Various anaerobic bacteria are capable of H2 production during fermentation. Typical Bio-H2 yields from fermentation are 2 mol of H2 per mol of glucose with typically 1.3 mol H2 per mol glucose being achieved [14], [37] and it has been suggested that in order for Bio-H2 to become commercially viable yields must increase to >6 mol H2 per mol of glucose [7]. Heterologous production of hydrogenases, the enzymes responsible for the majority of biological H2 production (and oxidation), from other H2-producing organisms is an attractive strategy to improve Bio-H2 yields since non-native hydrogenases that have alternative catalytic properties, cellular localisations or substrate specificities may offer advantages.
The two main classes of bacterial hydrogenases are named [FeFe] and [NiFe] according to the metals in their respective active sites. In both cases, the Fe ions carry additional CO and CN− ligands [18]. Also in both cases, additional accessory proteins are required for the biosynthesis of the metal cofactors with their non-proteinaceous ligands [8], [12]. [FeFe]-hydrogenases typically have H2-production activities that are 10–100 times greater than [NiFe]-hydrogenases and are therefore attractive candidates for projects aimed at studying H2 production [1], [19]. For biosynthesis of the active site of an [FeFe]-hydrogenase at least three accessory proteins (HydE, HydF and HydG) are required [8]. Thus, for producing [FeFe]-hydrogenases in a non-native host such as Escherichia coli, heterologous co-production of these accessory proteins is essential for recovery of active enzyme [2], [30], [42].
E. coli can perform a mixed-acid fermentation and under these growth conditions, where respiratory electron acceptors are limited, the cell faces the challenge of re-cycling NAD+ from the NADH generated by glycolysis. E. coli normally tackles this problem by producing an alcohol dehydrogenase (AdhE), which combines two activities into a single protein: acetyl CoA-dependent aldehyde dehydrogenase and alcohol dehydrogenase [22]. Both reactions utilise NADH as reductant and the resultant ethanol is excreted from the cell. An adhE mutant is therefore severely compromised in its growth under strict fermentative conditions [13], [15], [24], [35]. In other biological systems it is possible to link cofactor cycling directly to H2 metabolism. Ralstonia eutropha (re-named Cupriavidus necator), for example, produces a soluble complex between a diaphorase and a [NiFe]-hydrogenase that allows NADH production in a H2-dependent manner [10]. Caldanaerobacter subterranus subsp. tengcongensis (formerly Thermoanaerobacter tengcongensis) is a thermophilic Gram-negative bacterium [58] that produces a cytoplasmic [FeFe]-hydrogenase originally reported to produce H2 with NADH as sole electron donor [52]. The prospect of an [FeFe]-hydrogenase biased towards H2 production, linked directly to NADH oxidation, mades the Ca. subterranus enzyme very attractive for potential Bio-H2 applications. The Ca. subterranus enzyme comprises a complex of four subunits, HydA-D [52]. The HydA subunit is an [FeFe]-hydrogenase predicted to contain an active site ‘H’-cluster as well as four other Fe-S clusters; the HydB subunit is predicted to be a flavin-containing diaphorase subunit with three additional Fe-S clusters; and HydC and HydD are small electron-transferring proteins each predicted to harbour a single Fe-S cluster. All four proteins have been co-purified in a single complex [52].
In recent years, electron-bifurcating hydrogenases, which direct electrons from H2 oxidation to two different acceptors, and electron-confurcating hydrogenases, which simultaneously receive electrons from two different sources [49], have been described in various biological systems [9], [50], [51], [56]. In the example of an electron-confurcating enzyme an [FeFe]-hydrogenase receives electrons from NADH and reduced ferredoxin, which together drive H2 production and recycling of NAD+ [51]. The source of reduced ferredoxin varies between biological systems, but could be linked to pyruvate::ferredoxin oxidoreductase (POR) [56]. Despite the initial report that the Ca. subterranus [FeFe]-hydrogenase receives electrons only from NADH [52], this enzyme shares considerable overall sequence identity (43–56%) with the subunits of the [FeFe]-hydrogenase from Thermotoga maritima that has been characterised as an electron-bifurcating enzyme [51].
In this work, the overall aim was to engineer an NADH-dependent [FeFe]-hydrogenase into E. coli central energy metabolism. Although from a thermophilic bacterium, the Ca. subterranus NADH-dependent [FeFe]-hydrogenase was a very attractive candidate given its probable bias towards H2 production and its diaphorase activity linked directly to its hydrogenase activity [52]. To this end, a synthetic version of the Ca. subterranus NADH-dependent [FeFe]-hydrogenase was designed, constructed and activated. An E. coli strain was then constructed where the synthetic operon encoding the Ca. subterranus enzyme replaced adhE at its native chromosomal locus. This E. coli engineered strain was tested for H2 production, but no gas production was evident. However, co-production of Th. maritima ferredoxin and POR in the engineered strain was found to able to induce low but detectable amounts of hydrogen production. Further genetic experiments led to the conclusion that the Ca. subterranus NADH-dependent [FeFe]-hydrogenase could likely operate in vivo as an electron-confurcating enzyme. This work provides first proof-of-concept evidence that an active NADH-linked [FeFe]-hydrogenase can be produced in E. coli, and that this enzyme has the potential to be further engineered for bioenergy applications.
2. Experimental procedures
2.1. Bacterial strains and growth conditions
Strains constructed in this work are listed in Table 1. The FTD147h3 strain, which carries a synthetic operon encoding the Ca. subterranus NADH-dependent [FeFe]-hydrogenase in place of adhE, was constructed as follows: a DNA fragment of approximately 500 bp upstream of the adhE gene, including all regulatory elements, was amplified by PCR and cloned into pBluescript (AmpR) as a KpnI/EcoRI fragment. Next, a 500 bp fragment covering the adhE stop codon and downstream sequence was amplified and cloned as a HindIII/SalI fragment in the same vector, thus resulting in a pBluescript-encoded ΔadhE allele. Next, this plasmid was digested with EcoRI/HindIII and the synthetic hydC-tte0891-hydD-hydB-hydA operon inserted. The new ΔadhE::(hydC-tte0891-hydD-hydB-hydA) allele was then transferred to pMAK705 and on to the chromosome of FTD147 as described [25]. The strain Teatotal1 was constructed by moving the unmodified ΔadhE allele from pBluescript onto pMAK705 and from there onto the chromosome of FTD147. The FTD147h3 strain was further modified by the addition of an ΔiscR allele to yield strain CLK001. Strains CPD152E and CPD159F were constructed by introducing the Keio Collection ΔpflA::kan allele [4] or a pflA::Tn5 transposon insertion [23] into strains CLK001 and FTD147h3, respectively by P1kc mediated transduction [48].
Table 1.
Strains and plasmids utilized in this work.
| Strain | Relevant genotype | Source |
|---|---|---|
| MC4100 | E. coli K-12. F−, λ−, [araD139]B/r, Δ(argF-lac)U169, e14-, flhD5301, Δ(fruK-yeiR) 725(fruA25), relA1, rpsL150(StrR), rbsR22, Δ(fimB-fimE)632(::IS1), deoC1 | [11] |
| K38 | E. coli K-12. HfrC, phoA4, pit-10, tonA22, ompF627, relA1 | [36] |
| FTD147 | as MC4100, ΔhyaB, ΔhybC, ΔhycE | [16] |
| Teatotal1 | as FTD147, ΔadhE | this work |
| FTD147h3 | as FTD147, ΔadhE::(hydC-tte0891-hydD-hydB-hydA) | this work |
| CPD159F | as FTD147h3, pflA::Tn5 | this work |
| CLK001 | as FTD147h3, ΔiscR | this work |
| CPD152E | as CLK001, ΔpflA::Kan | this work |
| PK4854 | E. coli K-12. F−, λ−, ilvG−, rfb-50, rph-1, ΔiscR | [21] |
| Plasmid | Relevant genotype | Source |
|---|---|---|
| pGP1-2 | encoding T7 polymerase (KanR) | [53] |
| pUNI-PROM | as pT7.5 (AmpR, PT7, Ptat) | [28] |
| pUNI-Tte-Hyd | as pUNI-PROM, synthetic operon encoding Ca. subterranus HydC, Tte0891, HydD, HydB, HydA | this work |
| pUNI-Tte-HydΔA | as pUNI-Tte-Hyd, ΔhydA | this work |
| pUNI-Tte-HydΔB | as pUNI-Tte-Hyd, ΔhydB | this work |
| pUNI-Tte-HydΔAB | as pUNI-Tte-Hyd, ΔhydAB | this work |
| pUNI-Tte-HydΔC | as pUNI-Tte-Hyd, ΔhydC | this work |
| pUNI-Tte-HydΔD | as pUNI-Tte-Hyd, ΔhydD | this work |
| pUNI-Tte-HydΔ0891 | as pUNI-Tte-Hyd, Δtte0891 | this work |
| pUNI-Tte-HydhisC | as pUNI-Tte-Hyd, with hexa-His affinity tag sequence in hydC | this work |
| pUNI-Sh-EFG | as pUNI-PROM, natural Sh. oneidensis SO_3923 (hydG); SO_3924 (hydX); SO_3925 (hydE); and SO_3926 (hydF) operon | this work |
| pUNI-Tm-POR-Fd | as pUNI-PROM, natural Th. maritima tm0015-tm0018 operon (encoding POR), and tm0927 (ferredoxin) | this work |
| pSU-PROM | as pSU40 (KanR, Ptat) | [28] |
| pSU-Sh-EFG | as pSU-PROM with insert fragment from pUNI-Sh-EFG | this work |
| pACYCDuet-1 | as pACYC184 (CmR, PT7lac-1, PT7lac-2, lacI+) | Novagen |
| pDuet-Sh-GX-EF | as pACTC-Duet-1, with natural Sh. oneidensis hydGX and hydEF cloned under separate T7 promoters | this work |
| pSU23 | CmR, PT7lac | [5] |
| pSU23-Sh-GX-EF | as pSU23 with insert fragment from pDuet-SH-GX-EF | this work |
| pSUtat-Sh-GX-EF | As pSU23-Sh-GX-EF with E. coli tat promoter | this work |
2.2. Plasmid construction
A list of the key plasmids studied in this work is provided in Table 1. For the construction of the synthetic operon encoding the Ca. subterranus NADH-dependent [FeFe]-hydrogenase, the primary amino acid sequences of the products of hydC, tte0891, hydD, hydB, and hydA were back-translated into DNA sequence, which was then codon optimised using the OPTIMZER software [43] with codon adaptation indices of between 0.7–0.8. Appropriate restriction enzyme sites were chosen and inserted and a strong ribosome binding site and linker sequence analysed using RBS CALCULATOR [46] and inserted before each gene. This final sequence was then synthesized as a service by Biomatik Corp (USA). The synthetic operon was sub-cloned into the E. coli production vector pUNI-PROM (AmpR; PT7; Ptat) [28] as a BamHI and SalI fragment resulting in the plasmid pUNI-Tte-Hyd. To delete individual hydrogenase genes, thus allowing facile identification of produced gene products, pUNI-Tte-Hyd was digested with: SpeI (pUNI-Tte-HydΔC); BglII (pUNI-Tte-HydΔ0891); SacI (pUNI-Tte-HydΔD); SphI (pUNI-Tte-HydΔB); XhoI (pUNI-Tte-HydΔA); and SphI and XhoI (pUNI-Tte-HydΔAB).
To fuse an N-terminal hexa-Histidine tag to HydC, the hydC gene was amplified using the oligonucleotides TtehydCNTermHisfor (GCGCACTAGTAGGAGGAAAAAAAAATGCACCATCACCATCACCATCAAGGTATGAAAGAGGCG) and TtehydCNTermHisrev (GCGCACTAGTTTATTCGAACTTTTTCAGGATTTCG) and subsequently digested with SpeI and cloned into SpeI-digested pUNI-Tte-Hyd, resulting in pUNI-Tte-HydhisC.
To construct plasmids that would encode accessory genes required for [FeFe]-hydrogenase biosynthesis, the four gene operon containing SO_3923 (hydG); SO_3924 (hydX); SO_3925 (hydE) and SO_3926 (hydF) was amplified by PCR from Shewanella oneidensis genomic DNA, digested with BglII/HindIII and cloned into BamHI/HindIII-digested pUNI-PROM and pSU-PROM (KanR; Ptat) to give the plasmids pUNI-Sh-EFG and pSU-Sh-EFG. To delete sections of the Sh. oneidensis hydGXEF operon to allow identification of produceed genes, pUNI-Sh-EFG was digested with: SacI (pUNI-Sh-EFGSacI); ClaI (pUNI-Sh-EFGClaI); XhoI (pUNI-Sh-EFGXhoI); and NcoI (pUNI-Sh-EFGNcoI).
In order to separate each half of the Sh. oneidensis hydGXEF operon, hydGX and hydEF were amplified using the oligonucleotides DRGXfor (GCGCGAATTCAGGAGGAAAAAAAAATGAGCACACACGAGC), DuetShGtoXrev (CGCGAAGCTTTCATCTGTTAAACCC) and DREFfor (GCGCAGATCTAGGAGGAAAAAAAAATGATCACTCGCCCTAGC), newDuetShEtoFrev (CGCGGACGTCCTATTGCTGAGGATTGCGG), respectively, before the hydGX EcoRI/HindII and hydEF BglII/AatII fragments were subsequently cloned into pACYCDuet1 resulting in pDuet-Sh-GX-EF. The hydGX-hydEF fragment was then subcloned into the production vector pSU23 and an EcoRI/HindIII, fragment, resulting in pSU23-Sh-GX-EF. Finally, in order to insert the constitutive promoter of the E. coli tat operon (Ptat) upstream of hydG, the tat promoter was amplified using the following oligonucleotides: tatfor (GCGCGAATTCTGTCGGTTGGCGCAAAACACGC) and tatrev (GCGCGAATTCCTGTGGTAGATGATGATTAAACAAAGC), which was then digested with EcoRI and cloned into pSU23-Sh-GX-EF, resulting in pSUtat-Sh-GX-EF.
The Thermotoga maritima gene tm0927 (encoding a ferredoxin), and the operon tm0015-tm0018 (encoding the γ, δ, α and β subunits of a pyruvate-ferredoxin oxidoreductase), were amplified by PCR using Th. maritima genomic DNA as template (a gift from the group of Michael Adams, University of Georgia) and cloned into pUNI-PROM, yielding pUNI-Tm-Fd6 and pUNI-Tm-POR, respectively. The pUNI-Tm-POR plasmid was further modified by the addition of the tm0927 gene, yielding a plasmid that would encode all five genes from Th. maritima (pUNI-Tm-POR-Fd).
All constructs made during this study were sequenced on both strands to ensure that no undesired mistakes had been introduced during the amplification procedure.
2.3. Protein methods
For gene product synthesis tests E. coli strain K38/pGP1-2 [53] was transformed with the required plasmid. Synthesis of plasmid-encoded gene products was induced by heat shock and followed by labelling with 35S-Methionine as described previously [53]. Samples were separated by SDS-PAGE (12% w/v acrylamide) after which gels were fixed in 5% (v/v) acetic acid, 10% (v/v) methanol, dried, and proteins visualised by autoradiography.
Isolation of the synthetic histidine-tagged hydrogenase complex was carried out by Immobilised Metal Affinity Chromatography (IMAC). 5 L of LB supplemented with 0.4% (w/v) glucose, 2 mM cysteine and 2 mM ferric ammonium citrate and antibiotics was inoculated and grown anaerobically at 37 °C for 16 h. All buffers used throughout purification were saturated with N2 to remove O2 and cell pellets, cell-containing buffers, or crude extracts were flushed with argon to protect from O2. Cells were harvested by centrifugation and pellets resuspended in either 50 ml of B-PER® solution (Thermo Scientific), which is a detergent-based cell lysis cocktail, or 50 ml of 50 mM Tris.HCl pH 7.5, 1 mM DTT, 2 mM flavin mononucleotide, 150 mM NaCl and 25 mM imidazole. B-PER lysis was achieved by the addition of lysozyme and DNAse I followed by agitation at room temperature for 1 h. Sonication was also preceded by the addition of lysozyme and DNAse I and the following conditions were used to lyse the cells using a 102-C sonication horn (Branson) and Digital 450 Digital Sonifier (Branson): 20% amplitude; 5 s pulse on/off; and lysis duration of 20 min (40 min total). Following either method of lysis, unbroken cells were removed by centrifugation and resultant crude extracts were applied to 5 ml HisTrap HP affinity columns (GE Healthcare) at a flow rate of 0.5 ml min−1. The columns had been previously equilibrated with N2-saturated Ni-purification buffer A (50 mM Tris.HCl pH 7.5, 1 mM DTT, 150 mM NaCl and 25 mM imidazole). A linear gradient of 0–100% buffer B (50 mM Tris.HCl pH 7.5, 1 mM DTT, 150 mM NaCl and 1 M imidazole) was then applied to the column to elute bound proteins.
Size exclusion chromatography coupled with multi-angle laser light scattering (SEC-MALLS) was performed using a Dionex Ultimate 3000HPLC system, a MAbPac SEC–1 (Dionex) column, an inline miniDAWN TREOS (Wyatt) multi-angle laser light scattering detector and a T-rEX (Optilab) refractive-index detector. After equilibration with 1.5 column volumes of SEC buffer (50 mM Tris.HCl pH 7.5, 150 mM NaCl), 500 μl of protein was applied to the column at a flow rate of 0.5 ml min−1. ASTRA v6.0.0.108 (Wyatt) software was used to determine molecular mass, polydispersity and radius of the enzyme complex.
SDS–PAGE was performed as described [33], and Western blotting was according to [54]. Monoclonal penta-His antibody was obtained from Qiagen. Protein identification was performed by tryptic peptide mass fingerprinting (Fingerprints Proteomics Service, University of Dundee).
2.4. Enzymatic assays
H2-dependent reduction of benzyl viologen (BV) was assayed by monitoring the reduction of BV at A600 as described [39]. H2-production using methyl viologen (MV) as electron donor was monitored in a Clark-type electrode modified to measure H2. Typically, 2 ml of anaerobic buffer (100 mM sodium phosphate pH 6.0 or 6.8) was added to the reaction chamber together with 12.5 mM MV and 650 μM sodium dithionite and allowed to equilibrate. The reaction was initiated by the addition of enzyme or cell extract and recorded at 37 °C.
2.5. Gas and metabolite quantification
Strains for H2 and metabolite analysis were grown anaerobically in either a supplemented M9 medium containing M9 salts [48], 2 mM MgSO4, 0.1 mM CaCl2, 0.2% (w/v) casamino acids, 3 μM thiamine hydrochloride, trace element solution SL-A [26], 0.8% (w/v) glucose or in TGYEP, pH 6.5 containing 0.8% (w/v) glucose [6].
In order to determine hydrogen content in the headspace of anaerobically grown cultures using gas chromatography, Hungate tubes were initially filled with 5 ml of medium and the headspace (approx. 10 ml) was flushed with nitrogen. After 17 h of growth 500 μl aliquots of the gas in the headspace was analysed on a Shimadzu GC-2014 gas chromatograph. Pure nitrogen was used as the carrier gas with a flow of 25 ml min−1, and the amount of hydrogen in the headspace was calculated based on a standard curve.
For organic acid analysis, the cell supernatants were passed through a 0.22 μM sterile filter and 10 μl applied to an Aminex HPX-87H (300 × 7.8 mm) ion exchange column. The flow was 0.5 ml min−1 at 50 °C and 5 mM sulfuric acid used as mobile phase on an Ultimate 3000 LC system. Organic acid retention peaks were recorded using Chromeleon 6.8 software (Dionex) and quantified by comparison with absorption of known amounts of standard of the organic acids.
3. Results
3.1. Design, construction and characterization of a synthetic [FeFe]-hydrogenase operon
The soluble, thermostable NADH-dependent [FeFe]-hydrogenase enzyme of Ca. subterranus [52] was chosen as a good candidate for a hydrogenase biased towards hydrogen production that uses a universal reductant (NADH) as a substrate. Although the Ca. subterranus enzyme would have a temperature optimum far above that of the normal growth conditions for E. coli, it was considered that any engineered enzyme could be further optimised following the initial characterisation. The constituent parts chosen to build a synthetic operon encoding this enzyme were the four proteins HydA-D as well as the hypothetical protein Tte0891, which is encoded within the native operon [52]. The primary amino acid sequences were back-translated into DNA sequence, codon optimised for E. coli using OPTIMIZER software [43], before a synthetic RBS and spacer sequence was included upstream of each synthetic gene: 5′-AGGAGGAAAAAAA-3′. This sequence, together with the sequence upstream of the RBS and spacer and the coding sequence itself, was then analysed using the RBS CALCULATOR software [47], which allows the efficiency of translation initiation to be predicted. The five synthetic sequences were then brought together to form a synthetic operon in which the natural gene-order was maintained (hydC, tte0891, hydD, hydB, hydA). Finally, unique restriction site sequences were chosen to separate each gene (Fig. 1A). The complete 5104 bp synthetic operon was then synthesised and cloned resulting in the vector pUNI-Tte-Hyd, which also contains a constitutive tat promoter (from E. coli) and a T7 promoter upstream of the synthetic genes (Table 1).
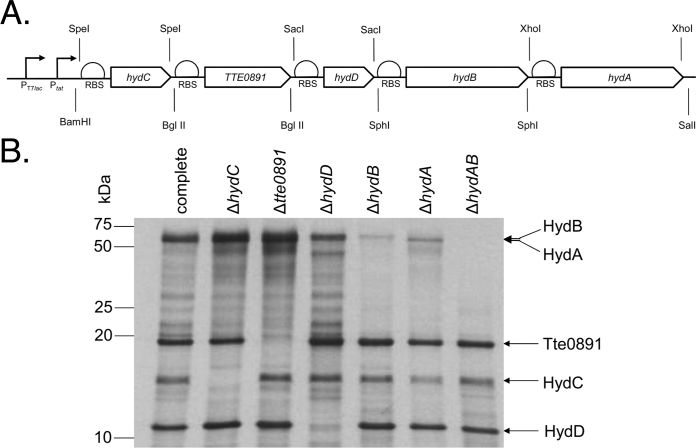
Fig. 1.
The products of a synthetic operon encoding an [FeFe]-hydrogenase are synthesised in E. coli. (A) The predicted structure of the synthetic operon encoding Ca. subterranus NADH-dependent [FeFe]-hydrogenase. Restriction sites and promoter regions are indicated. (B) The E. coli strain K38/pGP1-2 was transformed with plasmids: pUNI-Tte-Hyd (‘complete’); pUNI-Tte-HydΔC (‘ΔhydC’); pUNI-Tte-HydΔ0891 (‘Δtte0891’); pUNI-Tte-HydΔD (‘ΔhydD’); pUNI-Tte-HydΔB (‘ΔhydB’); pUNI-Tte-HydΔA (‘ΔhydA’); and pUNI-Tte-HydΔAB (‘ΔhydAB’), grown in M9 minimal medium lacking cysteine and methionine, and labelled by the addition of 35S-methionine. Protein samples were then separated by SDS–PAGE (12% w/v polyacrylamide), fixed, and visualised by autoradiography.
In order to validate that each gene in the synthetic operon was being correctly transcribed and translated the engineered restriction sites were used to further modify the pUNI-Tte-Hyd plasmid. A bank of six derivatives were constructed each carrying specific gene deletions in each of the five synthetic genes, as well as a ΔhydAB double deletion version (Table 1). The seven synthetic constructs were next used in 35S-methionine radiolabelling experiments. E. coli strain K38 (containing plasmid pGP1-2, a plasmid that encodes T7 polymerase) was transformed separately with pUNI-Tte-Hyd and the six deletion derivatives. Following pulse-labelling, SDS–PAGE and autoradiography protein products could be visualised and assigned to each gene product (Fig. 1B). In each case, the gene products migrated close to their theoretical mass by SDS–PAGE (Fig. 1B). This technique established that transcription and translation of this synthetic operon, the DNA sequence of which does not exist in nature, was possible in an E. coli host and results in the synthesis of apparently stable protein products.
3.2. Design, construction and optimization of a synthetic [FeFe] cofactor assembly operon
A set of accessory proteins is required in order to assemble the special ‘H-cluster’ found in the active site of [FeFe]-hydrogenases [8]. Most biological systems utilize the activity of three accessory proteins – HydE, -F, and -G. Both HydE and HydG are radical SAM (S-adenosyl methionine) enzymes, while HydF is predicted to be a GTPase that also has a scaffolding role for the immature cofactor [8]. Recently, the hydGXEF genes from Shewanella oneidensis have been utilised in heterologous production studies of an [FeFe]-hydrogenase in E. coli [32]. The γ-Proteobacterium Sh. oneidensis is closely related to E. coli but, unusually for this family of prokaryotes, it encodes a periplasmic [FeFe]-hydrogenase in its genome. Initially, the putative four-gene operon containing SO_3923 (hydG); SO_3924 (hydX); SO_3925 (hydE) and SO_3926 (hydF) was amplified by PCR from Sh. oneidensis genomic DNA and cloned directly to give pUNI-Sh-EFG and pSU-Sh-EFG (Table 1). To examine the translational efficiency of the cloned genes, the pUNI-Sh-EFG plasmid was then used in a radiolabelling experiment in E. coli (Fig. 2B). In this case, not all the predicted gene products could be confidently identified using this method (Fig. 2B). To improve the expression of all the necessary genes it was decided to clone both ‘halves’ of the operon (hydGX and hydEF) separately into the dual production vector pACYCDuet-1 (Fig. 2A and Table 1), where hydE would contain a synthetic RBS. The pACYCDuet-1 vector has two multiple cloning sites both under the control of separate PT7lac promoters and also encodes LacI in cis. Each half-operon was amplified by PCR and cloned with the initial gene in each half sharing the same RBS and spacer sequence (5′-AGGAGGAAAAAAA-3′) (Fig. 2A). The resultant plasmid, pDuet-Sh-GX-EF, was then analysed by 35S-Methionine radiolabelling experiments. In this case, all the three accessory proteins HydE, HydF and HydG were found to be transcribed and translated (Fig. 2B). Production of the HydX protein (23.9 kDa) was not detected (Fig. 2B).
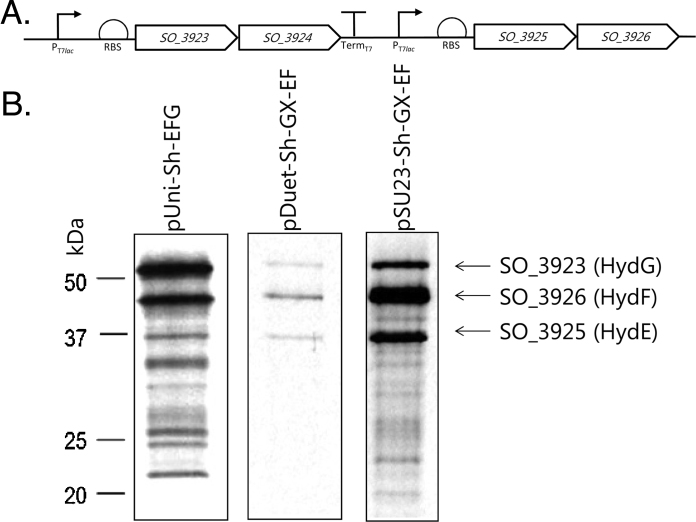
Fig. 2.
A construct for production of accessory genes required for [FeFe]-hydrogenase activity. (A) A construct encoding two bicistronic operons for Sh. oneidensis hydGX and hydEF was designed. The locations of promoters and engineered ribosome binding sites are shown. (B) The Sh. oneidensis hydG and hydEF genes are transcribed and translated. E. coli strain K38/pGP1-2 was transformed with plasmids: pUNI-Sh-EFG; pDuet-Sh-GX-EF; and pSU23-Sh-GX-EF and cultured in M9 minimal medium lacking cysteine and methionine and, where appropriate supplemented with 1 mM IPTG to de-repress LacI encoded on the pDuet-Sh-GX-EF plasmid. Cells were pulse-labelled with 35S-methionine and protein samples were then separated by SDS–PAGE (14% w/v polyacrylamide), fixed, and visualised by autoradiography.
Next, the entire PT7lac-RBS–spacer-hydGX- PT7lac-hydEF DNA fragment from pDuet-Sh-GX-EF was subcloned into pSU23 [5], which, unlike pACYC-Duet, does not encode the LacI repressor. This lead to increased production levels relative to pDuet-Sh-GX-EF as observed by radiolabelling experiments (Fig. 2B). Finally, as repression or careful induction of production of S. oneidensis operon was not thought to be required, the E. coli tat promoter (Ptat) was introduced upstream of the first gene to yield pSUtat-Sh-GX-EF. This removed the need to co-produce with T7 polymerase.
3.3. Purification and characterization of a synthetic hydrogenase complex
To facilitate in vitro characterisation of the [FeFe]-hydrogenase, the plasmid pUNI-Tte-HydhisC was constructed, which is identical to the pUNI-Tte-Hyd vector except that it encodes HydCHis. The E. coli strain PK4854 (as MG1655 ΔiscR) was chosen as the host strain since it is de-regulated for Fe-S cluster assembly [32]. PK4854 (ΔiscR) was co-transformed with pSUtat-Sh-GX-EF and pUNI-Tte-HydhisC and grown anaerobically with additional glucose, cysteine and ferric ammonium citrate. Nitrogen-saturated buffers were used during an initial immobilized metal affinity chromatography (IMAC) step. The eluted fractions exhibited a deep brown colour, and the UV/Vis absorption spectrum pointed to the presence of Fe-S clusters (Supp. Fig. S1). Analysis of the eluted peak fractions by SDS–PAGE revealed several proteins of the expected molecular masses of the NADH-dependent [FeFe]-hydrogenase (Fig. 3A). The identity of HydA, HydB and HydD was established by tryptic peptide mass-fingerprinting. The His-tagged HydC protein was located by Western immunoblot (Fig. 3B).
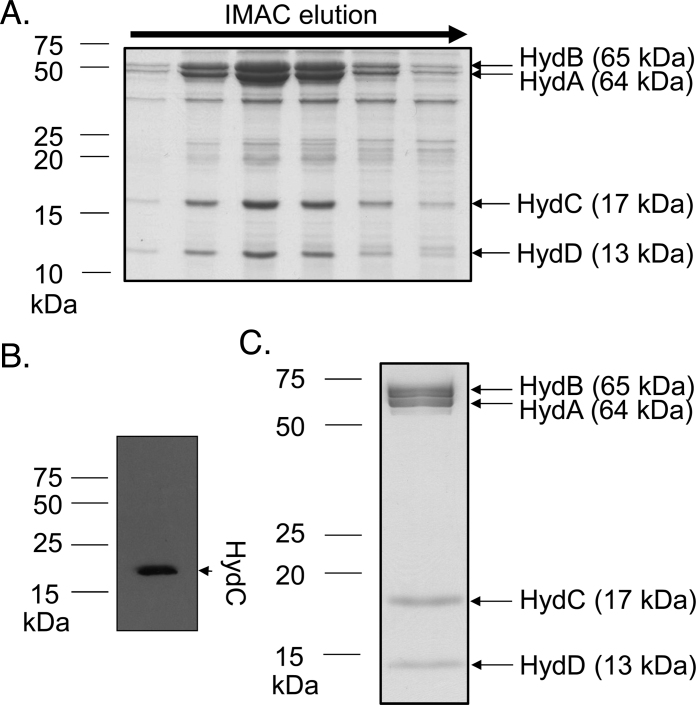
Fig. 3.
Isolation of a recombinant [FeFe]-hydrogenase. (A) E. coli strain PK4854 (ΔiscR) was transformed with plasmids: pUNI-Tte-HydhisC and pSUtat-Sh-GX-EF and cultured in LB supplemented with 0.4% (w/v) glucose, 2 mM cysteine, 2 mM ferric ammonium citrate. Cells were harvested and lysed by sonication. Crude cell extract was loaded onto a HisTrap™ HP column and eluted by an imidazole gradient and fractions (‘IMAC elution’) were collected and separated by SDS–PAGE (14% w/v acrylamide). Each subunit was identified using tryptic peptide mass fingerprinting. (B) Identification of HydCHis by Western immunoblotting. (C) SDS–PAGE analysis of the synthetic enzyme following SEC-MALLS. The peak fraction from SEC-MALLS analysis was collected, concentrated and separated by SDS-PAGE (12% w/v polyacrylamide), followed by staining with Instant Blue™.
To assess the molecular mass of the [FeFe]-hydrogenase complex Size-Exclusion Chromatography–Multi-Angle Laser Light Scattering (SEC-MALLS) experiments were carried out on the purified enzyme. Upon elution from the SEC-MALLS column the purified enzyme appeared to form a stable large monodisperse complex containing each of the four subunits (established by SDS–PAGE; Fig. 3C) with an apparent molecular weight of 325 (±0.1%) kDa and a hydrodynamic radius of 11.9 (±3.3%) nm.
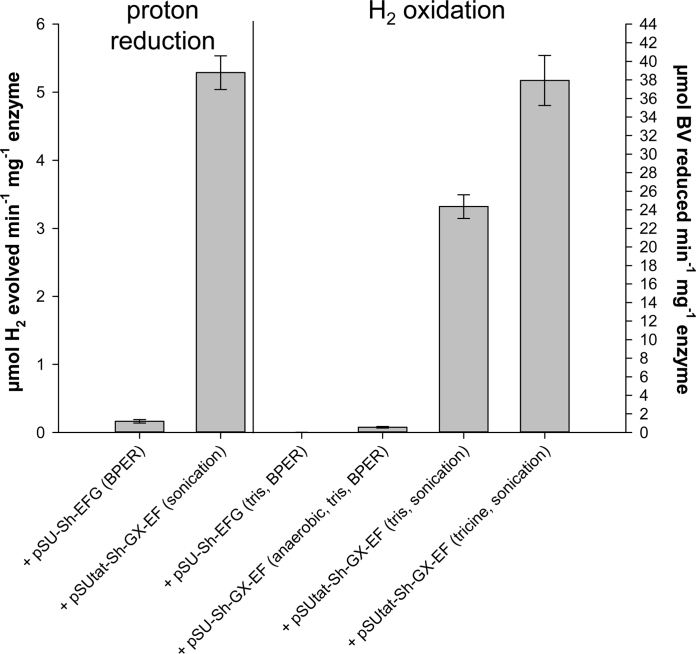
For hydrogen oxidation assays benzyl viologen (BV) was chosen as the electron acceptor because its standard reduction potential (E0′ −348 mV) is more positive than that of the H+/½ H2 redox couple (E0′−420 mV). The His-tagged enzyme purified from cells co-producing pSUtat-Sh-GX-EF clearly catalysed the reduction of BV with H2 as the electron donor at 37 °C (Fig. 4). Surprisingly, enzyme prepared from cells lysed by sonication displayed BV-linked activity >40 times greater than that isolated using a detergent-based chemical cocktail (Fig. 4).
Fig. 4.
Purified synthetic [FeFe]-hydrogenase displays hydrogenase activity in vitro. E. coli strain PK4854 was co-transformed with pUNI-Tte-Hydhisc and one of three accessory plasmids: pSU-Sh-EFG; pSU23-Sh-GX-EF; or pSUtat-Sh-GX-EF, as indicated. Harvested cells were lysed either by sonication or using a chemical cocktail (BPER, Thermo Scientific) as indicated and enzyme isolated by IMAC. ‘Proton reduction’ activity involved methyl viologen-dependent H2 production measured in a modified Clark-type electrode. The reaction was initiated by the addition of 10 μg of purified enzyme. ‘H2 oxidation’ assays involved H2-dependent benzyl viologen reduction monitored at 578 nm in a UV-vis spectrometer. The reaction was started by the addition of 20 μg of purified enzyme and recorded at 37 °C. Error bars represent the standard error of three independent experiments.
For hydrogen evolution experiments methyl viologen (MV) was chosen as the electron donor (E0′ = −443 mV). The His-tagged [FeFe]-hydrogenase purified using the sonication method demonstrated H2-evolution activity with reduced MV as the artificial electron donor (Fig. 4). Again, performing cell lysis with a chemical cocktail apparently inactivated the enzyme (Fig. 4).
3.4. Towards an engineered strain for Bio-H2 production
Having established that the [FeFe]-hydrogenase could be assembled in E. coli, the next step was to move away from antibiotic resistance-encoding multicopy plasmids and integrate the synthetic operon into the chromosome. The E. coli strain FTD147 (ΔhyaB, ΔhybC, ΔhycE) was chosen as a host since it contains no endogenous hydrogenase activity [44]. Using homologous recombination, the precise replacement of the adhE gene by the synthetic [FeFe]-hydrogenase operon was achieved and the new strain was called FTD147h3 (Table 1). In this strain the synthetic operon retains the constitutive tat promoter, but is also correctly positioned to be driven by the native adhE promoters.
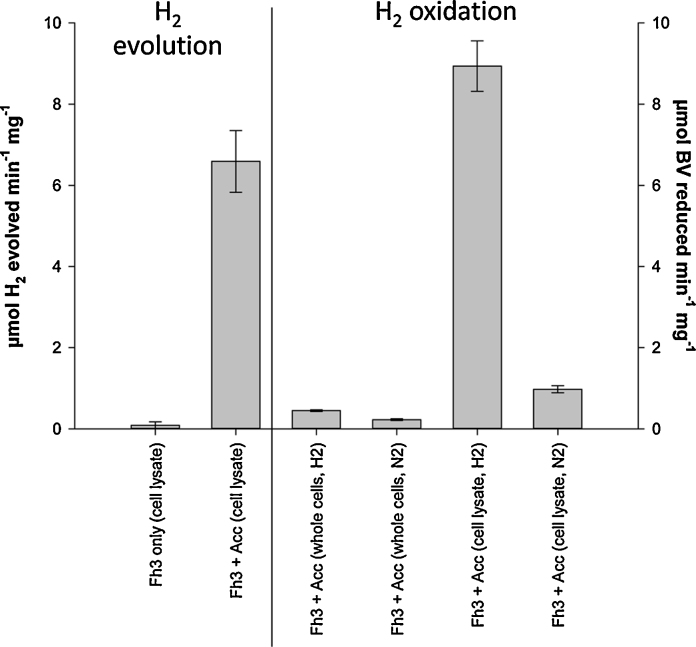
To establish whether the [FeFe]-hydrogenase was produced and active in FTD147h3 under physiological conditions for the E. coli host, the strain was transformed with pSUtat-Sh-GX-EF and anaerobic cultures prepared that had been supplemented with 0.4% (w/v) glucose, 2 mM cysteine and 2 mM ferric ammonium citrate. Cell pellets were flushed with argon throughout to maintain anaerobic conditions and cells were lysed by sonication. The FTD147h3 + pSUtat-Sh-GX-EF crude cell extract catalysed the reduction of BV with H2 as the electron donor at 37 °C (Fig. 5B). Assays were repeated in N2-saturated buffer to confirm H2-specific BV reduction (Fig. 5B). Intact whole cells were also used as negative controls (Fig. 5B), as oxidised BV is probably impermeable to the inner membrane [17]. The FTD147h3 + pSUtat-Sh-GX-EF crude cell extract also demonstrated H2-evolution activity with reduced MV as the artificial electron donor at 37 °C, and this activity was dependent upon co-production of the Sh. oneidensis hydEXFG accessory genes (Fig. 5A).
Fig. 5.
The engineered FTD147h3 strain displays hydrogenase activity. FTD147h3 (‘Fh3’) alone, or the strain transformed with pSUtat-Sh-GX-EF encoding accessory genes (‘Acc’), was cultured anaerobically in 0.4% (w/v) glucose, 2 mM cysteine, 2 mM ferric ammonium citrate. Cells were lysed by sonication resulting in a crude cell lysate, which was assayed for hydrogenase activity. ‘H2 evolution’ activity refers to methyl viologen-dependent H2 production measured in a modified Clark-type electrode. ‘H2 oxidation’ activity refers to H2-dependent benzyl viologen reduction monitored at 578 nm. N2-saturated buffer (‘N2’) and unbroken intact cells (‘whole cells’) were used as controls. Error bars represent the standard error of three independent experiments.
Having established that the chromosomally-encoded [FeFe]-hydrogenase is assembled and active in E. coli the next step was to assess Bio-H2 production in vivo. The FTD147h3 + pSUtat-Sh-GX-EF was grown anaerobically in rich medium. The culture headspace was then tested for the presence of H2 using gas chromatography, however no H2 production was observed.
4. Genetic evidence for an electron-confurcating mechanism for the Ca. subterranus [FeFe]-hydrogenase
Although originally reported as being able to utilise NADH as the sole electron donor for H2 production [52], the subunits of the Ca. subterranus [FeFe]-hydrogenase complex under investigation here share sequence identity with the Th. maritima [FeFe]-hydrogenase, which exhibits an electron-confurcating mechanism of hydrogen production involving reduced ferredoxin in combination with NADH as electron donors [51]. In the case of the Th. maritima system, reduced ferredoxin is a product of the oxidation of pyruvate (Em −500 mV) catalysed by pyruvate ferredoxin oxidoreductase (POR):
| pyruvate + CoA + 2Fdox ↔ acetyl-CoA + CO2 + H+ + 2Fdred |
Therefore, given the increasing body of research highlighting bifurcating or confurcating mechanisms for cytoplasmic [FeFe]-hydrogenases [9], [27], [45], [50], it was considered possible that the Ca. subterranus [FeFe]-hydrogenase under experimentation here was also an electron-bifurcating/confurcating enzyme.
To address this hypothesis directly, the Th. maritima gene tm0927 (encoding a ferredoxin) and the tm0015-tm0018 operon (encoding the γ, δ, α and β subunits of a pyruvate-ferredoxin oxidoreductase) [31] were cloned both separately and together in production vectors. Successful heterologous production of the Th. maritima ferredoxin and the POR subunits was established using 35S-methionine radiolabelling (Supp. Fig. S2).
Next, the FTD147h3 strain was co-transformed with a plasmid encoding accessory genes (pSUtat-Sh-GX-EF) and a plasmid encoding Th. maritima POR and ferredoxin (pUNI-Tm-POR-Fd). The strain was grown anaerobically in a supplemented minimal medium containing casamino acids and 0.8% (w/v) glucose. A minimal medium was chosen to allow further analysis of metabolites in the growth medium by HPLC, however supplementation of the medium with casamino acids was required to obtain some growth in the absence of adhE. Following incubation for 17 h the culture headspace was assayed for the presence of H2 by gas chromatography (Table 2). Compared to the control strains that do not produce H2, co-production of POR and Fd with the activated [FeFe]-hydrogenase in FTD147h3 increased in vivo H2 production to detectable levels, reaching 7.4 ± 4.1 nmols H2 OD600−1 ml−1 (Table 2). Please note that this is a single-end-point assay that determines the total amount of H2 in the culture headspace following growth. The values are normalised for cell growth and culture size to allow for comparison between different strains, however it is not possible to determine biochemical reaction rates from such a single-point assay.
Table 2.
Hydrogen production and metabolite analysis after anaerobic growth in supplemented M9 media.
| straina |
Relevant genotype |
hydrogenb |
pyruvatec |
succinatec |
lactatec |
formatec |
acetatec |
OD600nm |
|---|---|---|---|---|---|---|---|---|
| nmol OD600 nm−1 ml culture−1 | mM OD600−1 | |||||||
| MC4100 | – | 8232 ± 620 | 0.62 ± 0.02 | 3.9 ± 0.2 | 5.9 ± 0.2 | 10 ± 1.3 | 15 ± 0.4 | 1.25 |
| FTD147 | ΔhyaB, ΔhybC, ΔhycE | <1 | 0.41 ± 0.04 | 2.2 ± 0.1 | 6.9 ± 0.4 | 21 ± 2.2 | 13 ± 0.3 | 1.30 |
| Teatotal1 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE | <1 | 0.05 ± 0.01 | 2.5 ± 0.1 | 2.9 ± 0.0 | 3.8 ± 0.1 | 12 ± 0.5 | 0.49 |
| FTD147h3 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD | <1 | 0.03 ± 0.03 | 2.9 ± 0.5 | 3.7 ± 0.8 | 3.6 ± 0.6 | 11 ± 0.5 | 0.47 |
| FTD147h3 + pd | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD | 7.4 ± 4.1 | 0.2 ± 0.17 | 3.2 ± 0.9 | 5.8 ± 1.4 | 6.1 ± 2.6 | 8.2 ± 0.4 | 0.34 |
| CPD159F | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, pflA | <1 | 0.68 ± 0.03 | 2.4 ± 0.1 | 26 ± 0.1 | 1.2 ± 0.3 | 4 ± 0.1 | 1.03 |
| CPD159F + pd | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, pflA | 5.5 ± 1.5 | 0.73 ± 0.03 | 2.2 ± 0.2 | 24 ± 3 | 3.3 ± 0.5 | 3.3 ± 0.0 | 0.81 |
| CLK001 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR | <1 | 0.07 ± 0.05 | 2.9 ± 0.3 | 4.7 ± 0.3 | 3.3 ± 0.4 | 9.5 ± 0.4 | 0.45 |
| CLK001 + pd | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR | 9.0 ± 4.0 | 0.06 ± 0.03 | 2.7 ± 0.2 | 5.3 ± 1.2 | 4 ± 0.4 | 7.8 ± 0.1 | 0.49 |
| CPD152E | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR, ΔpflA | <1 | 1 ± 0.17 | 3 ± 0 | 27 ± 0.8 | 1.3 ± 0.2 | 4.4 ± 0.3 | 0.97 |
| CPD152E + pd | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR, ΔpflA | 7.9 ± 0.2 | n.d. | n.d. | n.d. | n.d. | n.d. | 0.77 |
The mean and standard deviation of at least three independent measurements are shown.
n.d.—not determined.
Cultures were grown anaerobically in supplemented M9 in closed Hungate tubes at 37 °C for 17 h.
H2 content was determined from headspace gas phase by gas chromatography.
Metabolites were determined from cell-free culture supernatants by HPLC.
‘+p’ denotes addition of accessory genes (pSUtat-Sh-GX-EF) and a plasmid encoding Th. maritima POR and ferredoxin (pUNI-Tm-POR-Fd).
In an attempt to increase this amount of H2 production a mutation in pflA was incorporated into the FTD147h3 strain to give CPD159F (Table 1). PflA is the pyruvate formatelyase (PflB) activating enzyme, which is required for anaerobic metabolism of pyruvate to generate formate and acetyl CoA. Here, the hypothesis was that prevention of disproportionation of pyruvate by the natural route should force more pyruvate towards POR and the synthetic hydrogenase system. Unfortunately, H2 production was not further boosted by adding a pflA mutation (Table 2), instead the mutation has the effect of inducing lactate production in the growth medium as assayed by HPLC, presumably by induction or activation of lactate dehydrogenases (Table 2).
Next, a version of FTD147h3 was prepared that was devoid of the iscR gene. IscR is the key regulator (repressor) of genes required for Fe-S cluster biosynthesis and some other metalloenzymes [21]. The genetic inactivation of iscR de-regulates the Fe-S cluster assembly machinery and, as a result, the new CLK001 strain (Table 1) would be expected to be more efficient at Fe-S protein biosynthesis [32]. The CLK001 strain was co-transformed with a plasmid encoding accessory genes (pSUtat-Sh-GX-EF) and a plasmid encoding Th. maritima POR and ferredoxin (pUNI-Tm-POR-Fd) and the culture headspace assayed for H2 gas following growth in casamino acid-supplemented minimal medium (Table 2). Again, compared to control strains, co-production of POR and Fd in CLK001 increased the amount of H2 produced in vivo to detectable levels, reaching 9.0 ± 4.0 nmols H2 OD600−1 ml−1 (Table 2), which is not significantly different to the H2 levels recorded for the FTD147h3 strain (Table 2). Further modification of CLK001 by incorporation on a pflA mutation to yield strain CPD152E (Table 1) did not increase the final amount of H2 produced (Table 2) but again led to an increase in lactate found secreted into the growth medium (Table 2).
Finally, the four engineered strains were transformed with the accessory plasmid and the plasmid encoding POR and ferredoxin before being cultured anaerobically in the rich medium TGYEP (Table 3). Growth in such rich medium means that metabolite quantification of the spent media by HPLC is not practical, however headspace analysis by gas chromatography was still possible (Table 3). In this case, all of the engineered strains ceased H2 production except those carrying an iscR modification (Table 3). Here, co-production of POR and Fd in CLK001 resulted in the amount of H2 produced in vivo elevated to 28.7 ± 5.1 nmols H2 OD600−1 ml−1 (Table 3).
Table 3.
Hydrogen production after anaerobic growth in rich media
| straina |
genotype |
hydrogenb |
OD600nm |
|---|---|---|---|
| nmol OD600−1 ml−1 | |||
| MC4100 | – | 5394 ± 190 | 1.71 |
| FTD147 | ΔhyaB, ΔhybC, ΔhycE | <1 | 1.87 |
| Teatotal1 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE | <1 | 1.00 |
| FTD147h3 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD | <1 | 0.40 |
| FTD147h3 + plasmidsc | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD | <1 | 0.59 |
| CPD159F | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, pflA | <1 | 1.70 |
| CPD159F + plasmidsc | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, pflA | <1 | 1.53 |
| CLK001 | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR | <1 | 0.63 |
| CLK001 + plasmidsc | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR | 28.7 ± 5.1 | 0.48 |
| CPD152E | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR, ΔpflA | <1 | 1.59 |
| CPD152E + plasmidsc | ΔhyaB, ΔhybC, ΔhycE, ΔadhE::hydABCD, ΔiscR, ΔpflA | 8.4 ± 3.3 | 1.35 |
The mean and standard deviation of at least three independent measurements are shown.
Cultures were grown anaerobically in TGYEP, pH 6.5 in closed Hungate tubes at 37 °C for 17 h.
H2 content was determined from headspace gas phase by gas chromatography.
denotes addition of accessory genes (pSUtat-Sh-GX-EF) and a plasmid encoding Th. maritima POR and ferredoxin (pUNI-Tm-POR-Fd).
5. Discussion
5.1. Characterization of a synthetic [FeFe]-hydrogenase.
In this work, a synthetic approach to molecular genetics has been partnered with a more traditional biochemical characterization of the products of those synthetic genes. A synthetic operon was designed, which does not exist in nature, and shown to encode all expected polypeptides in radiolabelling experiments. In turn, those polypeptides assemble into an active [FeFe]-hydrogenase when co-expressed with accessory genes from Sh. oneidensis. In the course of this work, attempts were made to utilise homologs of the hydEFG genes from Ca. subterranus instead. This was complicated by the fact that the Ca. subterranus genome does not contain an obvious hydEFG operon. Instead four separate genes were identified including tte1885 (encoding a protein most like HydE from C. acetylicum); tte0814 (encoding a GTPase most like HydF from C. acetylicum); tte1568 (most like HydG from C. acetylicum); and tte1073 (most like the structurally-defined HydE from Th. maritima, but also related to HydG from C. actylicum and Tte1885). A synthetic operon encoding these gene products was designed and assembled, however none of the polypeptides were expressed and so no [FeFe]-hydrogenase activation was observed.
Using the Sh. Oneidensis biosynthetic machinery allowed the isolation of enzyme that had hydrogen oxidation and proton reduction activities with redox dyes, and with a mass of 325 kDa the enzyme was demonstrated to adopt a (HydABCD)2 conformation. Moreover, the synthetic operon encoding the enzyme could be stably integrated into the E. coli chromosome, thus taking the first step towards generating a synthetic strain freed from mobile genetic elements carrying antibiotic resistance.
5.2. Re-wiring E. coli metabolism for H2 production.
Engineering bacterial hydrogen production has been a long-standing aim for some microbial biotechnologists. Common approaches have been to direct natural electron and carbon flux towards the native hydrogenases of E. coli by genetic and metabolic engineering [37], [40], [55]. In addition, overproduction of heterologous hydrogenases in E. coli has also been attempted with various degrees of success. For example, simple production of the genes encoding the large and small subunits of the Tat-dependent, O2-tolerant, respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus was sufficient to increase H2 production [29], even though the source of reductant was not clear and the metallocofactor-deficient BL21 strain was used as a chassis [41].
The deliberate connection of a hydrogenase directly to a source of electrons, and in particular a source of reductant involved in central energy metabolism, is an attractive proposition. In this work, the NADH pool was chosen since recycling on NAD+ is critical for fermentative growth and NADH biochemistry is linked directly to H2 metabolism in many microbial systems. The cytoplasmic concentration of NADH cofactor is 1.4 nmoles/mg dry weight of E. coli grown under anaerobic fermentative conditions, and NAD+ is 4.8 nmoles/mg dry weight [34], [57]. Under anaerobic fermentative conditions this ratio is maintained primarily by the action of AdhE in E. coli [57]. In the absence of active AdhE, the synthetic [FeFe]-hydrogenase designed in this work was unable to generate hydrogen. This was initially surprising since in an earlier study the isolated native enzyme was able to produce H2 in vitro with NADH as electron donor [52]. Moreover, the soluble NADH-dependent [NiFe]-hydrogenase from Ralstonia eutropha was, following engineering, found to be activatable in E. coli and was successful in generating increased H2 in an adhE mutant [20]. In comparison, these data suggest that the synthetic [FeFe]-hydrogenase under investigation here was not operating at all efficiently as a stand-alone NADH-linked hydrogenase, and may be lacking in some vital components for its activity in vivo.
5.3. Heterologous production of a putative electron-confurcating hydrogenase for engineering H2 production
In the original Ca. subterranus hydrogenase characterisation [52], 1 mM Ti(III) citrate was added during the NADH-dependent H2 production assays [52]. Ti(III) citrate is a powerful reducing agent (E0′ = -500 mV), and this perhaps gives an initial hint that an additional source of electrons was needed for this enzyme to operate correctly. However, it should be noted that the enzyme was capable of the reverse reaction, H2-dependent reduction of NAD(P)+, without a second electron acceptor present [52], although this reaction would be thermodynamically favourable under the conditions used. Moreover, Schut and Adams [51] characterised the NADH-linked [FeFe]-hydrogenase from Th. maritima, which is an enzyme closely related to the Ca. subterranus hydrogenase at the amino acid level. In Th. maritima the [FeFe]-hydrogenase accepts a second source of electrons, this time from a high potential reduced ferredoxin (Em = −453 mV) [51], that acts as a thermodynamic driver thus allowing oxidation of NADH linked to H2 production [51].
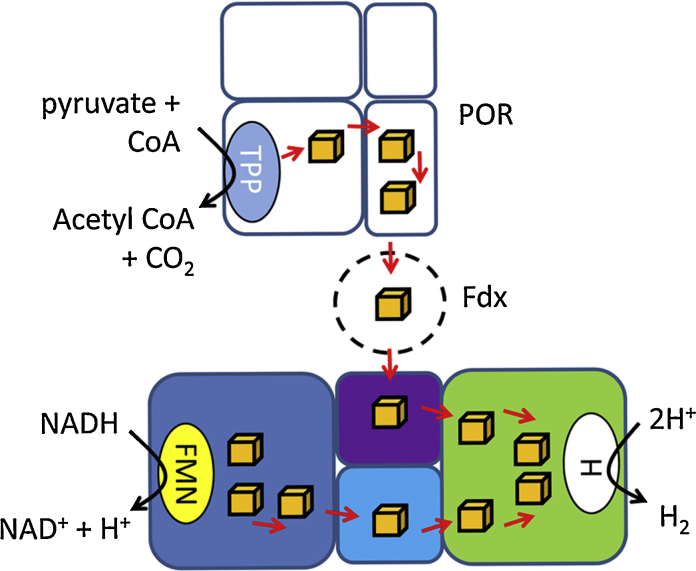
The possibility that the synthetic Ca. subterranus enzyme studied here also required a second input of electrons was tested directly here using a genetics-based in vivo approach. The genes encoding Th. maritima POR and ferredoxin were cloned onto a single vector and co-produced with the synthetic Ca. subterranus enzyme in E. coli. The engineered strain generated a small amount of H2 that could be increased by the addition of an iscR mutation (Table 2, Table 3). The amounts of hydrogen produced are small compared to what native E. coli can produce under similar growth conditions (Table 2, Table 3). For example, in rich media the essentially wild-type control strain produced around 5 μM H2 per OD unit per ml of culture (Table 3), which is around 180 times more than the engineered system. These results should be considered as proof-of-concept, highlighting that complex electron bifurcating systems can be engineered, but given the low amounts of H2 produced it is clear that the enzymes and strains involved will need further optimization to provide increased levels of Bio-H2. What these data clearly show is that the synthetic Ca. subterranus enzyme is most likely an electron confurcating/bifurcating system in vivo (Fig. 6), and this adds fresh backing to the original hypothesis on the physiological role of this enzyme in its native system [51]. While pyruvate oxidation has been linked directly to H2 production in a metallocofactor-deficient E. coli host previously [3], [41], this is, to our knowledge, the first synthetic engineering of an active electron confurcating/bifurcating [FeFe]-hydrogenase in a heterologous host. This system also allows a genetic system for the characterisation of such complex [FeFe]-hydrogenases, which may be of use to many other biological systems. The ability to connect additional H2-producing capabilities directly to central energy, carbon and cofactor metabolism may have the potential to be harnessed in future bioenergy research projects.
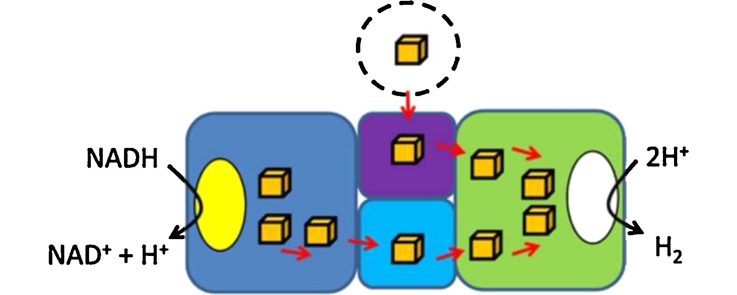
Fig. 6.
Hydrogen production by electron confurcation. Hypothetical model depicting the H2 production system engineered into E. coli based on collated evidence from several biological systems. A blue oval depicts the presence of a thiamine pyrophosphate (TPP) cofactor, the fellow oval represents flavin mononucleotide (FMN) and the white oval the ‘H’-cluster. Iron-sulfur clusters are represented by brown cubes. The hypothetical direction of electron flow is suggested by red arrows.
Acknowledgements
We thank Dr Florian Hauser and Dr Jen McDowall for performing some preliminary work on this project. Thanks also go to Dr Colin Hammond for assistance with SEC-MALLS and we acknowledge the expertise of the Fingerprints Proteomics Service at Dundee. The research was supported in part by the Northern Research Partnership (comprising the University of Dundee, Robert Gordon University and the University of Aberdeen) and in part through the Biotechnology and Biological Sciences Research Council awards BB/C516195/2, BB/H001190/1, BB/I02008X/1, BB/H003878-1 and BB/I022309-1, and PhD studentship funding to the University of Dundee. FAA and TP are recipients of Royal Society Wolfson Research Merit Awards.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2015.10.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Adams M.W. The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta. 1990;1020:115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- 2.Agapakis C.M., Ducat D.C., Boyle P.M., Wintermute E.H., Way J.C., Silver P.A. Insulation of a synthetic hydrogen metabolism circuit in bacteria. J. Biol. Eng. 2010;4:3. doi: 10.1186/1754-1611-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhtar M.K., Jones P.R. Construction of a synthetic YdbK-dependent pyruvate:H2 pathway in E. coli BL21(DE3) Metab. Eng. 2009;11:139–147. doi: 10.1016/j.ymben.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartolome B., Jubete Y., Martinez E., de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 6.Begg Y.A., Whyte J.N., Haddock B.A. Identification of mutants of Escherichia coli deficient in formate dehydrogenase and nitrate reductase activities using dye indicator plates. FEMS Microbiol Lett. 1977;2:47–50. [Google Scholar]
- 7.Benemann J. Hydrogen biotechnology: progress and prospects. Nat. Biotechnol. 1996;14:1101–1103. doi: 10.1038/nbt0996-1101. [DOI] [PubMed] [Google Scholar]
- 8.Broderick J.B., Byer A.S., Duschene K.S., Duffus B.R., Betz J.N., Shepard E.M., Peters J.W. H-Cluster assembly during maturation of the [FeFe]-hydrogenase. J. Biol. Inorg. Chem. 2014;19:747–757. doi: 10.1007/s00775-014-1168-8. [DOI] [PubMed] [Google Scholar]
- 9.Buckel W., Thauer R.K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim. Biophys. Acta. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorf T., Lenz O., Buhrke T., van der Linden E., Jones A.K., Albracht S.P., Friedrich B. [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 2005;10:181–196. doi: 10.1159/000091564. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban M.J., Cohen S.N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. (U.S.A.) 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casalot L., Rousset M. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 2001;9:228–237. doi: 10.1016/s0966-842x(01)02009-1. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham P.R., Clark D.P. The use of suicide substrates to select mutants of Escherichia coli lacking enzymes of alcohol fermentation. Mol. Gen. Genet. 1986;205:487–493. doi: 10.1007/BF00338087. [DOI] [PubMed] [Google Scholar]
- 14.Davila-Vazquez G., Arriaga S., Alatriste-Mondragón F., León-Rodríguez A., Rosales-Colunga L., Razo-Flores E. Fermentative biohydrogen production: trends and perspectives. Rev. Environ. Sci. Biotechnol. 2008;7:27–45. [Google Scholar]
- 15.de Graef M.R., Alexeeva S., Snoep J.L., Teixeira de Mattos M.J. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deplanche K., Calderari I., Mikheenko I.M., Sargent F., Macaskie L.E. Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia colimutant strains. Microbiology. 2010;156:2630–2640. doi: 10.1099/mic.0.036681-0. [DOI] [PubMed] [Google Scholar]
- 17.Dubini A., Pye R.L., Jack R.L., Palmer T., Sargent F. How bacteria get energy from hydrogen: a genetic analysis of periplasmic hydrogen oxidation in Escherichia coli. Int. J. Hydrogen Energy. 2002;27:1420–1423. [Google Scholar]
- 18.Fontecilla-Camps J.C., Volbeda A., Cavazza C., Nicolet Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 2007;107:4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 19.Frey M. Hydrogenases: hydrogen-activating enzymes. Chembiochem. 2002;3:153–160. doi: 10.1002/1439-7633(20020301)3:2/3<153::AID-CBIC153>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh D., Bisaillon A., Hallenbeck P.C. Increasing the metabolic capacity of Escherichia coli for hydrogen production through heterologous production of the Ralstonia eutropha SH operon. Biotechnol. Biofuels. 2013;6:122. doi: 10.1186/1754-6834-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giel J.L., Rodionov D., Liu M., Blattner F.R., Kiley P.J. IscR-dependent gene production links iron-sulfur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 22.Goodlove P.E., Cunningham P.R., Parker J., Clark D.P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 23.Goryshin I.Y., Jendrisak J., Hoffman L.M., Meis R., Reznikoff W.S. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 2000;18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 24.Gupta S., Clark D.P. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J. Bacteriol. 1989;171:3650–3655. doi: 10.1128/jb.171.7.3650-3655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton C.M., Aldea M., Washburn B.K., Babitzke P., Kushner S.R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hormann K., Andreesen J.R. Reductive cleavage of sarcosine and betaine by eubacterium-Acidaminophilum via enzyme-systems different from glycine reductase. Arch. Microbiol. 1989;153:50–59. [Google Scholar]
- 27.Huang H., Wang S., Moll J., Thauer R.K. Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J. Bacteriol. 2012;194:3689–3699. doi: 10.1128/JB.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack R.L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., Sargent F. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004;23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J.Y., Jo B.H., Cha H.J. Production of biohydrogen by heterologous production of oxygen-tolerant Hydrogenovibrio marinus [NiFe]-hydrogenase in Escherichia coli. J. Biotechnol. 2011;155:312–319. doi: 10.1016/j.jbiotec.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 30.King P.W., Posewitz M.C., Ghirardi M.L., Seibert M. Functional studies of [FeFe] hydrogenase maturation in an Escherichia colibiosynthetic system. J. Bacteriol. 2006;188:2163–2172. doi: 10.1128/JB.188.6.2163-2172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kletzin A., Adams M.W. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchenreuther J.M., Grady-Smith C.S., Bingham A.S., George S.J., Cramer S.P., Swartz J.R. High-yield production of heterologous [FeFe] hydrogenases in Escherichia coli. PloS one. 2010;5:e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.London J., Knight M. Concentrations of nicotinamide nucleotide coenzymes in micro-organisms. J. Gen. Microbiol. 1966;44:241–254. doi: 10.1099/00221287-44-2-241. [DOI] [PubMed] [Google Scholar]
- 35.Lorowitz W., Clark D. Escherichia colimutants with a temperature-sensitive alcohol dehydrogenase. J. Bacteriol. 1982;152:935–938. doi: 10.1128/jb.152.2.935-938.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons L.B., Zinder N.D. Genetic map of filamentous bacteriophage fl. Virology. 1972;49:45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- 37.Maeda T., Sanchez-Torres V., Wood T.K. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007;77:879–890. doi: 10.1007/s00253-007-1217-0. [DOI] [PubMed] [Google Scholar]
- 38.Manish S., Banerjee R. Comparison of biohydrogen production processes. Int. J. Hydrogen Energy. 2008;33:279–286. [Google Scholar]
- 39.Palmer T., Berks B.C., Sargent F. Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli. Methods Mol. Biol. 2010;619:191–216. doi: 10.1007/978-1-60327-412-8_12. [DOI] [PubMed] [Google Scholar]
- 40.Penfold D.W., Sargent F., Macaskie L.E. Inactivation of the Escherichia coliK-12 twin-arginine translocation system promotes increased hydrogen production. FEMS Microbiol. Lett. 2006;262:135–137. doi: 10.1111/j.1574-6968.2006.00333.x. [DOI] [PubMed] [Google Scholar]
- 41.Pinske C., Bonn M., Kruger S., Lindenstrauss U., Sawers R.G. Metabolic deficiences revealed in the biotechnologically important model bacterium Escherichia coliBL21(DE3) PLoS One. 2011;6:e22830. doi: 10.1371/journal.pone.0022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posewitz M.C., King P.W., Smolinski S.L., Zhang L., Seibert M., Ghirardi M.L. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem. 2004;279:25711–25720. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 43.Puigbo P., Guzman E., Romeu A., Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redwood M.D., Mikheenko I.P., Sargent F., Macaskie L.E. Dissecting the roles of Escherichia colihydrogenases in biohydrogen production. FEMS Microbiol. Lett. 2008;278:48–55. doi: 10.1111/j.1574-6968.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- 45.Rydzak T., McQueen P.D., Krokhin O.V., Spicer V., Ezzati P., Dwivedi R.C., Shamshurin D., Levin D.B., Wilkins J.A., Sparling R. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein production profiles and growth phase-dependent changes in protein production. BMC Microbiol. 2012;12 doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salis H.M. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- 47.Salis H.M., Mirsky E.A., Voigt C.A. Automated design of synthetic ribosome binding sites to control protein production. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 49.Schink B. Electron confurcation in anaerobic lactate oxidation. Env. Microbiol. 2015;17:543. doi: 10.1111/1462-2920.12568. [DOI] [PubMed] [Google Scholar]
- 50.Schuchmann K., Muller V. A bacterial electron-bifurcating hydrogenase. J. Biol. Chem. 2012;287:31165–31171. doi: 10.1074/jbc.M112.395038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schut G.J., Adams M.W. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 2009;191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soboh B., Linder D., Hedderich R. A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology. 2004;150:2451–2463. doi: 10.1099/mic.0.27159-0. [DOI] [PubMed] [Google Scholar]
- 53.Tabor S., Richardson C.C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive production of specific genes. Proc. Natl. Acad. Sci. (U.S.A.) 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. (U.S.A.) 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran K.T., Maeda T., Wood T.K. Metabolic engineering of Escherichia colito enhance hydrogen production from glycerol. Appl. Microbiol. Biotechnol. 2014;98:4757–4770. doi: 10.1007/s00253-014-5600-3. [DOI] [PubMed] [Google Scholar]
- 56.Wang S., Huang H., Kahnt J., Thauer R.K. A reversible electron-bifurcating ferredoxin- and NAD-dependent [FeFe]-hydrogenase (HydABC) in Moorella thermoacetica. J. Bacteriol. 2013;195:1267–1275. doi: 10.1128/JB.02158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wimpenny J.W., Firth A. Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J. Bacteriol. 1972;111:24–32. doi: 10.1128/jb.111.1.24-32.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue Y., Xu Y., Liu Y., Ma Y., Zhou P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 2001;51:1335–1341. doi: 10.1099/00207713-51-4-1335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.