Abstract
Large-scale genomic investigations have just begun to illuminate the molecular genetic contributions to major psychiatric illnesses, ranging from small-effect-size common variants to larger-effect-size rare mutations. The findings provide causal anchors from which to understand their neurobiological basis. Although these studies represent enormous success, they highlight major challenges reflected in the heterogeneity and polygenicity of all of these conditions and the difficulty of connecting multiple levels of molecular, cellular, and circuit functions to complex human behavior. Nevertheless, these advances place us on the threshold of a new frontier in the pathophysiological understanding, diagnosis, and treatment of psychiatric disease.
Genetic findings are set to illuminate the causes and to challenge the existing nosology of psychiatric conditions, some of which, until recently, were purported to have a non-biological etiology (1). After decades of false starts, we now have confirmed associations between genetic variants that increase the risk of schizophrenia (SCZ), autism spectrum disorder (ASD) (2), major depression, and bipolar disorder (BPD), and in some cases the underlying gene(s) have been identified (3–8). These achievements were not necessarily a forgone conclusion. Despite evidence for the relatively high heritability of some psychiatric disorders, claims of successful genetic mapping followed by replication failure (9, 10), along with doubts about the biological validity of the inherently syndromic categorical psychiatric diagnoses, suggested that behavioral disorders would prove to be less tractable to molecular genetic dissection.
The recent discoveries in psychiatric genetics follow technological advances in molecular biology and conceptual advances in the genetics of complex disorders (11, 12). By interrogating genetic variation at millions of single-nucleotide polymorphisms (SNPs) in the genome using microarrays, one can efficiently perform genome-wide association studies (GWASs) in thousands of individuals. Sufficiently large sample sizes have enabled the robust detection of association between disease status and common alleles (“common variants,” population frequencies usually greater than 5%) (13). In the majority of cases, loci identified through GWASs lie in regulatory regions of the genome (14) and do not unequivocally implicate a specific gene. However, because many regulatory regions lie close to their cognate genes (15), investigators typically report the closest gene as responsible (in the absence of functional data, we follow that tradition here but recognize its limitations). Microarrays have also permitted the detection of multiple rare structural chromosomal variants referred to as copy number variation (CNV; the gain or loss of DNA >1 kb in size) that contribute to a variety of psychiatric disorders, including ASD and SCZ (16). Last, advances in genome sequencing have made it possible to obtain the complete protein coding sequence [whole-exome sequencing (WES)] of tens of thousands of individuals (17), with whole-genome sequences at a similar scale on the horizon. The identification of rare mutations in protein-coding domains (“rare variants,” frequency usually <0.1%) via WES has become a standard approach, exemplified by the findings that rare protein-disrupting variants contribute to the risk of ASD (4) and SCZ (18, 19). Although these advances do not yet deliver a complete picture of the genetic architecture (the number of loci and relative contribution from different forms of genetic variation) for any psychiatric disorder (Fig. 1), there is sufficient information to draw some general conclusions.
“Genetic findings blur not only psychiatric disease boundaries but also the boundaries between disease state and normal variation….as risk genes are identified…”
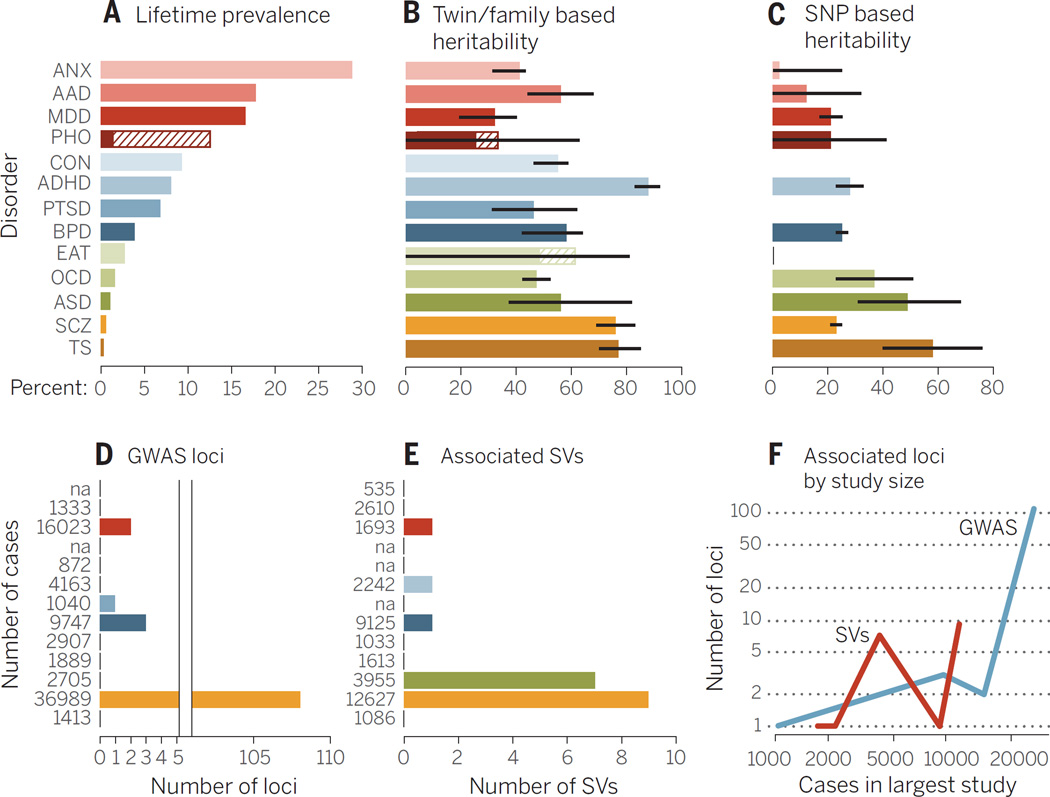
Fig. 1. Summary of genetic analyses performed on 13 psychiatric disorders.
(A) Highest lifetime (point for ASD) prevalence in percentages. The discontinuous bar in phobias represents the range in different forms. (B) Heritability estimates; bars, standard error (SE). (C) SNP-based heritability estimates; bars, SE. (D) Number of genome-wide significant loci. The x axis is discontinuous because of the large difference of associated loci between disorders. (E) The number of associated structural variants (SVs) that either reach genome-wide significance or have been replicated with P ≤ 0.01 in another study. (F) The y axis shows associated GWAS loci (blue) and SVs (green) by the number of cases (x axis) in the largest study for that disorder. The number of cases in the largest study for GWAS (D) and SV studies (E) is reported next to each disorder. Abbreviations are as follows: ANX, any anxiety disorder; AAD, alcohol abuse disorder; MDD, major depressive disorder; PHO, = any phobia; CON, conduct disorders; PTSD, post traumatic stress disorder; EAT, eating disorders; TS, Tourette syndrome. The order of disorders and their color coding are maintained throughout the bar plots. See table S1 for underlying data and references amalgamated from many sources.
The polygenicity of psychiatric illness
In addition to finding specific genes, molecular genetics can provide information about the heritability of psychiatric disease, an approach that has led to some important insights about the genetic architecture of psychiatric illness. The degree of SNP sharing among disease cases estimates the common, inherited portion of a trait (20). Such SNP heritability estimates can be used to test hypotheses about the extent to which heritability arises from many loci of small effect (“polygenicity”). Using this approach, a large proportion of the genetic contribution to psychiatric disease is found to consist of common variants at a large number of loci, although each variant has only a small effect on disease risk, consistent with findings from other common, complex diseases (21). Thus, a major component of risk of psychiatric illness is polygenic. At the same time, the SNP heritability does not explain all of the estimated additive heritability, suggesting that other, as-of-yet–unmeasured factors such as rare variants, also contribute.
Polygenicity, the small effects of individual loci, and the rarity of large-effect loci mean that the most critical requirement for successful genetic dissection of a psychiatric disorder is the availability of sufficiently large clinical cohorts, emphasized by the recent discoveries of the Psychiatric Genomics Consortium (PGC), whose collaborative large-scale approach has had a major impact on the field (3). However, there is an inherent practical tradeoff between sample size and the depth of phenotyping. Disorders with large environmental risk factors, such as anxiety and depression, may benefit from more attention to clinical phenotyping. Rigorous attention to the phenotype and screening for known and putative risk factors may increase the power to detect genetic effects, as evidenced by the recent CONVERGE GWAS in depression (22).
Major-effect-size contributions
A common perception is that GWASs have returned few biological insights, and, consequently, that deeper insights will come from studying the effects of individual large-effect-size genes. Mutations that segregate in a Mendelian fashion are one example of being both necessary and sufficient to cause disease, but these are rare and hard to find in most psychiatric diseases. Analysis of CNV and WES provide another source of rare penetrant mutations.
Particularly convincing has been the recent discovery of the role of large-effect de novo (arising in gametes) mutations in ASD, where the first genome-wide studies of de novo mutations in psychiatric disorders revealed a role for rare (<0.1%) de novo CNVs in ASD (23, 24). Studies indicate that large (>500 kb) rare de novo gene–containing CNVs occur in ~5 to 7% of people with nonsyndromic ASD, versus ~1% of unaffected siblings (25). Several of these rare CNVs are recurrent, some inherited from apparently unaffected parents (such as 15q11-13 or 16p11.2), and none individually account for more than 1% of cases of ASD. Studies of smaller gene-disrupting CNVs also suggest a role for inherited CNVs with lower penetrance (26).
Several major-effect loci due to de novo or inherited CNV also increase the risk for SCZ. These loci display variable expressivity and incomplete penetrance (27, 28), and several are associated with other disorders, including ASD, epilepsy, and intellectual disability (29, 30). The role of large rare CNVs (rare or de novo) in BPD, major depression, substance abuse, obsessive-compulsive disorder (OCD), attention deficit hyperactivitity disorder (ADHD), or anxiety disorders is less clear and, with a few exceptions, have a smaller magnitude of contribution relative to ASD or SCZ. In ADHD, sample sizes have been relatively small, and the greatest signal resides in those with comorbid intellectual disability (31). Studies of parent-child trios in SCZ and BPD (32) have observed odds ratios (ORs) for carrying large rare de novo CNVs of about five for both, although if de novo CNVs <500 kb are considered, the OR is higher, albeit with wide confidence intervals. Similar to ASD, the contribution from de novo events comes mostly from sporadic rather than familial cases, where the rate of de novo CNVs is closer to that of controls.
Finding a rare causal mutation via WES is challenging, because variants changing protein-coding sequences are common (33); identifying sufficient recurrent mutations to confirm the candidacy of a specific gene requires resequencing of thousands of individuals (34) and parents if the goal is to define causal or contributory de novo mutations. Success varies by disorder. In ASD, WES in nearly 5000 probands across two large studies has demonstrated the contribution of rare de novo protein-disrupting mutations to disease risk, identifying 33 genes that are recurrently mutated and therefore highly likely to be pathogenic (4, 35). An additional several hundred mutant genes are observed only once in probands, each with an estimated 40% chance of being contributory, based on the frequency of similar events in controls (4). Overall, current estimates from families containing a single affected individual suggest that up to about 30% of cases harbor large-effect de novo coding mutations, due to either single-nucleotide variants or structural variants (4), each of which is rare in the population. As is the case with CNV (26, 36), inherited rare single-nucleotide variants also play a role in ASD (37), although their contribution warrants further refinement.
Similar success evades genetic analysis of other psychiatric disorders, for which we have very few large-effect genes that have been independently replicated. Work on SCZ is closest to identifying mutations in specific genes. Initial studies identified increased rates of de novo mutations (38), observing that the number of loss-of-function events in cases is almost three times higher than in control family trios (8.7% compared to 2.9%) (39). However, studies with larger sample sizes (18, 19) failed to confirm an overall enrichment of de novo mutations, and identification of individual susceptibility genes via WES has eluded researchers. Enrichment of mutations was found, but only when analysis was restricted to sets of hypothesis-driven candidate genes (19).
On the other end of the spectrum, relatively rare variants (SNPs with frequencies less than 5%) are predicted to account for 21% of the heritability in Tourette syndrome but none of the heritability of OCD (40). Still, for Tourette syndrome, only a single very rare dominant mutation has been identified in one family (41). For other disorders, ranging from major depression to substance abuse and anxiety disorders, for which there is strong evidence of heritable polygenic risk, we lack significant evidence of rare large-effect-size variant contributions, consistent with differences in genetic architecture across psychiatric diseases, although study design and small cohort sizes may also contribute.
Cross-disorder overlap: Genetics as a tool for nosology
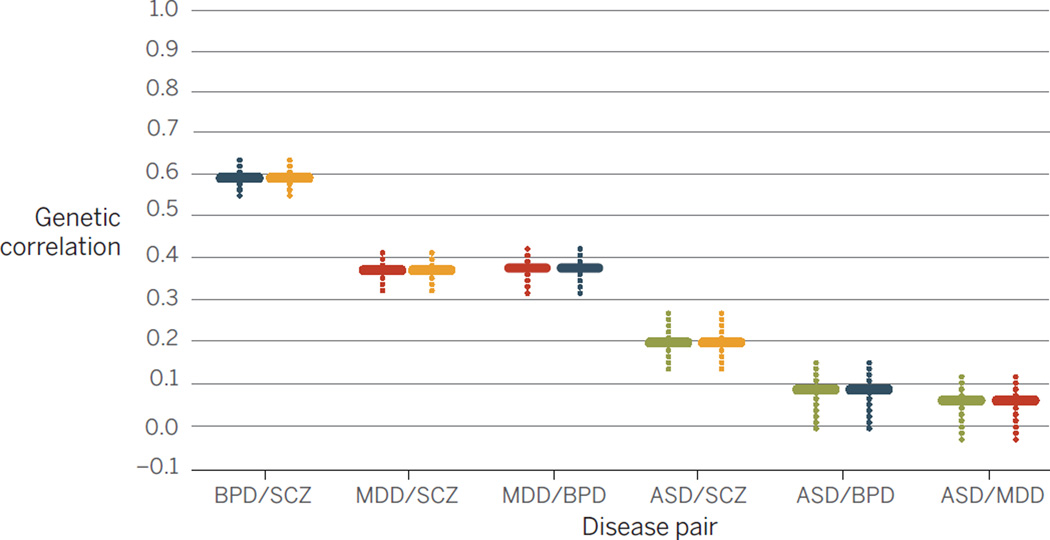
SNP data can also be used to estimate the genetic correlation (tagged by common variation) between disorders (42) (Fig. 2). Some disorders clearly share genetic risk: Between BPD and SCZ the correlation is 0.68; between BPD and major depressive disorder, the correlation is 0.47. Six genome-wide significant loci are associated with a combined BP+SCZ phenotype (42). However, SNPs for both BP and SCZ were genome-wide significant in CACNA1C, ANK3 and ITIH3-ITIH4, but not in MHC, ODZ4, TCF4 and other loci that were genome-wide significant for either disorder separately. Thus, both polygenic risk scores (42) and GWAS hits (42) discriminate between disorders, and both overlapping and disease specific genetic risk factors can be identified (43).
Fig. 2. Pairwise genetic correlations for four psychiatric disorders.
Plotted on the vertical axis are BPD, SCZ, MDD, and ASD (2). The horizontal colored lines mark the mean of the genetic correlation based on SNP sharing for each pair of illnesses, and the dotted vertical colored lines are the SEs of the estimates. Data are from Maier et al. (69).
A similar view of diagnostic overlap and specificity is provided by rare, penetrant mutations that are risk factors for multiple psychiatric disorders. Mutations in evolutionarily constrained, fetal-brain expressed genes, many of whose RNAs are bound by the Fragile X mental retardation protein (FMRP) (4, 18, 44, 45), are associated with ASD, SCZ, and intellectual disability (ID), as well as epilepsy. Similarly, few large CNVs are disease-specific, and the most common such mutation, the 22q11-13 deletion, predisposes to both ASD and SZ, as do others (29, 30). The observed variable expressivity is consistent with the hypothesis that large-effect mutations that disrupt highly evolutionarily constrained genes do not lead to a specific clinically defined disorder, but rather increase risk for a range of developmental disorders associated with ID via disruption of the highly canalized process of brain development (46). From this perspective, clinically defined disorders may represent either the limited repertoire or our limited measurements of behavioral responses to the insult. Moreover, the complexity of brain function and structure is not reflected in recognized in current psychiatric disease nosology. This view provides impetus to the Research Domain Criteria initiative (RDOC) (47) in which psychiatric classification would be replaced with assessment of neurobiological mechanisms, informed largely by genetic discoveries (Fig. 3) (48).
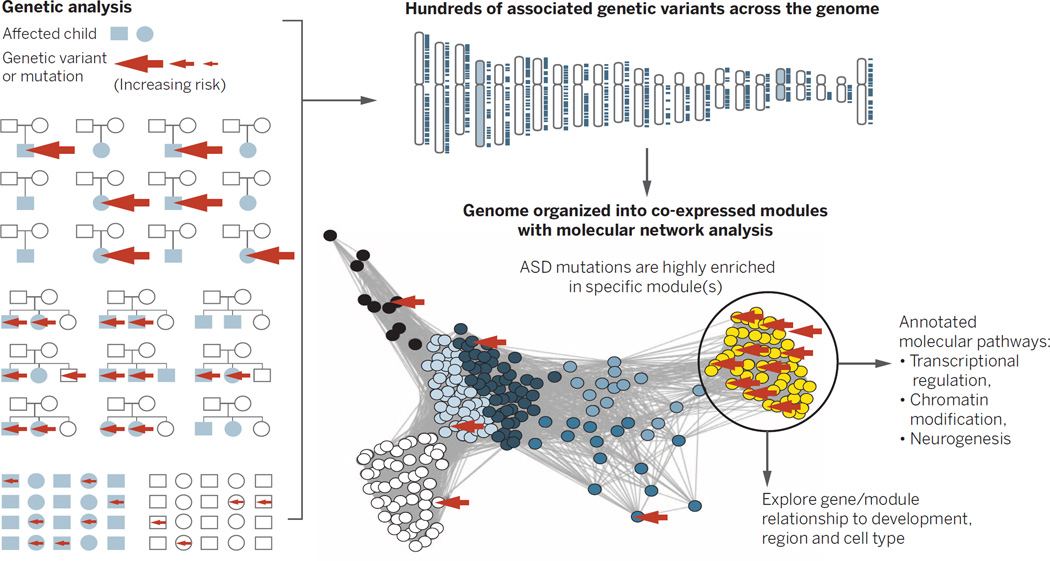
Fig. 3. Heterogeneous genetic risk factors converge in biological networks.
Different study designs, such as trios, multiplex affected families, or case-control (shown at far left) identify different forms of genetic risk in cases (the arrow size indicates the relative effect size). By integrating these data with biological network data, one can assess in a genome-wide manner whether disease-associated risk variants are enriched in specific biological networks (46). Here, for illustration, we depict rare de novo variants associated with ASD, enriched in the yellow module. The function of this module of co-regulated genes can be further annotated using gene ontology, which implicates these large-effect ASD-associated variants in chromatin remodeling, transcriptional regulation, and neurogenesis. Networks can be subsequently mapped onto developmental time points, brain regions, circuits, or cells.
Phenotype definition
Genetic findings blur not only psychiatric disease boundaries but also the boundaries between disease state and normal variation. One recent study suggests that polygenic risk for BPD and SCZ contributes to creativity (49). Unaffected individuals within families harboring major psychiatric illness often harbor quantitative traits shared with affected individuals, but below the diagnostic threshold, so-called intermediate phenotypes or endophenotypes (50). Yet despite many attempts to break psychiatric diseases into simpler intermediate components, the use of quantitative or qualitative endophenotypes in genetic studies has had mixed success (51–53). Severity or age of onset may be useful for risk stratification in some cases (22, 54) but not in others. Additionally, many potential endophenotypes, ranging from cognitive, to behavioral, to anatomical, although highly heritable, appear nearly as genetically complex as the disorders with which they are associated, as is the case for structural neuroimaging phenotypes (55). Still, as risk genes are identified, studying genotype-intermediate phenotype relationships should greatly inform our understanding of disease mechanisms (48).
Neurobiological mechanisms via genetically guided disease modeling
A major reason for identifying the genetic basis of a psychiatric disorder is to develop platforms for understanding disease mechanisms at a cellular-molecular level and accelerate therapeutic development. Despite the potential limitations of studying psychiatric disease in model organisms, mouse models of large-effect-size or Mendelian risk genes for ASD appear promising (56, 57). Many show large behavioral or cognitive deficits relative to wild-type littermates, as well as cellular or physiological phenotypes that may underlie disease pathophysiology. In parallel, advances in stem cell biology now make it possible to generate and study human neurons and their development in vitro (58), providing a platform for drug discovery and phenotypic screening. However, significant challenges exist, including the potential for in vitro artifacts, rigorous definition of cell types, or matching to in vivo brain development. The few studies examining monogenic forms of psychiatric disease via induced pluripotent stem cell–derived neurons are very encouraging (59–61), but consist of relatively small sample sizes. Integrating in vivo modeling in model organisms with in vitro modeling based on tissues derived from human stem cells will help balance the limitations of each system alone.
Additionally, there is an inherent tension between the study of individual genes and the emerging genetic architecture of psychiatric illness, which implicates potentially thousands of genes in each disorder. If psychiatric disorders are a collection of rare conditions, then detailed individual investigation is the most direct route forward. The evidence supports this hypothesis at least in part for ASD and other childhood-onset disorders, in which we now know that the effects of different rare major–effect-size alleles account for some of the phenotypic complexity. Yet there is remarkably little evidence that this is true for SCZ, BPD, major depression, substance abuse, and the anxiety disorders. The apparent specificity of drug action in some of these conditions might be interpreted to imply shared mechanisms among responders. Yet one would not assume shared etiologies among patients with infectious or rheumatologic diseases, whose fevers symptomatically respond to aspirin. The existence of only a few central switches that determine vulnerability to illness is challenged by the extreme polygenicity and apparent genetic heterogeneity in these disorders. We need to understand in an unbiased manner whether there is a convergence of these multiple complex genetic factors on a relatively constrained set of biochemical pathways (62).
Systems genetics approaches
The highly polygenic nature of psychiatric disease and the failure of genome-wide studies to support the role of candidate genes (with a few exceptions) suggest that generalizable mechanistic insight is unlikely to be obtained from analysis of a single dysfunctional molecule in isolation. Genes do not act in isolation, but most models only account for a few features at a time. A systems genetics approach that considers function at a network level permits us to methodically approach the daunting task of connecting heterogeneous genetic risk factors to brain mechanisms (46) (Fig. 4).
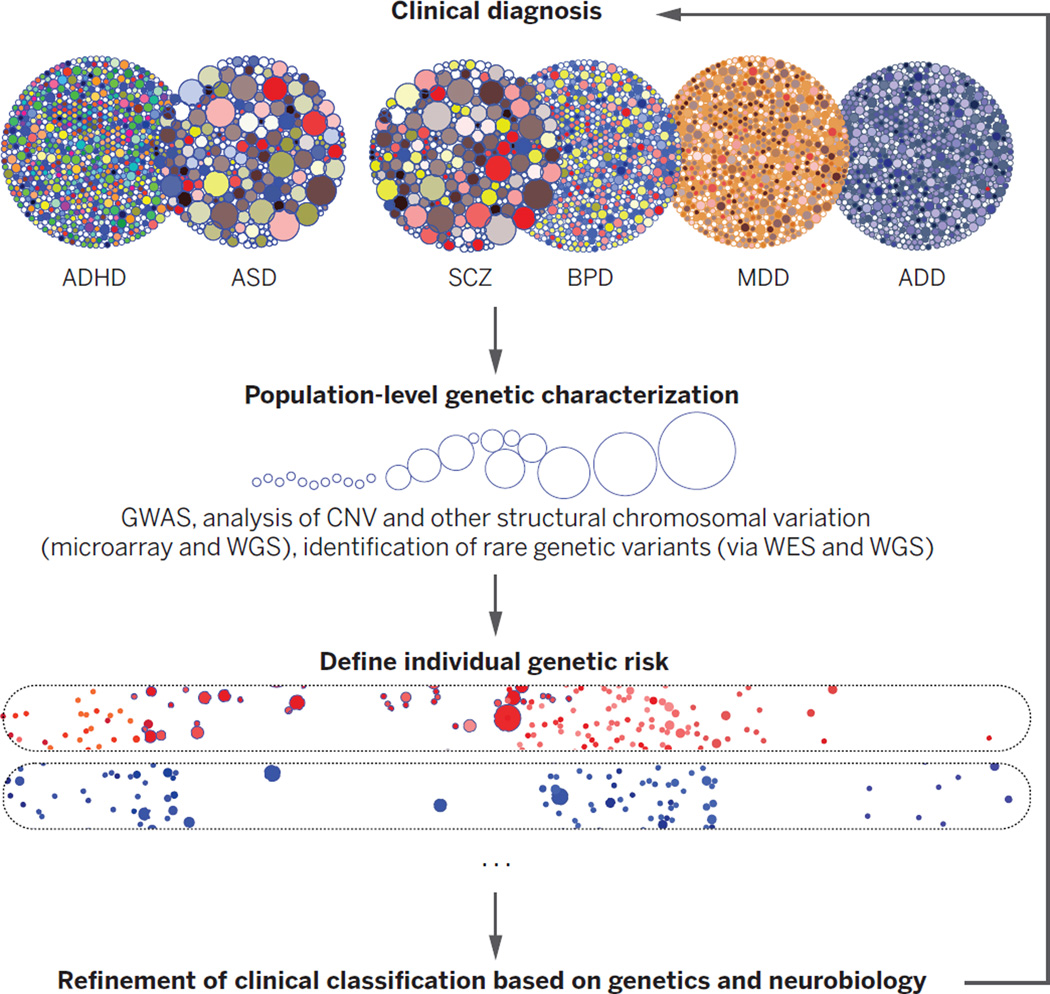
Fig. 4. Refining diagnoses based on genetic susceptibility.
Clinical disorders (abbreviations are as in Fig. 1) and their overlap, represented by the big circles. The smaller dots within each circle represent contributing genetic or environmental risk factors. Once genetic risk is defined in population studies, it can be used to define factors underlying disease risk in individuals, identifying distinct (or overlapping) entities, two of which are represented by the elongated ovals at the bottom, grounded in causal mechanistic understanding. These subtypes should more clearly inform prognosis and treatment than do current categorical disease entities. The sizes of the dots within the circles represent the relative effect sizes of variants.
Several recent genome-wide network studies in ASD and SCZ do indeed suggest that the disorder risk converges on shared molecular pathways, where currently identified genetic variation is enriched (46). In ASD, these pathways involve the regulation of transcription and chromatin structure during neurogenesis, and subsequent processes of synaptic development and function during early fetal cortical development. Alternative approaches based on protein interactions alone, and the integration of protein, gene expression, or phenotype data, identify similar pathways or show the convergence of multiple ASD risk loci on similar biological processes or networks. A network study of genetic variation underlying SCZ implicates similar risk stages during the development of the prefrontal cortex (63), which is consistent with neuroanatomical and physiological studies.
Although there is evidence that both common and rare disease susceptibility loci are likely to converge on specific molecular and biological pathways in ASD and potentially SCZ (64), many issues remain. The pathways as currently defined are broad and should be refined at the level of protein function and cellular signaling to obtain more specific insight into disease pathogenesis. Furthermore, knowing how these pathways reflect genetic risk at the level of individuals is necessary to develop a mechanistic understanding of disease.
Moving forward
We now have a reasonable framework for understanding the basic genetic architecture underlying psychiatric disease, yet the number of susceptibility loci so far identified accounts for only a small percentage of the variance in liability to disease (65). With the exception of SCZ and ASD, so few replicated loci have been identified that the need to identify more is undeniable. Still, the field is at an appropriate juncture to consider when it would be reasonable to stop looking for genes and focus entirely on studying their function. Certainly, a more complete catalog of genes and mutations would provide a clearer indication of cross-disorder overlap and disease-specific biochemical and circuit convergence. One important justification for the creation of a more complete catalog is the need to address disease heterogeneity and to understand composite genetic risk in individuals. A measure of success would be the ability to genotype individuals at known risk loci and classify their disease based on a neurobiological framework (47); this is a feasible goal for the next decade (Fig. 3). Leveraging electronic medical records, remote data gathering, and electronic media, coupled with forthcoming populationlevel clinical whole-genome studies, should accelerate these efforts.
In the meantime, comparative studies are needed to understand the shared and distinct phenotypic effects of rare large-effect alleles in humans and model systems. To some extent, the phenotypes of psychiatric disease must represent the common outcome of multiple different pathways at the level of brain systems that underlie their shared behavioral and cognitive phenotypes (29, 62). Decades of research have identified ASD in multiple rare genetic syndromes with specific constellations of multi-organ phenotypes, including Timothy syndrome, tuberous sclerosis, Potocki-Lupski syndrome, cortical dysplasia focal epilepsy syndrome, and fragile X syndrome. New syndromes identified via WES are no different, suggesting that the phenotypic complexity of ASD may in part be explained by the effects of different rare major-effect-size alleles (66). Such inverse mapping or genotype-first approaches are at early stages in SCZ and BPD (48). Additionally, it is not known what genetic or environmental factors influence the diversity of clinical outcomes in people harboring most of the major-effect loci. Understanding the mechanisms of such variable expressivity will undoubtedly provide critical pathophysiological clues.
Rigorous attempts to define intermediate phenotypes that represent components of the underlying disease neurobiology are also necessary to develop a more therapeutically meaningful nosology, but this area has proven very challenging (67). One potential avenue is using more data-driven approaches in humans, such as genetic risk scores and Mendelian randomization based on the joint effects of multiple risk loci to refine causal relationships between disease and intermediate phenotypes (68). Moreover, specificity of action is a prerogative of neural development and neuronal circuits, not genes. Technologies that have brought activity in neuronal circuits under exogenous control repeatedly demonstrate the existence of circuits that are responsible for specific behaviors. Circuits and diseases that can be modeled in genetically tractable organisms permit hypothesis testing that draws on genetic results to propose genes for functional testing.
One key piece of basic biological information that is missing is knowledge of cell type diversity and its molecular basis in the brain. To fully interpret the mechanisms of genetic variants it is necessary to have a more complete molecular and cellular parts list of the developing and mature brain. Moreover, as most common variants lie in presumptive regulatory regions, we need to integrate emerging knowledge about cells and circuits with an unbiased understanding of mechanisms of gene regulation across neurodevelopmental stages. All of these goals will be hastened by organized efforts such as the Brain Initiative (braininitiative.nih.gov/), the Allen Institute (http://alleninstitute.org), psychEncode (http://psychencode.org), Genotype-Tissue Expression (www.gtexportal.org), and the PGC.
We conclude by emphasizing the transformational role that genetic insights can play when investigating conditions for which nothing is currently known about their underlying biology. The idea that psychiatric disease might be purely “functional” can no longer be entertained. With ASD, we are entering an era where genetic dissection informs phenotypic heterogeneity and where biological insights are starting to emerge. The next few years will undoubtedly see a radical transformation of our understanding of the biological origins of all neuropsychiatric disorders.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Ruzzo, V. Leppa, G. Coppola, M. Gandal, and J. Stein for their helpful comments on the manuscript and for their contributions to figure content and design and to table S1. Figure 1 was produced by D.H.G. and V. Leppa based on data in table S1; Fig. 2 was produced by J.F.; Fig. 3 was produced by D.H.G. and N. Parikshak; and Fig. 4 was produced by G. Coppola and D.H.G. We gratefully acknowledge funding support from the National Institute of Mental Health (grants 5R01 MH100027 and 5R01 MH094714 to D.H.G.), the Simons Foundation (grant 206744 to D.H.G.), and the Wellcome Trust (J.F.) for our research. We apologize to our colleagues whose primary work we were not able to cite due to space limitations and instead refer to reviews where many of the primary references can be found. D.H.G. has consulted for Synapdx and Roche.
Footnotes
REFERENCES AND NOTES
- 1.Bettelheim B. The Empty Fortress: Infantile Autism and the Birth of the Self. New York: Free Press; 1972. [Google Scholar]
- 2.Stefansson H, et al. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ripke S, et al. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I, et al. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rubeis S, et al. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talkowski ME, et al. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelsoe JR, et al. Nature. 1989;342:238–243. doi: 10.1038/342238a0. [DOI] [PubMed] [Google Scholar]
- 10.Levinson DF, et al. Science. 2002;296:739–741. doi: 10.1126/science.1069914. [DOI] [PubMed] [Google Scholar]
- 11.Kruglyak L. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 12.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy MI, et al. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 14.Maurano MT, et al. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Schizophrenia Consortium. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tennessen JA, et al. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell SM, et al. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fromer M, et al. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, et al. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehret GB, et al. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CONVERGE Consortium. Nature. 2015 [Google Scholar]
- 23.Jacquemont ML, et al. J. Med. Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebat J, et al. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders SJ, et al. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucan M, et al. PLOS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levinson DF, et al. Am. J. Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vacic V, et al. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geschwind DH. Trends Cogn. Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-De-Luca D, et al. Am. J. Hum. Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams NM, et al. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malhotra D, et al. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacArthur DG, et al. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Roak BJ, et al. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geschwind DH, State MW. Lancet Neurol. 2015 doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinto D, et al. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krumm N, et al. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girard SL, et al. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 39.Xu B, et al. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis LK, et al. PLOS Genet. 2013;9:e1003864. doi: 10.1371/journal.pgen.1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ercan-Sencicek AG, et al. N. Engl. J. Med. 2010;362:1901–1908. doi: 10.1056/NEJMoa0907006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SH, et al. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruderfer DM, et al. Mol. Psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell JC, et al. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irimia M, et al. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parikshak NN, Gandal MJ, Geschwind DH. Nat. Rev. Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Insel T, et al. Am. J. Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 48.Stefansson H, et al. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 49.Power RA, et al. Nat. Neurosci. 2015;18:953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 50.Gottesman II, Gould TD. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 51.Greenwood TA, et al. Am. J. Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowe JK, Werling DM, Constantino JN, Cantor RM, Geschwind DH. Am. J. Psychiatry. 2015;172:266–275. doi: 10.1176/appi.ajp.2014.14050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaste P, et al. Biol. Psychiatry. 2015;77:775–784. doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walsh T, et al. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 55.Hibar DP, et al. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nestler EJ, Hyman SE. Nat. Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen JA, Peñagarikano O, Belgard TG, Swarup V, Geschwind DH. Annu. Rev. Pathol. 2015;10:111–144. doi: 10.1146/annurev-pathol-012414-040405. [DOI] [PubMed] [Google Scholar]
- 58.Dolmetsch R, Geschwind DH. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paşca SP, et al. Nat. Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugathan A, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shcheglovitov A, et al. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geschwind DH. Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gulsuner S, et al. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Nat. Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaugler T, et al. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeste SS, Geschwind DH. Neurology. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bearden CE, Freimer NB. Trends Genet. 2006;22:306–313. doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Evans DM, Davey Smith G. Annu. Rev. Genomics Hum. Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 69.Maier R, et al. Am. J. Hum. Genet. 2015;96:283–294. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.