Abstract
Autism spectrum disorder is typical of the majority of neuropsychiatric syndromes in that it is defined by signs and symptoms, rather than by aetiology. Not surprisingly, the causes of this complex human condition are manifold and include a substantial genetic component. Recent developments in gene-hunting technologies and methods, and the resulting plethora of genetic findings, promise to open new avenues to understanding of disease pathophysiology and to contribute to improved clinical management. Despite remarkable genetic heterogeneity, evidence is emerging for converging pathophysiology in autism spectrum disorder, but how this notion of convergent pathways will translate into therapeutics remains to be established. Leveraging genetic findings through advances in model systems and integrative genomic approaches could lead to the development of new classes of therapies and a personalised approach to treatment
Introduction
The term autism spectrum disorder (ASD) defines a neurodevelopmental syndrome that affects about one in 100 people worldwide, and is characterised by early dysfunction in social interactions and the presence of repetitive and restrictive behaviours.1 ASD is consistently reported to be more prevalent in boys than in girls.1People with ASD have a broad range of phenotypes and commonly also have intellectual disability (35%), language delay (50%), or epilepsy (5–15%). Until a decade ago, little was known about the causes and pathogenic mechanisms of ASD. Although, in general, strong evidence indicates that the causes include both genetic and environmental factors,2 genetic research over the past decade has taken the leading role in the elucidation of pathophysiology. Strikingly, ASD is one of a relatively small number of neuropsychiatric conditions for which substantial progress in gene discovery has been made during this period. This progress has been a result of conceptual and technological advances in the study of genetics of human disease, highly effective advocacy and philanthropy that has led to large study cohorts and widespread data sharing, and an evolving and maturing view of the role of common and rare genetic variation in human disease.3 Advances in the genetics of ASD have therefore provided both insight into the neurobiology of a complex disorder and a template for understanding the genetics and genomics of other common neuropsychiatric conditions.
In this Series paper, we describe basic concepts in the genetic architecture of common human disease, contrasting the concept of rare, penetrant mutations with common genetic variation. We review progress in the identification of ASD genes, including studies of genetic linkage, analysis of copy number variants (CNVs), whole-genome association, and exome sequencing to identify rare transmitted and de novo variation. We also discuss the endophenotype concept and its use in gene discovery. Finally, we consider the promise of these genetic findings for understanding the pathophysiology of ASD, and highlight some of their important clinical implications.
Genetic variation and human disease
Sequencing of the human and other mammalian genomes over the past decade has provided a foundational set of tools to begin to understand human genetic variation (panel 1). Because most living humans are the progeny of rapid expansion of the human population from a relatively small number of remote ancestors (about 10 000 years ago), we share large segments of our genomes and hence a substantial amount of genetic variation with other humans.4 This so-called common variation is formally defined as a change in the sequence or structure of DNA from a reference ancestral genome that is observed at a population frequency of more than 1%. From an evolutionary standpoint, such common variation would be predicted to be under strong selection pressure, making it unlikely that common variation would have strong or even moderate negative effects on early survival or fecundity (reproductive fitness).
Panel 1: Types of human genetic variation.
Any two individuals will differ from each other at only approximately 1% of the 3 billion nucleotides that comprise the human genome. To the extent that risk for a disorder is conferred to a subset of individuals by genetics, it is the portion of their genome that varies from others that accounts for this risk. The understanding of the nature of genetic variation has advanced substantially in the past 15 years, due in no small measure to the (initial) completion of the human genome project in 2001. It is now appreciated that any two human genomes vary both with regard to the sequence of DN A and with regard to the submicroscopic structure of chromosomes. Genetic variation can be usefully divided into various classes or types that have relevance for gene discovery, which are summarised in the definitions below.
Common variation: any variation from the reference genome that is present in more than 1% of individuals
Rare variation: any variation from the reference genome that is present in the population at a frequency of 1% of the genome or less
Copy number variation: a variation in submicroscopic chromosomal structure of greater than 1000 nucleotides (an arbitrary cutoff; see “in-del” below). Copy number variants (CNVs) can be either common or rare, transmitted or de novo
Transmitted variation: any variation from the reference genome that is present in every cell of the parent (or parents) and is passed to an offspring
De novo variation: a variation that is new; a spontaneous mutation in either sperm or egg will lead to the variant being present in every cell of the offspring (also known as germlinede novo variation) but not detectable in either parent
Single-nucleotide polymorphism (SNP): a variation in DNA sequence that is common in the population
Single-nucleotide variant (SNV): a variation in the DNA sequence that is rare in the population
Insertion-deletion (in-del): a variation in the DNA that involves the insertion or deletion of a small number of nucleotides (the boundary between a small CNV and an in-del is arbitrary)
Coding variant: a variation from the reference genome that is present in the approximately 1% of the genome that codes for the transcription of proteins
Non-coding variant: a variation that is found outside the portion of the genome that codes for proteins, including introns, regulatory elements, and intergenic segments
By contrast, rare genetic variation (<1% frequency), which also includes very rare (private, <0·01%) and de novo (non-inherited) variation, may have large biological effects. For rare, or very rare, transmitted variation that carries this risk, the hypothesis is that allele frequencies are kept low as a consequence of purifying (negative) selection. Mutations survive in the population as a consequence of recessive inheritance, incomplete penetrance, balancing selection, or spontaneous mutation. De novo mutation (mutations arising in the germline that are not present in the parental somatic genome) is typically very rare or private, because each event in a new generation has not had time to propagate within a population and is subject to strong purifying selection.5 Similarly, dominant or recessive mendelian disorders are caused by highly penetrant rare mutations, consistent with the low frequency of these conditions in most populations.
Several noteworthy exceptions to this predictable relationship between allele frequency and disease effect size have been identified. For example, the ε4 allele of APOE occurs at an average human population frequency of 14%, but carries a greater than four-times increase in the risk for Alzheimer’s disease.6 Similarly, the major common risk variant for macular degeneration carries a greater than 2·5-times increase in risk.7 These figures are not surprising because these disorders generally have a late age of onset and, consequently, would be predicted to have little effect on reproductive fitness. However, why such variation is maintained at fairly high rates in the population has not been fully explained. For example, this stability might simply be a consequence of the risk allele truly escaping selective pressure, or some as-yet-unknown benefit to carrying the ε4 allele of APOE might exist. Nonetheless, in general, common variants with large disease effect sizes (>2·0) are exceptional: most known common risk alleles have been shown to increase susceptibility by less than 1 · 5–2 · 0 fold.
The combination of this understanding of the human genetic architecture with the prevalence of many of the diseases seen frequently in the population led to the common disease–common variant (CDCV) hypothesis.8,9This theory holds that disease susceptibility in common disorders arises as a consequence of common alleles and is on a continuum of normal phenotypic variation. In the case of ASD, despite the plausible effect of autism-related variation on fecundity, the CDCV hypothesis became the dominant view of the allelic architecture within the psychiatric genetics community.10 The CDCV hypothesis in this case suggests specifically that many variants of small effect conspire in a given individual to place that person on a population distribution above a (somewhat arbitrary) threshold for the categorical diagnosis, and that the same variants that increase risk for social dysfunction or language dysfunction in ASD would also modulate these phenotypes in the general population.11Strong evidence supports a substantial contribution of common genetic variation to ASD,12,13 but for reasons we will discuss, progress in gene discovery so far has resulted almost exclusively from the investigation of rare, not common, variation.
The completion of the human genome project followed by rapid advances in microarray technology made whole-genome assessment of common variation efficient and inexpensive, and resulted in a wave of studies aimed at testing the CDCV theory, particularly after the turn of the millennium. These analyses took the form of both linkage and association studies. The same technology has since enabled the detection of a subset of rare mutations: the submicroscopic changes in chromosomal structure known as CNVs. Genome-wide assessment of rare variation at single-base resolution, enabled by the development of next-generation sequencing, lagged about 10 years behind genome-wide genotyping and was only introduced to the field about 5 years ago.
Gene discovery in ASD
Linkage studies
In view of the strong evidence for the heritability of ASD, early genetic studies focused largely on ascertaining multiply affected (multiplex) pedigrees to support genetic linkage analysis aimed at identifying chromosomal regions co-inherited by those with the disorder.14 This approach to the search for heritable variation within families is supported by the more than ten-times increased risk to a sibling of a proband with autism versus the population risk, and the observation of sub-threshold autistic-like traits in first-degree relatives of those with ASD, referred to as the broader ASD phenotype.15–18 Non-parametric linkage studies (those that were not based on the proposal of an a priori model of intra-generational transmission) began with samples of 100–200 families. Although thought to be sufficiently powered at the time, few studies yielded genome-wide significant loci. These early studies used coarse marker sets with incomplete information content and, coupled with the small sample sizes, had very low power. Most loci that were identified, even those exceeding genome-wide significance thresholds, were not replicated in subsequent studies.14,19 To date, only two ASD linkage peaks have been replicated at levels that can be considered genome-wide significant: one on chromosome 7q35 and the other mapping to chromosome 20pl3.14,20
In view of the higher prevalence of ASD in males, which averages 3–4-times that of females in most genetic studies (but ranges in epidemiological studies from about 1 5-times to 16-times that of females),1 various approaches have been used to try to identify chromosomal loci that contribute to the sex differential. However, no significant X linkage has been identified, and when families have been stratified by sex of the affected children, most loci identified are autosomal.21 Again, no male-related or female-related loci have been formally replicated, but some have reached genome-wide significance, including a locus on chromosome 8.20Importantly, these and other linkage regions have subsequently been carefully assessed by dense typing of common single-nucleotide polymorphisms (SNPs).22These analyses should plausibly identify the contribution of a common risk variant mapping within the linkage interval, as they have been powered to detect effect sizes of more than 1 2. However, no common variation has been identified or even reached the threshold of regional significance (correcting for multiple testing based on only those SNPs within the linkage region), perhaps with the exception of CNTNAP2 (contactin associated proteinlike 2) and NGF (nerve growth factor).23,24 Additionally, no association signals have been found that can account for the linkage peaks in these regions, which suggests that even within segments shared by affected families, rare genetic variation is likely to be playing a significant part, consistent with the expectations of small individual-allele effect sizes from common variants.25
Intermediate phenotypes and endophenotypes
To tackle phenotype heterogeneity in ASD, researchers have explored the use of endophenotypes, which can be defined as heritable, simple components of a complex neuropsychiatric condition.26–28 By using a quantitative endophenotype (eg, language scores or head circumference) rather than a qualitative endophenotype (eg, diagnosis), analyses can include individuals without ASD, who are presumed on the basis of family studies of the broader autism phenotype to carry some of the same risk alleles. This approach has been applied to other complex neuropsychiatric disorders29 and has had mixed results in ASD.
Alarcón and colleagues28 identified a quantitative trait locus (QTL) on chromosome 7q35 associated with language delay in ASD. Subsequently, evidence was reported that the CNTNAP2 gene contributes to this QTL.11 Consistent with the idea that common variation exerts its effects broadly in the population, others have shown that this locus might contribute to specific language impairment and language-related neurodevelopmental disorders in several other populations;30,31 however, these studies have provided varying degrees of evidence and none has shown genome-wide significance.
The social responsiveness scale (SRS) captures autistic-like social impairment using a quantitative questionnaire, the scores on which are highly heritable.32,33 Social responsiveness therefore has promise as a quantitative trait representing social endophenotypes in ASD. Several groups have used the SRS to identify QTL in linkage studies, but most have relied on small sample sizes34,35and have identified peaks that have not been replicated. Recently, using five-times larger samples, and correcting for sex differences, Lowe and colleagues22 identified two peaks with a logarithm of the odds ratio (LOD) score of more than 4·0 on chromosome 8. This work showed that the use of this quantitative scale rather than the qualitative diagnosis of ASD provided significant additional power. Similar to the qualitative linkage peaks described here, no detectable role for common variation under these peaks was identified.22
In summary, only two regions can be considered genome-wide significant for qualitative linkage to ASD: chromosome 20pl3 and chromosome 7q35. Promising QTL analyses have also identified genome-wide significant peaks for the SRS. Much more work is needed to develop and refine relevant, heritable ASD endophenotypes by studying multiplex families and unaffected relatives. Although endophenotype analysis is unlikely to supplant increasing sample size as a way to detect genetic risk factors, analysis of endophenotypes will be important for understanding the components of ASD to which small to moderate effect-size alleles contribute. Such analyses might also help in understanding the mechanism by which common risk genes contribute to clinically distinct disorders, such as ASD, attention deficit hyperactivity disorder, or schizophrenia.
Substantial progress has been made in the identification of rare variants contributing to ASD, relying largely on categorical diagnoses and focusing on so-called simplex families (those with only a single affected individual) in an effort to enrich for de novo mutations. These approaches—as opposed to pedigree studies that use quantitative phenotyping approaches based on modern cognitive neuroscience—are not likely to be adequate for, and are certainly not well suited to, assessment of phenotypic subgroups or QTLs in ASD.26,36 Additionally, sample sizes for studies of inherited forms of ASD need to be much larger than they are at present to have sufficient power to identify and replicate loci and individual common alleles at genome-wide significance (a level of significance that reflects correction for the large number of multiple comparisons made in genome-wide studies).
Genetic association
Before the development of genome-wide methods, common-allele candidate-gene studies dominated ASD genetics, as they did across all of medicine. With hindsight, these studies were clearly not powered to detect the effects of common alleles contributing to common disease, were unable to control adequately for cryptic differences in ancestry in cases versus controls, and relied on overly permissive, nominal thresholds for statistical significance. As a consequence, widespread difficulties have been encountered in the replication of findings and in the interpretation of most candidate-gene results reported in the literature.
When the search for common variants contributing to common disease initially turned to genome-wide association studies (GWAS), the assumption was made that some individual variants would impart increased risks of between 1·5 and 2·0 times, and power calculations indicated that such variants could be found with a few thousand cases and controls. The empirical data subsequently showed that the effect sizes of common variants were, in general, smaller than anticipated, and that the genetic makeup of common neuropsychiatric disorders, including ASD, was highly heterogeneous. As a consequence, studies so far remain largely underpowered. At present, although some genome-wide significant findings exist, none has been consistently replicated across studies. In retrospect, this is not surprising, as the largest cohorts have had of the order of 5000 cases and controls.36–38 Ten-times larger cohorts have been needed in other common diseases to identify replicable genome-wide findings.39 In schizophrenia, for example, when the cohorts increased from 10 000 participants to 50 000 participants, ten-times the number of loci were identified.40 Thus, to identify common variation in ASD, which is estimated to comprise 40–60% of the genetic risk for ASD, much larger family-based cohorts are needed.12
In a recent study by Chaste and colleagues,36 ten plausible substitute quantitative and categorical phenotypes were assessed in simplex ASD families, including intelligence quotient, quantified restrictive and repetitive behaviours, several measures of autism severity, insistence on sameness, and sensory sensitivity. The researchers found that dividing the Simons Simplex Collection (SSC; a research cohort comprising families with two unaffected parents, a single affected offspring and, in most cases, at least one unaffected sibling) on the basis of these characteristics had a very modest impact on genetic homogeneity and could not substitute for dramatically increased sample size to detect common variation contributing to ASD. As noted, one important observation is that the SSC is relatively depleted in familial risk, including selection against sharing of quantitative social impairment as measured by the SRS, so it might not have the power present in multiplex cohorts that segregate the broader autism phenotype. Nonetheless, overall, increased sample sizes are clearly needed.
Rare and de novo mutations
The first clues that very rare and de novo mutations play an important part in ASD began to be appreciated at the turn of the millennium. Convincing evidence had emerged from well-controlled studies that several monogenic intellectual disability syndromes were associated with increased rates of ASD (panel 2). As these findings gained acceptance, it followed that FMR1 (fragile X mental retardation 1), PTEN (phosphatase and tensin homologue), and TSC1 (tuberous sclerosis 1) were the first ASD genes to be cloned. At the same time, several laboratories began to focus on rare and recessive mutations in more typical or idiopathic ASD pedigrees. For example, in 2003, Jamain and colleagues41 followed up on the observation of X chromosome deletions mapping to a single interval in a small number of affected girls, sequenced the interval in a group of affected families, and identified a de novo loss-of-function (LoF) mutation in the gene NLGN4X (neuroligin 4, X-linked) in an unaffected mother, transmitted to two affected boys. They then sequenced a second member of the neuroligin family, NLGN3, and found a single-base missense substitution at a highly conserved aminoacid position in NLGN3 in a second small family. The importance of the former finding was reinforced in short order by the linkage mapping of a similar but distinct rare truncating mutation in NLGN4X in an extended pedigree showing X-linked inheritance of both intellectual disability and autism. This finding was followed oon thereafter by the identification of rare de novo, apparently LoF, mutations in SHANK3 (SH3 and multiple ankyrin repeat domains 3) in individuals with ASD,42 and rare recessive mutations in CNTNAP2 linked to autism and intractable epilepsy in consanguineous Amish families.43
Panel 2: The role of monogenic conditions in understanding autism spectrum disorders.
Although it has been recognised for more than 25 years that rare monogenic, medical genetic conditions cause autism spectrum disorder (ASD), it was not until rare mutations were discovered by sequencing in idiopathic ASD cases that this idea developed broad traction.41,42 In the early days of ASD genetics, well-recognised neurodevelopmental syndromes, such as fragile X syndrome (caused by mutations in FMR1) and tuberous sclerosis (mutations in TSC1 and TSC2), were not considered to be mainstream causes of ASD. With the advent of whole-exome sequencing, it is now predicted that at least 25% of people with ASD carry rare protein-disrupting mutations that have a major role in their risk for the disorder. Most of these disorders are associated with intellectual disability, and many recognised causes of ASD, including mutations in FMR1, TSC1, and TSC2, and deletions in the 16p11.2 region, have a penetrance of no more than 25% for autistic social impairment. Nonetheless, these monogenic forms of ASD provide many opportunities to further our understanding of the causes and pathogenic mechanisms of autism: (1) the potential to identify risk modifiers that might be influencing the expressivity of these large-effect mutations; (2) increased power relative to common variants (with small effect sizes) to study the neurobiological consequences of a mutation in model systems; (3) genetically homogeneous patient populations with ASD to increase the power of a variety of studies, including neuroimaging, neuropsychology, stem cell-based cell biological investigations, and mechanistically targeted treatment trials; and (4) cross-disorder study to identify common brain pathways shared by those with syndromic forms of ASD.
Although seminal findings, these discoveries relied either on the identification of rare, apparently mendelian transmission of a complex behavioural phenotype, or on the mapping of chromosomal abnormalities, which were present in only a tiny fraction of typical clinical samples. By 2004, the sequencing of the human genome, coupled with array technology, enabled the routine identification of rare CNVs (a gain [duplication] or loss [deletion] of chromosomal material of greater than 1000 bp) across the genome in both affected and unaffected populations.44,45 The detection of CNVs turned out to be a watershed in the genetics of ASD.46,47 These variants were found in considerable (initially about ten-times) excess in simplex families compared with typical controls,47 and early studies showed that CNVs disrupted the locus containing SHANK3, NRXN1 (neurexin 1), and other promising candidate autism risk loci.48,49 These observations were closely followed by the recognition that some de novo CNVs clustered in specific regions of the genome, providing a robust method to identify at-risk intervals.50–52 These studies converge on an estimate that 5–7% of individuals with a diagnosis of ASD will carry a CNV conferring substantial risk.
De novo copy number variation and single-nucleotide variants
These early findings, cumulatively pointing to the importance of rare, de novo, and recessive mutations and to a broad range of phenotypes relating to even highly penetrant mutations, were a harbinger of the dramatic progress in ASD genetics seen during the past decade. As sample sizes expanded, multiple CNVs were identified that carried substantially increased risk for ASD.53,54 By contrast with common-variant candidate-gene studies, a convergence of results across large samples and laboratories quickly underscored the reliability of the findings. In addition, the development of approaches that established genome-wide thresholds for assessing the relevance of multiple de novo events mapping to the same locus helped to build consensus about the identification of risk regions and genes.54,55 So far, multiple loci have been replicated as carrying ASD risks, including CNVs at chromosomal regions 1q21, 7q11.23, 15q11–13, 16p11.2, and 22q11.2. Importantly, these regions, although carrying substantial risks for ASD, are not specific for social disability, as they have also been found to confer risks for a wide range of neurodevelopmental phenotypes, including schizophrenia, language impairment, mood disorders, epilepsy, and intellectual disability.53 Whether this phenomenon is stochastic or environmental, or due to modification by genetic background, or both, remains to be established.
With the emergence of whole-exome sequencing (obtaining the DNA sequence of the fraction [about 1·5%] of the genome that codes for all known proteins), the genome-wide study of de novo mutations was rapidly extended to single-base resolution. Many studies have rapidly converged on the following key, consensus findings:56–65 (1) an increased rate of de novo LoF or likely gene disrupting (LGD) mutations exists in simplex ASD cases versus controls; (2) in view of the very low base rate in the unaffected population, the observation of even a small number of these events mapping to the same gene in affected, unrelated individuals provides considerable power to assign relevance to an individual gene (table); (3) genes so far found to carry multiple de novo LoF or LGD mutations are functionally heterogeneous; (4) on the basis of overall mutation rates, the target size—ie, those genes conferring some ASD risk due to de novo LoF or LGD mutations—is estimated to be between 400 and 1000 genes;56,58,60,65 (5) the rate of de novo single-nucleotide variants (SNVs) increases with paternal age and those associated with ASD risk are most often present on the paternal chromosome;56–58,60–65 (6) de novo insertions and deletions that disrupt protein function carry very similar risks to de novo LoF SNVs;56–58 and (7) de novo missense mutations appear also to show a risk signal in aggregate, although not surprisingly, of smaller magnitude than for LoF mutations.56,58,65
Table.
Genes with two or more de novo or rare inherited variants in probands with autism spectrum disorder and their associated gene ontology terms
| Gene name | Gene ontology terms | |
|---|---|---|
| ADNP56,58 | Activity-dependent neuroprotector homeobox |
Cellular nitrogen compound metabolic process, chromatin binding, DNA binding, ion binding, negative regulation of neuron apoptotic process,transcription, DNA-templated |
| ANK256,58 | Ankyrin 2, neuronal | ATPase binding, ion channel binding, potassium channel regulator activity, bridging, protein localisation to M-band, protein localisation to T-tubule, regulation of cardiac muscle cell membrane potential, response to methylmercury, SA node cell action potential, spectrin binding, structural constituent of cytoskeleton |
| ANKRD1158 | Ankyrin repeat domain 11 | Bone development, face morphogenesis, in utero embryonic development, multicellular organism growth, odontogenesisof dentin-containing tooth, protein binding |
| ARID1B56,58 | AT rich interactive domain IB (SWIl-like) |
Anatomical structure development, cellular nitrogen compound metabolic process, chromatin remodelling, chromatin-mediated maintenance of transcription, DNA binding, protein binding transcription factor activity, transcription coactivator activity |
| ASXL356 | Additional sex combs like transcriptional regulator 3 |
Cellular nitrogen compound metabolic process, DNA binding, ion binding, metal ion binding, regulation of transcription, DNA-templated |
| BCLllA56 | B-cellCLL/lymphoma 11A (zinc finger protein) |
DNA binding, ion binding, metal ion binding, negative regulation of dendrite development, negative regulation of protein homo-oligomerisation, nucleic acid binding transcription factor activity, protein heterodimerisation activity, protein sumoylation, RNA polymerase II core promoter proximal region sequence-specific DNA binding |
| CACNA2D356 | Calcium channel, voltage- dependent, alpha 2/delta subunit 3 |
Calcium ion transport, ion binding, metal ion binding, regulation of ion transmembrane transport, transmembrane transport, transport, voltage-gated calcium channel activity |
| CHD258 | Chromodomain helicase DNA binding protein 2 |
Anatomical structure development, ATP binding, ATP-dependent DNA helicase activity, ATPase activity, cellular nitrogen compound metabolic process, cellular response to DNA damage stimulus, chromatin modification, core promoter sequence-specific DNA binding, DNA binding, DNA duplex unwinding, DNA metabolic process, helicase activity |
| CHD856,58 | Chromodomain helicase DNA binding protein 8 |
Armadillo repeat domain binding, ATP-dependent chromatin remodelling, beta-catenin binding, canonical Wnt signalling pathway, DNA helicase activity, histone binding, in utero embryonic development, methylated histone binding, negative regulation of fibroblast apoptotic process, p53 binding, positive regulation of transcription from RNA polymerase II promoter |
| CUL356 | Cullin 3 | Cyclin binding, cyclin catabolic process, embryonic cleavage, enzyme binding, mitotic metaphase plate congression, negative regulation of Rho protein signal transduction, POZ domain binding, protein heterodimerisation activity, trophectodermal cellular morphogenesis, ubiquitin protein ligase binding, ubiquitin-protein transferase activity |
| DIP2A58 | DIP2 disco-interacting protein 2 homologue A (Drosophila) |
Catalytic activity, metabolic process, multicellular organismal development, negative regulation of gene expression, regulation of apoptotic process, transcription factor binding |
| DSCAM58 | Down syndrome cell adhesion molecule |
Anatomical structure development, cell adhesion, dendrite morphogenesis, dendrite self-avoidance, locomotory behaviour, negative regulation of cell adhesion, positive regulation of phosphorylation, post-embryonic retina morphogenesis in camera-type eye, protein binding |
| DYRK1A56,58 | Dual-specificity tyrosine- (Y)-phosphorylation regulated kinase 1A |
Circadian rhythm, cytoskeletal identical negative regulation of DNA damage response, signal transduction by p53 class mediator, non-membrane spanning protein tyrosine kinase activity, peptidyl-serine phosphorylation, peptidyl- threonine phosphorylation, peptidyl-tyrosine phosphorylation, positive regulation of protein deacetylation, protein autophosphorylation |
| FOXP158 | Forkhead box Pl | Cardiovascular system development, chromatin binding, in utero embryonic development, lung development, motor neuron axon guidance, negative regulation of transcription, DNA-templated, nucleic acid binding transcription factor activity, positive regulation of immunoglobulin production, positive regulation of mesenchymal cell proliferation |
| GIGYF158 | GRB10 interacting GYF protein 1 |
Insulin-like growth factor receptor signalling pathway, protein binding |
| GRIN2B56,58 | Glutamate receptor, ionotropic, N-methyl-D- aspartate 2B |
Calcium ion transport, cation channel activity, cation transmembrane transport, cell-cell signalling, detection of mechanical stimulus involved in sensory perception of pain, fear response, glutamate receptor signalling pathway, glycine binding, in utero embryonic development, ion binding, ionotropic glutamate receptor signalling pathway, neurological system process |
| KATNAL256,58 | Katanin p60 subunit A-like2 |
ATP binding, ATP catabolic process, ATPase activity, catabolic process, cellular nitrogen compound metabolic process, cytoskeletal DNA metabolic process, DNA recombination, DNA repair, four-way junction helicase activity, helicase activity, ion binding, microtubule binding, microtubule severing, microtubule-severing ATPase activity, response to stress |
| KDM5B58 | Lysine (K) -specific demethylase 5B |
Cellular nitrogen compound metabolic process, cellular protein modification process, DNA binding, histone demethylase activity (H3-K4 specific), histone H3-K4 demethylation, negative regulation of transcription, DNA-templated, nucleic acid binding transcription factor activity, oxidation-reduction process, oxidoreductase activity |
| KDM6B58 | Lysine (K) -specific demethylase 6B |
Beta-catenin binding, cardiac muscle cell differentiation, cell differentiation, cellular protein modification process, cellular response to hydrogen peroxide, dioxygenase activity, endothelial cell differentiation, histone demethylase activity (H3-K27 specific), histone demethylation, inflammatory response, ion binding, mesodermal cell differentiation, metal ion binding |
|
KMT2C (MLL3)56 |
Lysine (K) -specific methytransferase 2C |
Cellular nitrogen compound metabolic process, cellular protein modification process, DNA binding, histone methylation, histone methytransferase activity (H3-K4 specific), ion binding, methylation, methyltransferase activity, poly(A) RNA binding, RNA binding, signal transduction, transcription, DNA-templated, zinc ion binding |
| KMT2E58 | Lysine (K)-specific methyltransferase 2E |
Cell cycle arrest, cell differentiation, cellular nitrogen compound metabolic process, cellular protein modification process, cellular response to retinoic acid, DNA metabolic process, DNA methylation, enzyme binding, erythrocyte differentiation, histone H3-K4 methylation, histone methyltransferase activity (H3-K4 specific), immune system process, ion binding |
| MED13L58 | Mediator complex subunit 13-like |
Cellular nitrogen compound metabolic process, protein binding transcription factor activity, regulation of transcription from RNA polymerase II promoter, RNA polymerase II transcription cofactor activity, transcription from RNA polymerase II promoter |
| NCKAP158 | NCK-associated protein 1 | Anatomical structure development, apoptotic process, basal protein localisation, cell migration involved in gastrulation, embryonic body morphogenesis, embryonic foregut morphogenesis, endoderm development, enzyme binding, establishment or maintenance of actin cytoskeleton polarity, Fc-gamma receptor signalling pathway involved in phagocytosis, immune system process |
| PHF258 | PHD finger protein 2 | Calcium ion transport, ion binding, metal ion binding, regulation of ion transmembrane transport, transmembrane transport, transport, voltage-gated calcium channel activity |
| POGZ56,58 | Pogo transposable element with ZNF domain |
Cellular component assembly, chromosome organisation, DNA binding, ion binding, kinetochore assembly, metal ion binding, mitotic sister chromatid cohesion, nucleic acid binding, protein binding |
| RIMS158 | Regulating synaptic membrane exocytosis 1 |
Cell–cell signalling, cellular component assembly, enzyme binding, enzyme regulator activity, glutamate secretion, intracellular protein transport, ion binding, ion channel binding, membrane fusion, membrane organisation, metal ion binding, poly(A) RNA binding, positive regulation of gene expression, positive regulation of inhibitory postsynaptic membrane potential |
| SCN2A56,58 | Sodium channel, voltage gated, type II alpha subunit |
Intrinsic apoptotic signalling pathway in response to osmotic stress, ion transport, membrane depolarisation during action potential, nervous system development, neuron apoptotic process, sodium ion transmembrane transport, sodium ion transport, transmembrane transport, transmembrane transporter activity, transport, voltage-gated sodium channel activity |
| SUV420H156 | Suppressor of variegation 4–20 homologue 1 (Drosophila) |
Anatomical structure development, cellular nitrogen compound metabolic process, cellular protein modification process, histone methylation, histone methyltransferase activity (H4-K20 specific), methyltransferase activity, muscle organ development, regulation of transcription, DNA-templated |
| SYNGAP156 | Synaptic Ras GTPase activating protein 1 |
Dendrite development, enzyme regulator activity, GTPase activator activity, negative regulation of neuron apoptotic process, negative regulation of Ras protein signal transduction, pattern specification process, positive regulation of GTPase activity, Ras protein signal transduction, receptor clustering, regulation of synapse structure and activity, SH3 domain binding |
| TBR156,58 | T-box, brain, 1 | Axon guidance, cellular nitrogen compound metabolic process, commitment of neuronal cell to specific neuron type in forebrain, DNA binding, nucleic acid binding transcription factor activity, positive regulation of transcription from RNA polymerase II promoter, regulation of gene expression, RNA polymerase II core promoter sequence-specific DNA |
| TCF7L258 | Transcription factor 7-like 2 (T-cell specific, HMG- box) |
Armadillo repeat domain binding, beta-catenin binding, bone mineralisation, canonical Wnt signalling pathway, catenin import into nucleus, cell cycle arrest, cell proliferation, cellular nitrogen compound metabolic process, cellular response to starvation, chromatin binding, chromosome organisation, DNA binding, DNA metabolic process |
| TNRC6B58 | Trinucleotide repeat containing 6B |
Fc-epsilon receptor signalling pathway, gene expression, gene silencing by RNA, immune system process, innate immune response, neurotrophin TRK receptor signalling pathway, Notch signalling pathway, nucleotide binding, phosphatidylinositol-mediated signalling, poly(A) RNA binding, positive regulation of nuclear-transcribed mRNA catabolic process |
| WAC58 | WW domain containing adaptor with coiled-coil |
Cellular nitrogen compound metabolic process, cellular protein modification process, cellular response to DNA damage stimulus, chromatin binding, enzyme binding, G1 DNA damage checkpoint, histone H2B conserved C-terminal lysine ubiquitination, negative regulation of proteasomal ubiquitin-dependent protein catabolic process |
| WDFY358 | WD repeat and FYVE domain containing 3 |
1-phosphatidylinositol binding, aggrephagy, autophagy, beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase activity, catabolic process, ion binding, lipid binding, metal ion binding, response to stress |
The list of genes associated with autism spectrum disorder (ASD) was compiled from two original research papers that present a comprehensive statistical analysis of essentially all published ASD exomes sequenced to date,56,58 which also includes data from earlier work.57,59–61,63–65 The list includes genes that harbour two or more rare, de novo splice-site, nonsense, or frameshift variants in ASD probands or those with a false discovery rate of less than 0·05, calculated using the TADA method,56 which combines data on rare inherited or de novo damaging missense, splice-site, nonsense, or frameshift variants. Note that KDM5B has an equal count of de novo variants in unaffected siblings.58 The gene ontology (GO) terms for these genes—standardised descriptions of gene products that indicate associated biological processes, cellular components, and molecular functions—were obtained from Ensembl BioMart. The resulting list was shortened using REViGO to obtain a representative subset of GO terms using an algorithm that clusters by semantic similarities, avoiding redundancy. The highest-ranked biological processes and highest-ranked molecular function GO terms are listed for each gene.
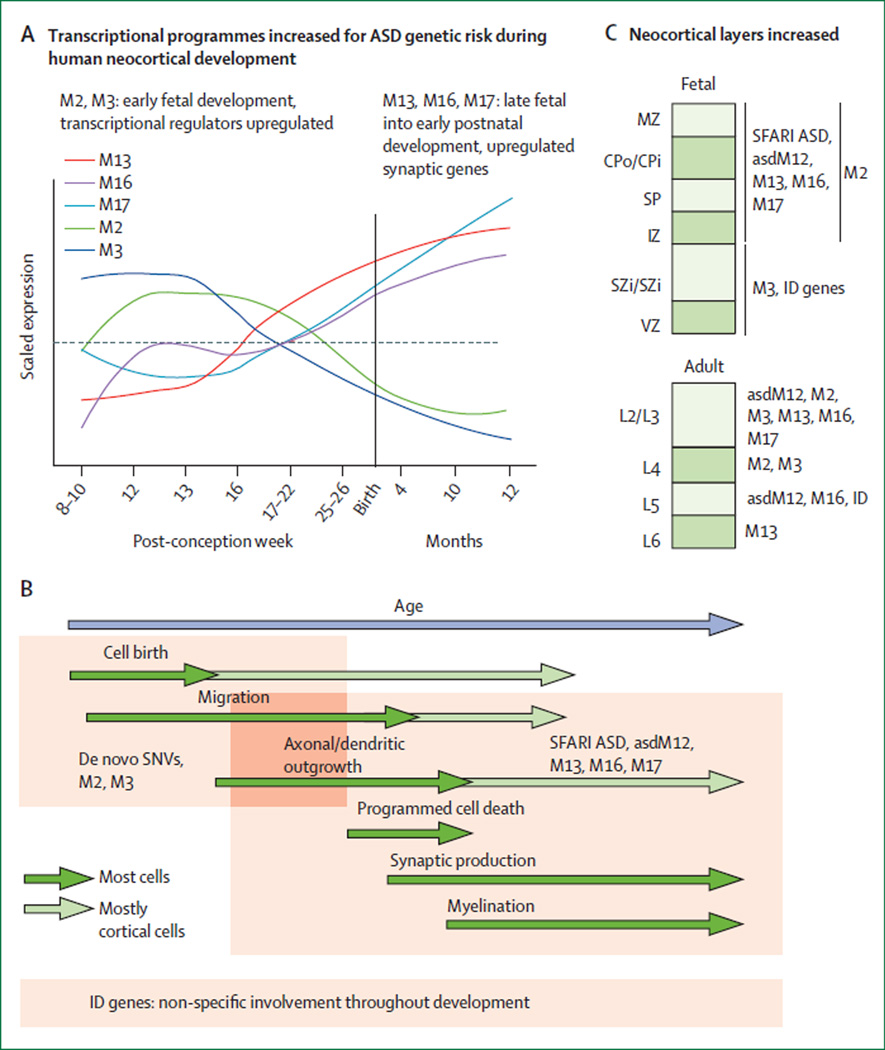
The growing set of risk genes identified by statistical methods, without reference to an a priori biological hypothesis, has also enabled relatively unbiased assessments of the representation of pathways or processes relevant to ASD (table, figure 1). On the basis of exome-sequencing gene-discovery efforts, a strong representation has been reported of genes associated with chromatin modification, synaptic function, targets of fragile X mental retardation proteins, targets of RBFOX splicing factors, and early embryonic development, among other things.56,58,63,68 Additionally, several recent studies have broadened the scope of analyses aimed at identifying functional clustering of ASD genes to include the question of when and where mutations might be having their effect.59,67,69 In this case as well, agreement has emerged that subsets of ASD risk genes seem to have particular relevance for mid-fetal cortical development, particularly in glutamatergic neurons (figure 1).
Figure 1. A model of the effects of gene sets implicated in autism spectrum disorder.
(A) Autism spectrum disorder (ASD) risk genes from several sources, including mutations identified by exome sequencing60,61,63,65 of candidate genes from the Simons Foundation Autism Research Initiative (SFARI) database and neuronal genes dysregulated in the post-mortem brains of people with ASD,66 were compiled as described in detail by Parikshak and colleagues.67 ASD risk genes were differentially enriched in five co-expression modules throughout development: M2, M3, M13, M16, and M17. (B) Early transcriptional regulators in M2/M3 are enriched for rare de novo variants (RDNVs), whereas the later-expressed synaptic genes in M13, M16, and M17 are associated with previously studied ASD candidate genes compiled in the SFARI database and dysregulated in the post-mortem brain tissue from patients diagnosed with an ASD. By contrast with ASD genes, more than 400 known mendelian intellectual disability (ID) risk genes compiled from multiple sources68 were not enriched for specific developmental trajectories. (C) By using publicly available gene expression data from specific cell types or cortical laminae, ASD risk genes were found to be more consistently associated with post-mitotic laminae during early fetal development (IZ, SP, CPo/CPi, and MZ) and upper cortical layers in adults (L2 or L3, and RDNV-associated genes in L4). Several gene co-expression modules that correspond to specific processes in brain development that are enriched for ASD genes are also strongly associated with markers of upper-layer glutamatergic neurons in adult cortex, which suggests that many ASD genes preferentially affect these cell types. Work from Willsey and colleagues59 identified a subset of genes enriched in lower-layer neurons, but overall supported enrichment in glutamatergic neurons. SNV=single-nucleotide variant. MZ=marginal zone. CPo=outer cortical plate. CPi=inner cortical plate. SP=subplate zone. IZ=intermediate zone. SZi=inner subventricular zone. VZ=ventricular zone. Adapted with permission from Parikshak and colleagues.67
Rare transmitted alleles
Considerable progress has been made via the study of rare transmitted variation. The examination of both inbred and outbred families has highlighted the contribution of rare recessive mutations to ASD, and several studies have highlighted the potential contribution of rare-variant detection in large-scale case–control samples.
Although studies of consanguineous families have not had the numerical yield of recent studies of de novo variation, the findings have proved to be crucial in the study of ASD pathophysiology. For example, Strauss and colleagues43 identified the rare homozygous truncating mutations in CNTNAP2 and their contribution to ASD and epilepsy through the study of inbred old-order Amish pedigrees. The authors of this report also noted abnormalities in glutamatergic pyramidal cell morphology and aberrant neuronal migration in the brains of mutation carriers undergoing epilepsy surgery.43Subsequent studies of the gene in model systems have provided additional insights into pathophysiology,70including a role for oxytocin neurons in the genesis of ASD.71 Of note, a recent large-scale analysis of rare heterozygote missense mutations in this gene found no association with autism,72 consistent with its recessive inheritance in the Amish and the lack of phenotype observed in the Amish mutation carriers, who are themselves heterozygotes. Although these findings provided little insight into the potential contribution of very rare and de novo LoF mutations affecting only one gene copy (in view of a paucity of these findings in more than 2000 probands), they point to a quite circumscribed role for CNTNAP2 mutations in the overall population risk of ASD. Importantly, a small contribution to population risk is not synonymous with a quantification of the overall importance of the gene; the result has no bearing on whether or not the biological insights gained by the study of homozygous LoF mutations might generalise to affected individuals who carry contributing mutations in other genes.3
Rare recessive mutations have also been identified in the genes SLC9A9 (solute carrier family 9, subfamily A [NHE9, cation proton antiporter 9], member 9)73 and BCKDK (branched chain ketoacid dehydrogenase kinase)74 in Middle Eastern consanguineous families with autism and epilepsy. SLC9A9 is an endosomal Na+– H+ exchanger,75 whereas BCKDK is an essential enzyme in branched chain aminoacid synthesis.76 The latter finding led to the exciting hypothesis, currently being tested, that branched chain aminoacid supplementation in affected children and newborn mutation carriers might mitigate some or all of the resulting phenotype, particularly in view of evidence for rescue in mouse models.74 Finally, several papers point to an excess of de novo mutations in outbred families with ASD, an observation that is likely to lead to specific gene discoveries as relevant sample sizes increase.77,78
Similarly, although less statistically powerful than findings from studies of de novo mutation, the calculation of the burden of rare heterozygous transmitted variations in cases versus controls has contributed to gene discovery in ASD. For example, Neale and colleagues61 recently showed that case–control exome sequencing can provide an important complement to studies of de novo mutation. Subsequently, He and colleagues79 presented a method to integrate evidence from de novo and transmitted rare variation into a single statistic that was used to maximise gene discovery in a large-scale exome study that included both trios and case–control samples.56
Towards a new neurology of ASD
The genetic advances outlined above make it clear that ASD is a common syndrome that comprises, at least in part, several hundred rare disorders. Similar heterogeneity must be expected for the role of common variants and rare, intermediate effect-size alleles that have yet to be found. Thus, from any perspective, understanding of ASD is complicated by extreme genetic complexity and heterogeneity. Moreover, substantial challenges to translating even the most robust genomic findings into an actionable understanding of ASD pathology have emerged along with recent success in gene discovery: the biological pleiotropy of many ASD genes; the highly dynamic molecular landscape of human brain development; and poor understanding of the molecular, cellular, and circuit-level diversity and function of the human CNS.3,80
Fortunately, despite these challenges, evidence has steadily accumulated that the highly heterogeneous and functionally diverse set of ASD genes identified so far converge on a smaller set of specific molecular pathways or brain circuits.11,81,82 Several lines of evidence support this idea of molecular convergence—most importantly, post-mortem gene expression in ASD compared with the typically developing brain, and gene expression and protein-network analysis56,58,59,63,66–69,83,84 (figure 1). Diverse approaches indicate that ASD risk genes interact in specific pathways and cell types. In addition to synaptic function, evidence exists for roles in a wide range of other cellular functions, from chromatin modification to receptor signalling to protein synthesis. Furthermore, a subset of large-effect ASD risk genes is enriched in cortical glutamatergic neurons. In post-mortem brains from people with ASD, coordinated downregulation of synaptic function genes and upregulation of microglial and neural immune genes have been reported,66 which when coupled with other findings, seems to signify a global dysregulation of cortical patterning and synaptic homoeostasis.66 In this context, the glial upregulation is not necessarily inflammatory, but probably reflects aberrant synaptic pruning.
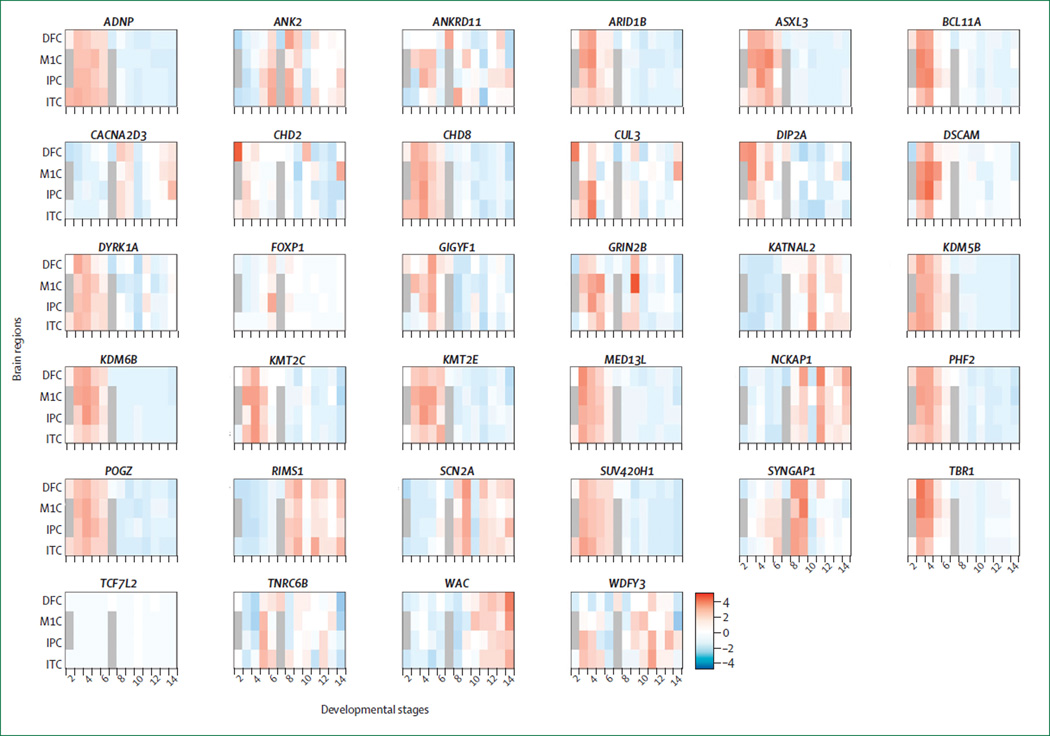
In view of the compelling evidence that most cases of autism have their onset in the first months of life,85 and the emerging evidence that at least a subset of ASD risk genes converge in time during fetal development (figure 2), a pressing question is how and whether effective treatments can be developed to target core symptoms. A priori, we cannot answer this question with certainty, but we look for guidance to recent findings from animal models and to clinical experience with other neurodevelopmental disorders. For example, adult reversal of putatively developmental behavioural phenotypes has been observed in several neurodevelopmental disorders.87 Moreover, epilepsy, which shows a high degree of co-morbidity with ASD, also has many forms, including those with neurodevelopmental causes. Remarkably, most cases of epilepsy, even neurodevelopmental epilepsy, respond in late childhood or adulthood to antiepileptic drugs, although variably. In summary, although far from definitive, the available data suggest that at least some forms of ASD involve time-specific developmental deficits as well as ongoing alterations in CNS functioning that might present targets for treatment, even well after the first emergence of symptoms.88 Furthermore, although many high-confidence ASD risk genes are most highly expressed during fetal brain development, a group of risk genes is involved in neuronal signalling, the expression of which coincides with neuronal maturation, providing another potential postnatal treatment window (figure 2).
Figure 2. Developmental and regional expression patterns of newly identified recurrently mutated risk genes in autism spectrum disorder.
Heat map depicting gene expression patterns in various brain regions throughout development for each of the recurrently mutated autism spectrum disorder (ASD) risk genes depicted in the table, using gene expression data adapted from Kang and colleagues.86 The colour code is scaled from red (high expression) to blue (low expression). Developmental stages range from post-conception week 4–8 (stage 1) and mid-gestation (stages 4–6), through birth to 6 months (stage 8), and into adulthood (stage 13 and beyond). Most genes have very circumscribed developmental gradients of expression, showing either high fetal expression (eg, chromatin and transcriptional regulatory genes) or a postnatal increase concomitant with neuronal maturation (eg, synaptic signalling genes). DFC=dorsolateral prefrontal cortex. M1C=primary motor (M1) cortex. IPC=inferior parietal cortex. ITC=inferior temporal cortex.
Clinical relevance
At present, rare ASD-related mutations are anticipated in 20–40% of clinical samples.58 An important related issue is whether and how genetic diagnoses will play a key part in clinical assessment and care. Clearly, at present, the identification of rare forms of autism can provide relevant clinical information. For example, the identification of FMR1 mutations in a proband will have obvious implications for family counselling, including increased vigilance for learning disability in related girls and the presence of fragile X-associated tremor/ataxia syndrome in older family members. Similarly, copy number variation at 22q11 has been associated with cardiac abnormalities, and 15q11–13 with sudden death, and both would warrant consultation, particularly if the first presentation is for early behavioural issues. Current expert consensus is that genetic testing is warranted in most individuals with ASD, starting with chromosomal microarray or testing of specific genes such as FMR1 or PTEN, depending on the patient’s sex and clinical presentation.89
Although most genetic findings would not dictate treatment at present, they might nonetheless have potential benefits for the patient and their family, in addition to the research community. In view of the many uncertainties that accompany a diagnosis of ASD, it is not at all surprising that some families are eager to learn specific information about potential molecular mechanisms, even if these will not alter the course of treatment. Furthermore, as noted previously, some genetic findings may have a bearing on family planning. Moreover, as clinical samples increase in size, it is likely that genetic testing, along with family history and clinical assessment, will help to predict prognosis in many cases.90,91
However, the limitations of current genetic testing should be recognised. For example, from the first observations of de novo CNVs contributing to ASD, it has been clear that these are not acting in a purely mendelian manner, and that great care must be taken with regard to estimating recurrence risk. For example, the families with 16p11.2 deletions and duplications in the first reports of association with ASD showed multiple affected siblings where only one carried the high-risk CNV,50–52similar to the high unaffected carrier frequency with NRXN1 deletions.92 A recent genome-wide sequencing study93 has reinforced this important point by showing that the identification of a de novo ASD mutation in one child does not necessarily indicate that the risk for recurrence for a future sibling would be at the population base rate. At present, the best predictor of sibling recurrence remains family history.
Conclusions
The combination of advances in ASD genetics, the rapidly declining costs of next-generation sequencing, and the widespread deployment of electronic medical records will enable novel and potentially powerful approaches to the use of accumulating clinical data to advance research. The potential for large-scale investigations of this type has been well demonstrated in data-mining studies based solely on clinical data in ASD,94 and one can easily imagine that the combination of routine clinical sequencing with expansive clinical databases will open new vistas in understanding the genetics, phenomenology, and comorbidities of ASD. However, many challenges remain, including the need to ensure the integrity and accuracy of clinical data that could be biased by a range of sociological, demographic, or economic issues. Furthermore, it is unlikely that research defining imaging or physiological biomarkers will be available in these large, biomedically based population cohorts. Still, the scale of these data and their connection with longitudinal medical records and related laboratory tests will probably enable significant and more efficient discovery in parallel with smaller, focused laboratory-based studies. Developing the resources and infrastructure necessary for these large-scale collaborative studies will be an essential first step on the path to more precise and personalised treatment in ASD, a necessity for this clinically and aetiologically diverse syndrome.
Search strategy and selection criteria.
We searched PubMed for papers published in English between Jan 1, 1995, and Feb 28, 2015, using the terms “autism” and “genetics”. Articles were also identified through searches of the reference lists of selected papers and the authors’ personal files. The final selection was based on relevance to the topics covered in this Series paper.
Acknowledgments
This work was made possible by funding from the National Institute of Mental Health (grant 5R01 MH100027 to DHG and MWS; 5P50 HD055784 to DHG; 5R01 MH094714 to DHG; 5R01 MH100028 to DHG and MWS; and U01 MH100239 to MWS), the Simons Foundation (grant 206744 to DHG and 274624 to MWS), and the Overlook International Foundation (to MWS).
Footnotes
Contributors
DHG and MWS planned the content, performed literature searches, wrote the initial draft, and edited the manuscript.
Declaration of interests
DHG has served on scientific advisory boards for Novartis and Synapdx Corporation; he has patents pending to Regents of the University of California for mTOR (case #2012–776) and genes dysregulated in autism (application #14/119,755), and a patent issued to Regents of the University of California for biomarkers for autism (US patent 8,173,369). MWS has served on scientific advisory boards for Pfizer and Synapdx Corporation.
For Ensembl BioMart see http://www.ensembl.org/biomart/
For REViGO see http://revigo.irb.hr/
For the Simons Foundation Autism Research Initiative (SFARI) database see http://sfari.org/
Contributor Information
Daniel H Geschwind, Neurogenetics Program, Department of Neurology, and Center for Autism Research and Treatment, Semel Institute, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Matthew W State, Department of Psychiatry, Langley Porter Psychiatric Institute, University of California, San Francisco, CA, USA.
References
- 1.Elsabbagh M, Divan G, Koh YJ, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YS, Leventhal BL. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol Psychiatry. 2015;77:66–74. doi: 10.1016/j.biopsych.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krystal JH, State MW. Psychiatric disorders: diagnosis to therapy. Cell. 2014;157:201–214. doi: 10.1016/j.cell.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiezun A, Pulit SL, Francioli LC, et al. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 2013;9:e1003301. doi: 10.1371/journal.pgen.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ringman JM, Coppola G. New genes and new insights from old genes: update on Alzheimer disease. Continuum (Minneap Minn) 2013;19:358–371. doi: 10.1212/01.CON.0000429179.21977.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarti A. Population genetics—making sense out of sequence. Nat Genet. 1999;21(1 Suppl):56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 10.Risch N, Spiker D, Lotspeich L, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klei L, Sanders SJ, Murtha MT, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sucksmith E, Roth I, Hoekstra RA. Autistic traits below the clinical threshold: re-examining the broader autism phenotype in the 21st century. Neuropsychol Rev. 2011;21:360–389. doi: 10.1007/s11065-011-9183-9. [DOI] [PubMed] [Google Scholar]
- 16.Constantino JN. The quantitative nature of autistic social impairment. Pediatr Res. 2011;69:55R–62R. doi: 10.1203/PDR.0b013e318212ec6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss LA, Arking DE Gene Discovery Project of Johns Hopkins & the Autism Consortium Daly MJ Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werling DM, Lowe JK, Luo R, Cantor RM, Geschwind DH. Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder. Mol Autism. 2014;5:13. doi: 10.1186/2040-2392-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone JL, Merriman B, Cantor RM, et al. Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet. 2004;75:1117–1123. doi: 10.1086/426034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe JK, Werling DM, Constantino JN, Cantor RM, Geschwind DH. Social responsiveness, an autism endophenotype: genomewide significant linkage to two regions on chromosome 8. Am J Psychiatry. 2015;172:266–275. doi: 10.1176/appi.ajp.2014.14050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu AT, Yoon J, Geschwind DH, Cantor RM. QTL replication and targeted association highlight the nerve growth factor gene for nonverbal communication deficits in autism spectrum disorders. Mol Psychiatry. 2013;18:226–235. doi: 10.1038/mp.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alarcón M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anney R, Klei L, Pinto D, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glahn DC, Knowles EE, McKay DR, et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:122–130. doi: 10.1002/ajmg.b.32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcón M, Cantor RM, Liu J, Gilliam TC, Geschwind DH Autism Genetic Research Exchange Consortium. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70:60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glahn DC, Williams JT, McKay DR, et al. Discovering schizophrenia endophenotypes in randomly ascertained pedigrees. Biol Psychiatry. 2015;77:75–83. doi: 10.1016/j.biopsych.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehouse AJ, Bishop DV, Ang QW, Pennell CE, Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011;10:451–456. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernes SC, Newbury DF, Abrahams BS, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiersen AM, Constantino JN, Volk HE, Todd RD. Autistic traits in a population-based ADHD twin sample. J Child Psychol Psychiatry. 2007;48:464–472. doi: 10.1111/j.1469-7610.2006.01720.x. [DOI] [PubMed] [Google Scholar]
- 33.Constantino JN, Davis SA, Todd RD, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33:427–433. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- 34.Coon H, Villalobos ME, Robison RJ, et al. Genome-wide linkage using the Social Responsiveness Scale in Utah autism pedigrees. Mol Autism. 2010;1:8. doi: 10.1186/2040-2392-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH. A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am J Psychiatry. 2007;164:656–662. doi: 10.1176/ajp.2007.164.4.656. [DOI] [PubMed] [Google Scholar]
- 36.Chaste P, Klei L, Sanders SJ, et al. A genome-wide association study of autism using the Simons Simplex Collection: does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.09.017. published online Sept 29. http://dx.doi.org/10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed]
- 37.Anney R, Klei L, Pinto D, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 44.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 45.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 46.Jacquemont ML, Sanlaville D, Redon R, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Autism Genome Project Consortium. Szatmari P, Paterson AD, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 51.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 53.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders SJ, Ercan-Sencicek AG, Hus V, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong S, Walker MF, Carriero NJ, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willsey AJ, Sanders SJ, Li M, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parikshak NN, Luo R, Zhang A, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peñagarikano O, Abrahams BS, Herman EI, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peñagarikano O, Lazaro MT, Lu XH, et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.3010257. 271ra278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murdoch JD, Gupta AR, Sanders SJ, et al. No evidence for association of autism with rare heterozygous point mutations in contactin-associated protein-like 2 (CNTNAP2), or in other contactin-associated proteins or contactins. PLoS Genet. 2015;11:e1004852. doi: 10.1371/journal.pgen.1004852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwede M, Garbett K, Mirnics K, Geschwind DH, Morrow EM. Genes for endosomal NHE6 and NHE9 are misregulated in autism brains. Mol Psychiatry. 2014;19:277–279. doi: 10.1038/mp.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Popov KM, Hawes JW, Harris RA. Mitochondrial alpha-ketoacid dehydrogenase kinases: a new family of protein kinases. Adv Second Messenger Phosphoprotein Res. 1997;31:105–111. [PubMed] [Google Scholar]
- 77.Yu TW, Chahrour MH, Coulter ME, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim ET, Raychaudhuri S, Sanders SJ, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He X, Sanders SJ, Liu L, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.State MW, Geschwind DH. Leveraging genetics and genomics to define the causes of mental illness. Biol Psychiatry. 2015;77:3–5. doi: 10.1016/j.biopsych.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 81.Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- 82.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 83.Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry. 2013;18:1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- 84.Ben-David E, Shifman S. Networks of neuronal genes affected by common and rare variants in autism spectrum disorders. PLoS Genet. 2012;8:e1002556. doi: 10.1371/journal.pgen.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones W, Klin A. Attention to eyes is present but in decline in 2-6-month-old infants later diagnosed with autism. Nature. 2013;504:427–431. doi: 10.1038/nature12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silva AJ, Ehninger D. Adult reversal of cognitive phenotypes in neurodevelopmental disorders. J Neurodev Disord. 2009;1:150–157. doi: 10.1007/s11689-009-9018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moreno-De-Luca D, Sanders SJ, Willsey AJ, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2013;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bernier R, Golzio C, Xiong B, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 94.Kohane IS. An autism case history to review the systematic analysis of large-scale data to refine the diagnosis and treatment of neuropsychiatric disorders. Biol Psychiatry. 2015;77:59–65. doi: 10.1016/j.biopsych.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]