Abstract
Vertebrate glycans constitute a large, important, and dynamic set of post-translational modifications that are notoriously difficult to manipulate and image. Although the chemical reporter strategy has been used in conjunction with bioorthogonal chemistry to image the external glycosylation state of live zebrafish and detect tumor-associated glycans in mice, the ability to image glycans systemically within a live organism has remained elusive. Here, we report a method that combines the metabolic incorporation of a cyclooctyne-functionalized sialic acid derivative with a ligation reaction of a fluorogenic tetrazine, allowing for the imaging of sialylated glycoconjugates within live zebrafish embryos.
Keywords: bioorthogonal chemistry, fluorescent probes, glycans, glycosylation, sialic acids
Cell-surface glycans mediate a variety of important biological processes, including inflammation, bacterial and viral infection, cardiovascular disease, cancer progression, and embryogenesis.[1] Owing to their complicated structures and non-template-driven synthesis, glycans are difficult to manipulate and interrogate compared to other biomacromolecules such as proteins and oligonucleotides. In particular, the biological importance of glycans has led to the need for in vivo imaging methods,[2] as imaging biomolecules in their native environments can provide a great deal of information on subcellular localization, interactions with neighboring cells and pathogens, and changes in expression resulting from various stimuli. Lectins and antibodies targeting specific glycan epitopes are ill-suited for in vivo imaging because of their poor tissue penetrance and low affinity for glycans.[3] As an alternative to these affinity-based methods, our laboratory developed the biorthogonal chemical reporter strategy to image and manipulate glycans;[4] in this strategy, a metabolic precursor to the sugar of interest armed with a bioorthogonal functional group is delivered to a population of cells or an organism by bathing, feeding, or injection, whereupon it is processed and appended to cell-surface glycans in lieu of the natural sugar. Subsequently, glycans that have incorporated the analogue can be visualized by a covalent ligation reaction with a probe molecule containing a complementary bioorthogonal functional group.
Our laboratory and others have previously employed the chemical reporter strategy to image a variety of glycan structures in developing zebrafish embryos,[5] which we chose to study because of the importance of the zebrafish as a model organism in developmental and infectious disease biology as well as its optical transparency.[6] In a typical experiment, an azido or alkynyl sugar is injected into the embryo during the 1–8 cell stage or added to the embryo’s medium during development. Then, the embryo is bathed in a solution of a fluorophore or affinity probe modified with the appropriate reaction partner for a copper-catalyzed or copper-free click ligation reaction. These experiments provide insight into the dynamics and localization of glycan expression during zebrafish development, but have been limited to studies of the enveloping layer, the outermost layer of the embryo analogous to skin. Exogenous fluorophores cannot access internal cells and tissues, and thus the method cannot be applied to study glycomic changes associated with most biological processes.
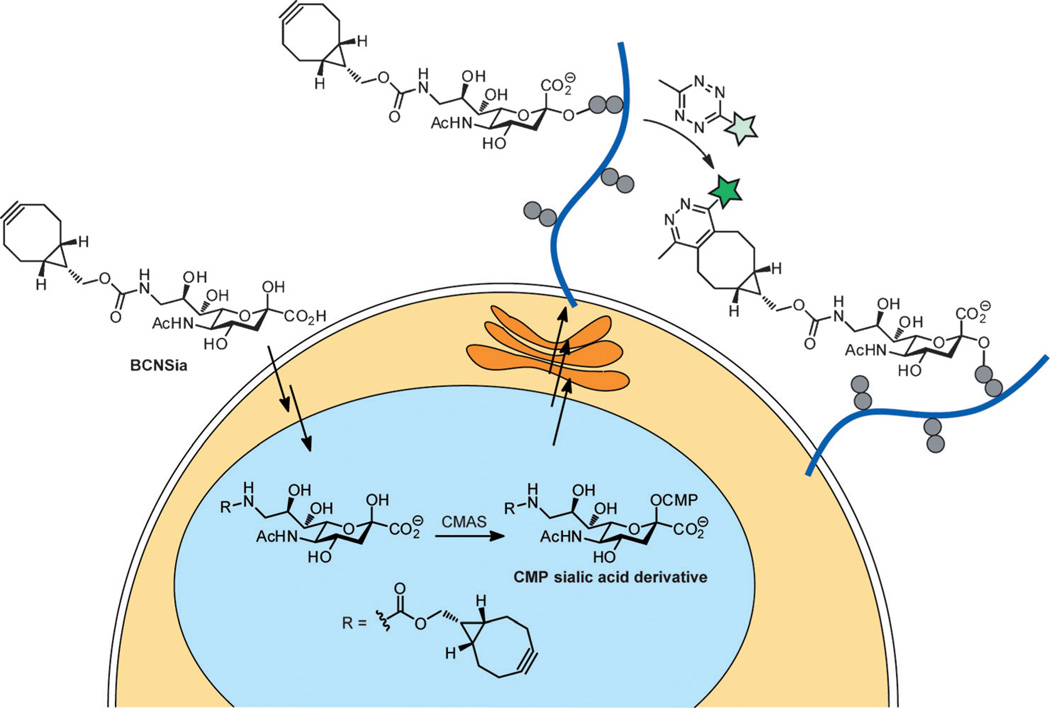
One possible approach to the system-wide labeling of cell-surface glycans is to introduce both the metabolic labeling substrate and, at a later stage, the imaging probe into the developing embryo by injection. However, unreacted probe would be trapped inside the organism, leading to high background fluorescence that would obscure signals from glycans of interest. This problem could be obviated by the use of a bioorthogonal fluorogenic probe whose fluorescence is activated in the course of the desired ligation reaction.[7] Although many such probes have been prepared in recent years, to the best of our knowledge, none has been used within a live animal thus far. Herein, we report that the metabolic incorporation of a bicyclononyne-functionalized sialic acid followed by reaction with a fluorogenic tetrazine enables the systemic imaging of sialylation during zebrafish embryogenesis (Figure 1).
Figure 1.
Expected metabolic pathway for the incorporation of BCNSia into cell-surface glycans followed by labeling with a fluorogenic tetrazine probe. CMAS = cytidine monophosphate N-acetylneuraminic acid synthetase, CMP = cytidine monophosphate.
Our selection of the cyclooctyne chemical reporter and tetrazine-based probe were driven by the following considerations: We sought bioorthogonal ligation reaction partners that would produce a large fluorescence enhancement, remain intact for up to several days during metabolic incorporation experiments, and react quickly at biologically relevant concentrations. Several groups have reported fluorogenic tetrazine probes that fluoresce upon reacting with strained alkenes and alkynes.[8] We were drawn to a recent study in which Devaraj and co-workers reported para-vinylenemethyltetrazines that exhibit up to 400-fold fluorescence turn-on;[9] we expected these tetrazines to be stable for at least several hours in serum,[10] making them good candidates for in vivo imaging experiments. Tetrazines are known to react with a variety of strained unsaturated hydrocarbons, including norbornene,[11] cyclopropene,[8c, 12] cyclooctyne,[8b,13] and trans-cyclooctene.[14] Of these choices, cyclooctyne stands out because it reacts more quickly than norbornene and cyclopropene,[15] but is not susceptible to the same in vivo instability as trans-cyclooctene, which isomerizes to relatively unreactive cis-cyclooctene within hours in serum.[16] Furthermore, several studies across different classes of fluorophores have shown that the pyridazine products resulting from alkyne–tetrazine cycloadditions are brighter than the dihydropyridazine products resulting from alkene–tetrazine cycloadditions.[9, 17]
Aside from its beneficial attributes, cyclooctyne has a liability as a chemical reporter: It is large relative to a monosaccharide substrate, unlike azides and terminal alkynes, the more common chemical reporters for glycan imaging. Indeed, unnatural monosaccharides containing modifications larger than a few atoms are generally poor substrates for the biosynthetic enzymes that enable their incorporation into glycans.[18] Sialic acid analogues are a fortuitous exception, however, in that large substituents at the C5 and C9 positions are tolerated by the enzymes involved in sugar activation and incorporation into mammalian cell-surface glycans.[19] Furthermore, the sialome is a very important sector of the glycome; sialylated glycans contribute to immune cell interactions, brain development, cancer progression, viral and bacterial infection, and many other glycobiological phenomena.[1] For these reasons, we chose to append a cyclooctyne to sialic acid for metabolic incorporation experiments, and prepared a bicyclononyne-functionalized sialic acid derivative (BCNSia, Figure 1) in a five-step synthesis from sialic acid (see the Supporting Information).[19b, 20]
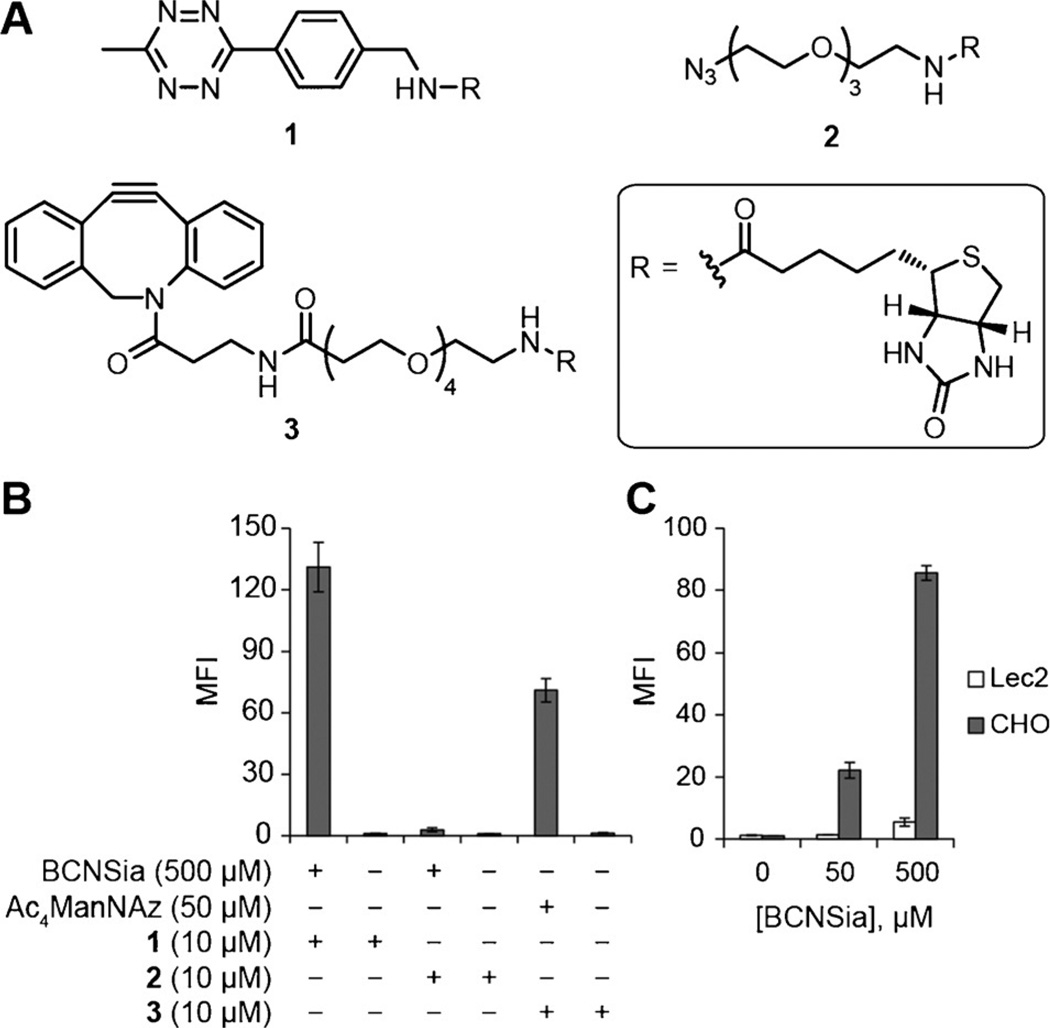
We first asked whether BCNSia could be metabolically incorporated into cultured mammalian cells. After growth in BCNSia-supplemented media for two days, human embryonic kidney 293 (HEK) cells exhibited robust fluorescence upon treatment with tetrazine-biotin probe 1 (Figure 2A) followed by avidin-488 detection and flow cytometry (Figure 2B). To assess the incorporation efficiency of BCNSia relative to that of commonly used peracetylated sugar derivatives, we compared the copper-free click reactions of BCNSia and peracetylated N-azidoacetylmannosamine (Ac4ManNAz), which is metabolized to an azide-bearing sialic acid derivative.[21] Cells were grown with either 500 µm BCNSia or 50 µm Ac4ManNAz for two days and then treated with azido-biotin probe 2 or dibenzoazacyclooctyne-biotin probe 3, respectively, and avidin-488. Although comparison of the fluorescence of these cell populations revealed that BCNSia was incorporated less efficiently than Ac4ManNAz (Figure 2B), use of the tetrazine ligation with BCNSia yielded adequate signal above background. To confirm that the low incorporation efficiency was not specific to HEK cells, we incorporated BCNSia into Jurkat cells and observed similar levels of signal above background that were dose-dependent up to 3 mm, the highest concentration that we tested (Supporting Information, Figure S1). Next, we verified that BCNSia was incorporated into mammalian cell-surface glycans in place of sialic acid by comparing the labeling of Chinese hamster ovary (CHO) cells and Lec2 cells. Lec2 is a CHO mutant that displays a 90% reduction in cell-surface sialylation owing to a deletion in CMPST, which transfers CMP sialic acid into the Golgi as a precursor to its addition to glycans.[22] Compared to CHO cells, Lec2 cells treated with BCNSia for two days displayed poor labeling with 1 (Figure 2C), consistent with the processing of BCNSia by CMP sialic acid synthetase and CMPST en route to cell-surface display. The small amount of labeling observed in Lec2 cells at high concentrations of BCNSia is consistent with the presence of a low basal level of sialylation. Taken together, these experiments establish that BCNSia can be incorporated into a variety of commonly used mammalian cell lines and is processed through the same metabolic pathway as sialic acid.
Figure 2.
BCNSia is incorporated into the cell-surface glycans of cultured mammalian cells. A) Structures of probe molecules used. B) HEK cells incubated with BCNSia or Ac4ManNAz and then treated with 1, 2, or 3 followed by avidin-488 display sugar-dependent labeling. C) Following treatment with BCNSia and detection with probe 1, Lec2 cells display significantly less BCNSia than do wild-type CHO cells. Error bars represent the standard deviation of three replicate experiments.
Next, we attempted to metabolically incorporate BCNSia into zebrafish embryos. Membrane-impermeable nucleotide sugars can be administered to zebrafish embryos by microinjection into the yolk during the 1–8 cell stage, a time period when the process of cytosolic streaming provides the sugar derivatives access to all daughter cells in the embryo.[5b] Given the high concentrations of BCNSia necessary for incorporation into mammalian cells and our desire to deliver the sugars to all cells of the developing embryo, we reasoned that microinjection would provide superior results to bathing embryos in sugar-containing media. We began by administering various amounts of BCNSia from 5 to 50 pmol. At 5 pmol, no adverse phenotype was visible, whereas at higher doses, the notochord became twisted and the embryos exhibited an extension defect (Figure S2).[23] Therefore, a 5 pmol dose of BCNSia was used in all subsequent experiments.
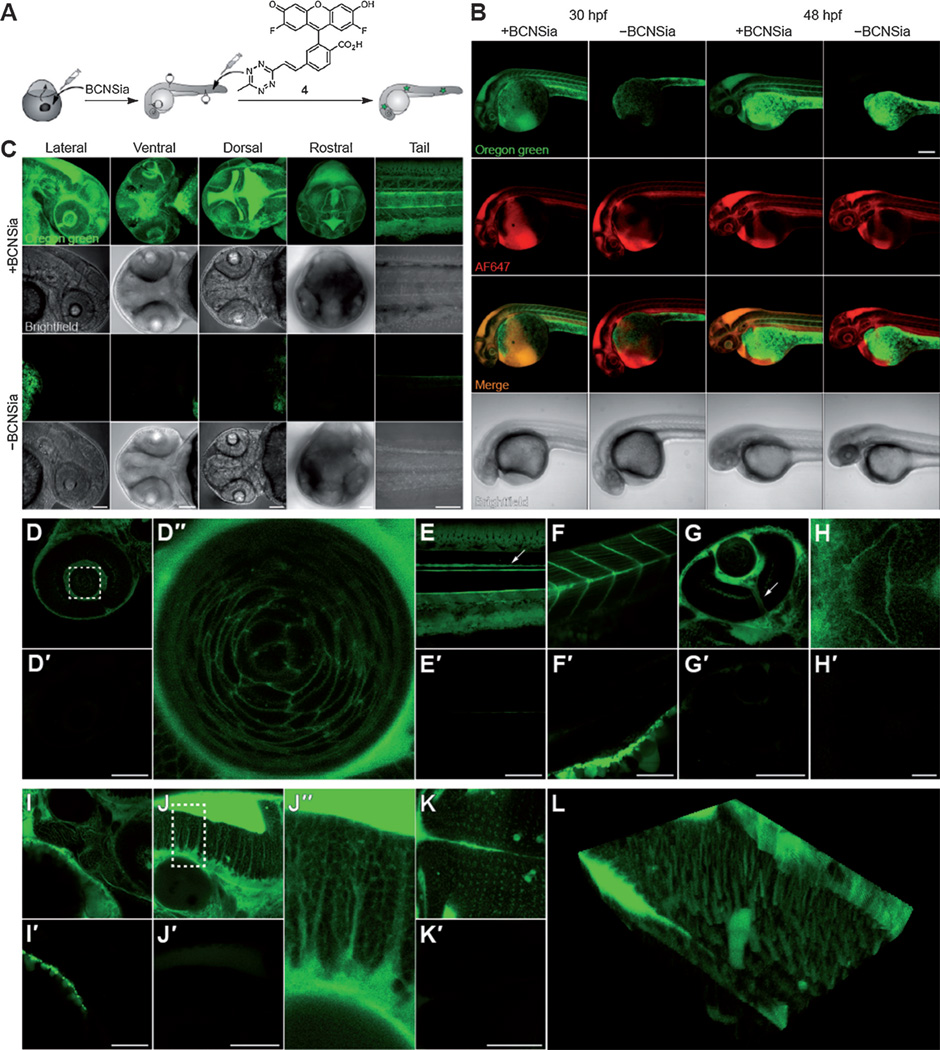
We next explored applications in molecular imaging by injection of fluorogenic tetrazine probe 4 into the caudal vein (Figure 3A). Probe 4 is virtually nonfluorescent, but successful reaction with a cyclooctyne yields a brightly fluorescent Oregon green moiety.[9] Embryos were injected with BCNSia at the 1–8 cell stage and were allowed to develop to various stages prior to imaging experiments. The caudal vein plexus is not fully formed until 30 hpf (hours post fertilization);[24] therefore, an equimolar mixture of 4 and Alexa Fluor (AF) 647 cadaverine, which we added as a nonspecific tracer of the vasculature, was injected into embryos at 30, 48, or 72 hpf. At 30 and 48 hpf, we observed robust BCNSia-dependent fluorescence from 4 throughout the interior of the embryo (Figure 3B); at 72 hpf, we observed little to no signal above background (Figure S3), likely because the dye quickly accumulated in the pronephric duct or because little BCNSia remained in the embryo after several days. The BCNSia-dependent signal observed at 30 and 48 hpf in areas of low perfusion is consistent with the labeling of cell-surface glycoconjugates by 4. We also observed BCNSia-independent green fluorescence in the yolk; colocalization in fixed embryos with a lipid droplet marker suggested that the yolk fluorescence was a result of encapsulation of 4 by lipid droplets (Figure S4),[25] perhaps followed by reaction with unsaturated fatty acids or sterols. As this phenomenon was confined to the yolk, we did not explore it further. Importantly, embryos injected with a non-fluorogenic tetrazine probe exhibited minimal BCNSia-dependent labeling (Figure S5), indicating the importance of using a fluorogenic ligation strategy.
Figure 3.
Zebrafish embryos treated with BCNSia and subsequently injected with 4 show robust systemic BCN-dependent labeling. A) Scheme showing injections of BCNSia and 4; embryos were injected with BCNSia or vehicle at the 1–8 cell stage and then injected with 4 in the caudal vein 1 h prior to imaging. B) Projection images of 30 and 48 hpf embryos treated with BCNSia or vehicle; 4 was co-injected with AF647-NH2 to map the vasculature. Scale bar: 200 µm. C) Projection images (20 Ö) of 48 hpf embryos from various viewpoints. D,D’) Lateral view of the eye and lens (D″). The dashed box in (D) indicates the field of view in (D″). E,E’) Lateral view of the tail with floor plate labeling (arrow). F,F’) Lateral view of the intermyotomal boundaries and muscle-fiber cells. G,G’) Ventral view of the optic nerve (arrow). H,H’) Ventral view of the developing mouth. I, I’) Lateral view of the developing jaw. J, J’) Lateral view of the hindbrain. The dashed box in (J) indicates the field of view in (J″). K,K’) Dorsal view of the hindbrain. L) Three-dimensional reconstruction of the dorsal view. Embryos were injected with BCNSia (D–L) or vehicle (D’–K’). All injections of 4 were performed at 48 hpf, except in (B). Scale bars for D–K: 100 µm.
Our initial experiments suggested that sialylation was abundant within several structures, which we explored in detail in embryos injected at 48 hpf with 4. The BCNSia dependence of labeling was particularly apparent in higher-magnification images of the head acquired from the lateral, ventral, dorsal, and rostral views, as well as in a lateral view of the tail (Figure 3C, Movies S1–S5, and Figure S6). In the eyes, both the retina and the surfaces of the onion-shaped fiber cells that make up the lens, which is known to heavily express the sialyltransferase St8SiaIII during development,[26] were sialylated (Figure 3D). Analysis of the labeling pattern within the lens showed that the fluorescence was more concentrated at intercellular contacts between three or more cells than at contacts between two cells (Figure 3D″). We also observed dense sialylation along the entire length of the floor plate, which is known to express the neural cell adhesion molecule, a developmentally important protein that displays polysialic acid (Figure 3E).[27] Also notable was the bright labeling along the intermyotomal boundaries as well as on the surface of muscle fiber cells in the myotomes, which also express St8SiaIII (Figure 3F).[26]
In addition to these structures that we had expected to be sialylated, we observed several novel sialylation patterns. A ventral view of the embryo revealed sialylation of the optic nerve (Figure 3G) as well as at the exterior of the developing mouth (Figure 3H); posterior to the mouth, we observed a tubular sialylation pattern from a lateral view (Figure 3I). In the hindbrain, we observed a columnar pattern of labeling along the dorsoventral axis of the embryo (Figure 3J). These cells displayed a neuron-like morphology, with a bulbous body and rod-like projections that traversed the height of the hindbrain until reaching the hindbrain ventricle (Figure 3J″). A dorsal view of this sialylation pattern showed the presence of more heavily sialylated cells arranged in a grid-like pattern extending laterally from the midline (Figure 3K). A three-dimensional reconstruction of these images (Figure 3L) indicated that the punctate labeling pattern observed in the dorsal view of the hindbrain (Figure 3K) indeed corresponds to the rod-like projections observed in the lateral view (Figure 3J).
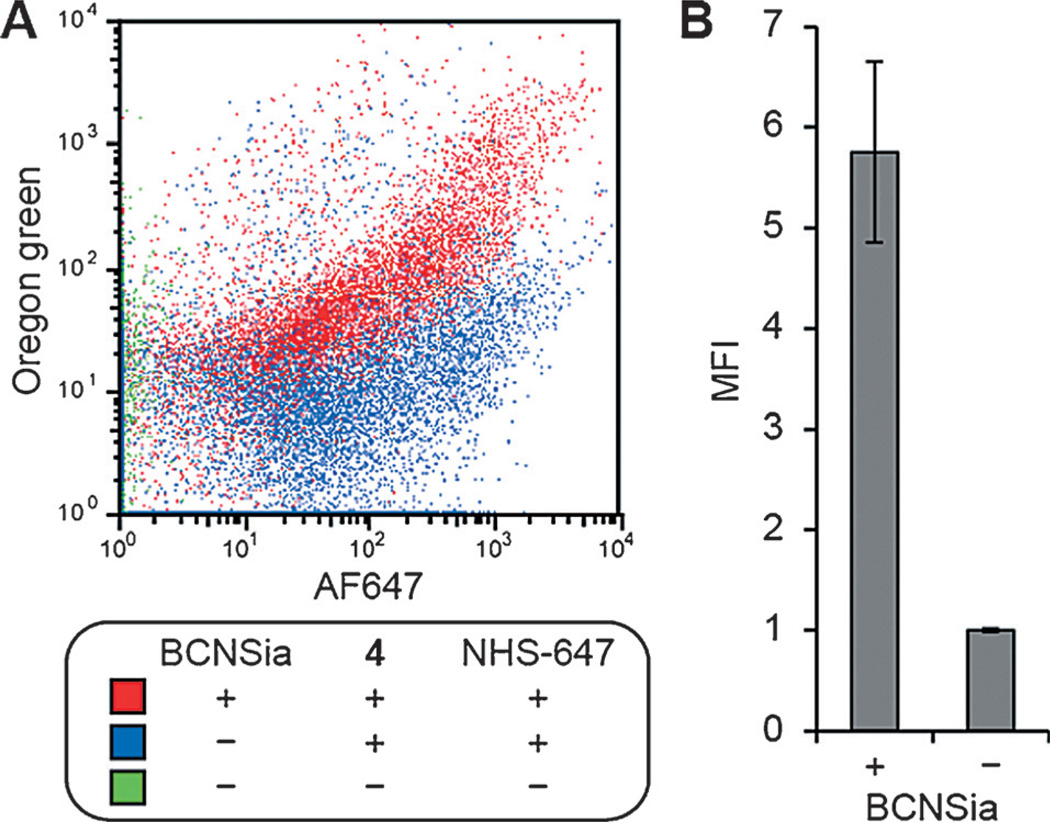
To independently confirm that injection of 4 into BCNSia-treated embryos resulted in covalent cell-surface labeling, we analyzed cells from dissociated embryos by flow cytometry. Embryos were microinjected with BCNSia, allowed to develop to 48 hpf, and injected with a mixture of 4 and NHS-AF647, a marker that was added to select for cells exposed to the vasculature. The embryos were then deyolked and dissociated into single cells by treatment with collagenase. Flow cytometry analysis indicated a BCNSia-dependent correlation between labeling with 4 and NHS-AF647 (Figure 4A). Analysis of cells displaying NHS-AF647 labeling showed robust tetrazine labeling that was dependent on BCNSia incorporation (Figure 4B), with minimal fluorescence arising from injection of 4 into embryos without BCNSia (Figure S7). Taken together, these data demonstrate both the successful display of BCNSia on the surfaces of internal zebrafish cells as well as our ability to covalently label BCNSia-containing glycoconjugates with our caudal vein injection method.
Figure 4.
Flow cytometry reveals that BCNSia-dependent fluorescence in zebrafish embryos arises from cell-surface labeling. Embryos were injected with BCNSia or vehicle at the 1–8 cell stage and then injected at 48 hpf with 4 and NHS-647, deyolked, dissociated, and analyzed by flow cytometry. A) Labeling with 4 as a function of labeling with NHS-647. B) Green fluorescence of cells from embryos injected with 4 and NHS-647, gated on cells displaying NHS-647 labeling. Error bars represent the standard deviation of three replicate experiments.
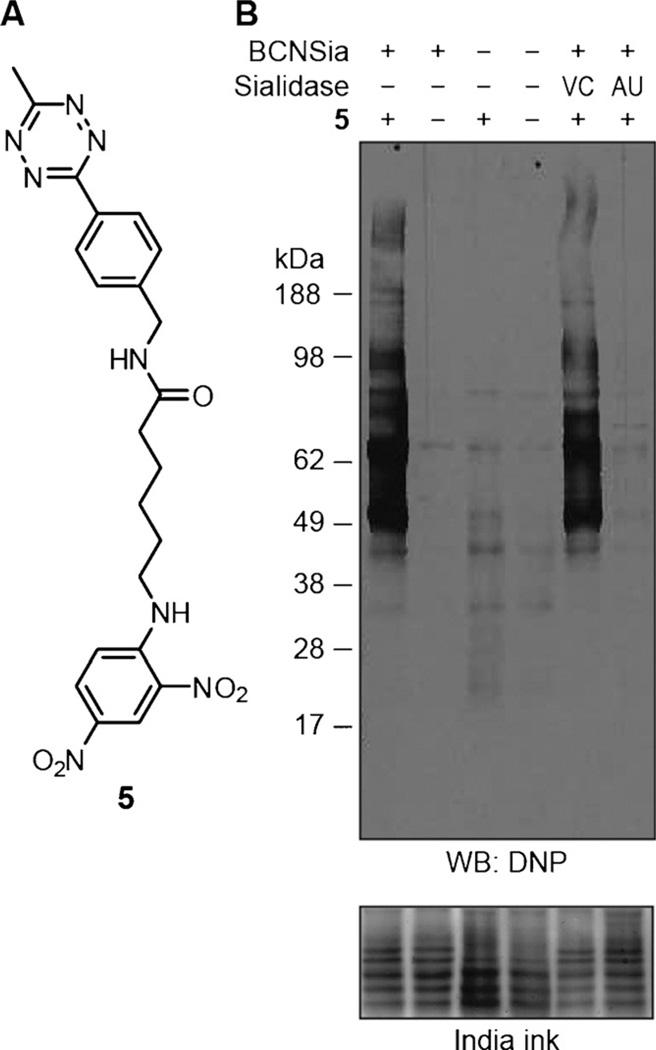
Next, we confirmed that BCNSia was incorporated into zebrafish sialoglycoproteins. We prepared lysates of deyolked embryos at 48 hpf, treated the lysates with tetrazine-dinitrophenyl conjugate 5 (Figure 5A), and analyzed the resulting products by Western blotting (Figure 5B). As expected, lysates from embryos injected with BCNSia and treated with 5 displayed robust labeling compared to uninjected and/or untreated embryos. Treatment with Vibrio cholerae sialidase had no effect, whereas treatment with Arthrobacter ureafaciens sialidase prevented subsequent tetrazine labeling. This finding indicates that the cyclooctyne was incorporated by the expected route as a sialoglycoconjugate rather than by metabolism of BCNSia into a cyclooctyne donor intermediate that could be linked to cell-surface biomolecules.
Figure 5.
Western blot analysis of zebrafish embryo lysates shows that BCNSia is present in sialoglycoproteins. A) Structure of affinity probe 5 used in Western blot experiments. B) Western blot of lysates. Embryos were injected with BCNSia or vehicle, grown to 48 hpf, deyolked, and lysed. The resulting lysates were treated with sialidase followed by 5 and then analyzed by Western blot with an anti-dinitrophenyl horse-radish peroxidase conjugate. AU = Arthrobacter ureafaciens, DNP = dinitrophenyl, VC = Vibrio cholerae.
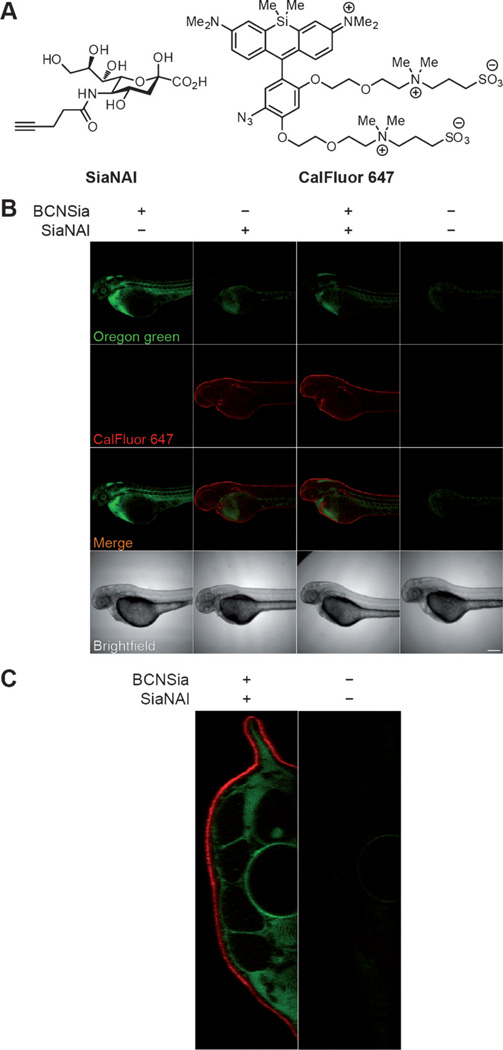
Finally, we showed that the tetrazine–BCNSia ligation can be carried out orthogonally to copper click chemistry in live zebrafish. We microinjected embryos with 5 pmol BCNSia and/or 50 pmol SiaNAl, the expected metabolic product of an N-pentynoyl mannosamine derivative that we have previously described.[28] At 48 hpf, the embryos were injected with 4 and then bathed in a copper click solution containing sodium ascorbate, BTTAA ligand,[29] and the azide probe CalFluor 647 for 30 min.[28b] We observed robust BCNSia-dependent labeling of interior cells and SiaNAl-dependent labeling of enveloping layer cells (Figure 6B). Analysis of embryo cross-sections confirmed that the signals from 4 and CalFluor 647 were confined to the interior and enveloping layers of the embryo, respectively (Figure 6C). Consistent with the very slow reaction rate of tetrazines with linear alkynes,[30] we observed in control experiments that embryos treated with SiaNAl were not labeled when bathed in media containing tetrazine 4 (Figure S8). Furthermore, although BCNSia should react with CalFluor 647, we observed no such labeling (Figure 6B), perhaps because BCNSia is not displayed on enveloping layer cells (Figure S8); however, in situations where cross-reactivity might present a problem, it would be straightforward to quench BCNSia on the enveloping layer with a sacrificial tetrazine reagent prior to and during the copper click reaction.
Figure 6.
Injection of BCNSia and SiaNAl at the 1–8 cell stage enables concurrent in vivo detection of spatially distinct glycoproteins using two different bioorthogonal ligation reactions. A) Structures of SiaNAl and CalFluor 647. B) Embryos were injected with BCNSia and/or SiaNAl or vehicle at the 1–8 cell stage, then injected with 4 and bathed in a copper click solution with CalFluor 647 at 48 hpf. Scale bar: 200 µm. C) A tail slice of an embryo treated with BCNSia and SiaNAl followed by 4 and CalFluor 647 as in (B) shows spatial separation of labeling.
In conclusion, we have demonstrated that the ability to metabolically incorporate a cyclooctyne-functionalized sialic acid, in conjunction with the use of a fluorogenic cyclooctyne-reactive probe, enables systemic in vivo fluorescence imaging of sialylation during zebrafish embryogenesis. Importantly, our method uses bioorthogonal chemistry to image glycosylation with sub-cellular resolution throughout the interior of a living organism, in contrast to previous in vivo imaging efforts that have been restricted to imaging enveloping layer cells or focused on the ability to detect the presence or absence of large masses of tumor cells.[17,31] This work opens up a number of promising avenues for further research. We have identified several new sialylated structures in the developing zebrafish, raising questions about tissue-specific sialyltransferase activity and the importance of sialylation in these structures for proper embryonic development. Additionally, one unique attribute of the cyclooctyne is its ability to engage in cycloaddition reactions with both tetrazines and azides. The latter functional group has been used by our laboratory to generate fluorogenic xanthene derivatives, including CalFluor 647, with emission maxima spanning the visible spectrum, potentially enabling multicolor labeling of temporally distinct classes of glycans.[32] Furthermore, methods to use fluorogenic ligation reactions in conjunction with metabolic engineering of glycans may find applications in cyclopropene–tetrazine ligations as cyclopropene can be metabolically incorporated into glycans by a variety of monosaccharide precursors.[33] Finally, we expect that our demonstration of the utility of bioorthogonal fluorogenic probes for in vivo imaging will motivate the use of such probes for the visualization of other classes of biomolecules.
Supplementary Material
Acknowledgements
We thank Prof. Sharon Amacher, Prof. Scott Laughlin, C. J. Cambier, Chelsea Gordon, Frances Rodriguez-Rivera, Lauren Wagner, and Nathan Yee for helpful discussions, and Prof. Christopher Chang for use of instrumentation. P.A. was supported by a National Science Foundation Graduate Research Fellowship. This work was supported by a grant from the National Institutes of Health (R37 GM059907). This work was performed in part at the UC Berkeley CRL Molecular Imaging Center, supported by the National Science Foundation (DBI-1041078) and the Gordon and Betty Moore Foundation.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201504249.
Contributor Information
Paresh Agarwal, Departments of Chemistry and Molecular and Cell Biology and Howard Hughes Medical Institute, University of California, Berkeley, B84 Hildebrand Hall, Berkeley, CA 94720 (USA).
Brendan J. Beahm, Departments of Chemistry and Molecular and Cell Biology and Howard Hughes Medical Institute, University of California, Berkeley, B84 Hildebrand Hall, Berkeley, CA 94720 (USA).
Peyton Shieh, Departments of Chemistry and Molecular and Cell Biology and Howard Hughes Medical Institute, University of California, Berkeley, B84 Hildebrand Hall, Berkeley, CA 94720 (USA).
Carolyn R. Bertozzi, Email: bertozzi@stanford.edu, Department of Chemistry and Howard Hughes Medical Institute, Stanford University, 333 Campus Drive, Stanford, CA 94305.
References
- 1.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.National Research Council (US) Committee on Assessing the Importance and Impact of Glycomics and Glycosciences. Transforming Glycoscience: A Roadmap for the Future. Washington, DC: National Academies Press; 2012. [PubMed] [Google Scholar]
- 3.Laughlin ST, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang PV, Bertozzi CR. Chem. Commun. 2012;48:8864–8879. doi: 10.1039/c2cc31845h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dehnert KW, Beahm BJ, Huynh TT, Baskin JM, Laughlin ST, Wang W, Wu P, Amacher SL, Bertozzi CR. ACS Chem. Biol. 2011;6:547–552. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. ChemBioChem. 2012;13:353–357. doi: 10.1002/cbic.201100649. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Möller H, Böhrsch V, Bentrop J, Bender J, Hinderlich S, Hackenberger CPR. Angew. Chem. Int. Ed. 2012;51:5986–5990. doi: 10.1002/anie.201108809. Angew. Chem.2012, 124, 6088–6092. [DOI] [PubMed] [Google Scholar]; g) Beahm BJ, Dehnert KW, Derr NL, Kuhn J, Eberhart JK, Spillmann D, Amacher SL, Bertozzi CR. Angew. Chem. Int. Ed. 2014;53:3347–3352. doi: 10.1002/anie.201310569. Angew. Chem.2014, 126, 3415–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieschke GJ, Currie PD. Nat. Rev. Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 7.Shieh P, Bertozzi CR. Org. Biomol. Chem. 2014;12:9307–9320. doi: 10.1039/c4ob01632g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Carlson JCT, Meimetis LG, Hilderbrand S, Weissleder R. Angew. Chem. Int. Ed. 2013;52:6917–6920. doi: 10.1002/anie.201301100. Angew. Chem.2013, 125, 7055–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J Am. Chem. Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang J, Šečkutė J, Cole CM, Devaraj NK. Angew. Chem. Int. Ed. 2012;51:7476–7479. doi: 10.1002/anie.201202122. Angew. Chem.2012, 124, 7594–7597. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wieczorek A, Buckup T, Wombacher R. Org. Biomol. Chem. 2014;12:4177–4185. doi: 10.1039/c4ob00245h. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Yang J, Šečkutė J, Devaraj NK. Angew. Chem. Int. Ed. 2014;53:5805–5809. doi: 10.1002/anie.201400135. Angew. Chem.2014, 126, 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karver MR, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2011;22:2263–2270. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. J Am. Chem. Soc. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 13.a) Sauer J, Heldmann DK, Hetzenegger J, Krauthan J, Sichert H, Schuster J. Eur. J. Org. Chem. 1998:2885–2896. [Google Scholar]; b) Chen W, Wang D, Dai C, Hamelberg D, Wang B. Chem. Commun. 2012;48:1736–1738. doi: 10.1039/c2cc16716f. [DOI] [PubMed] [Google Scholar]; c) Plass T, Milles S, Koehler C, Szymański J, Mueller R, Wießler M, Schultz C, Lemke EA. Angew. Chem. Int. Ed. 2012;51:4166–4170. doi: 10.1002/anie.201108231. Angew. Chem.2012, 124, 4242–4246. [DOI] [PubMed] [Google Scholar]
- 14.Blackman ML, Royzen M, Fox JM. J Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson DM, Nazarova LA, Prescher JA. ACS Chem. Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 16.Rossin R, van den Bosch SM, ten Hoeve W, Carvelli M, Versteegen RM, Lub J, Robillard MS. Bioconjugate Chem. 2013;24:1210–1217. doi: 10.1021/bc400153y. [DOI] [PubMed] [Google Scholar]
- 17.Neves AA, Stçckmann H, Wainman YA, Kuo JCH, Fawcett S, Leeper FJ, Brindle KM. Bioconjugate Chem. 2013;24:934–941. doi: 10.1021/bc300621n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Pouilly S, Bourgeaux V, Piller F, Piller V. ACS Chem. Biol. 2012;7:753–760. doi: 10.1021/cb200511t. [DOI] [PubMed] [Google Scholar]; b) Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 19.a) Oetke C, Brossmer R, Mantey LR, Hinderlich S, Isecke R, Reutter W, Keppler OT, Pawlita M. J Biol. Chem. 2002;277:6688–6695. doi: 10.1074/jbc.M109973200. [DOI] [PubMed] [Google Scholar]; b) Han S, Collins BE, Bengtson P, Paulson JC. Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 20.Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes FPJT, van Hest JCM, Lefeber DJ, Friedl P, van Delft FL. Angew. Chem. Int. Ed. 2010;49:9422–9425. doi: 10.1002/anie.201003761. Angew. Chem.2010, 122, 9612–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 22.a) Deutscher SL, Nuwayhid N, Stanley P, Briles EIB, Hirschberg CB. Cell. 1984;39:295–299. doi: 10.1016/0092-8674(84)90007-2. [DOI] [PubMed] [Google Scholar]; b) Eckhardt M, Gotza B, Gerardy-Schahn R. J Biol. Chem. 1998;273:20189–20195. doi: 10.1074/jbc.273.32.20189. [DOI] [PubMed] [Google Scholar]
- 23.a) Anderson C, Bartlett SJ, Gansner JM, Wilson D, He L, Gitlin JD, Kelsh RN, Dowden J. Mol. BioSyst. 2007;3:51–59. doi: 10.1039/b613673g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gansner JM, Gitlin JD. Dev. Dyn. 2008;237:3715–3726. doi: 10.1002/dvdy.21779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson ND, Weinstein BM. Nat. Rev. Genet. 2002;3:674–682. doi: 10.1038/nrg888. [DOI] [PubMed] [Google Scholar]
- 25.a) Flynn EJ, Trent CM, Rawls JF. J Lipid Res. 2009;50:1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McMenamin SK, Minchin JEN, Gordon TN, Rawls JF, Parichy DM. Endocrinology. 2013;154:1476–1487. doi: 10.1210/en.2012-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentrop J, Marx M, Schattschneider S, Rivera-Milla E, Bastmeyer M. Dev. Dyn. 2008;237:808–818. doi: 10.1002/dvdy.21451. [DOI] [PubMed] [Google Scholar]
- 27.Marx M, Rutishauser U, Bastmeyer M. Development. 2001;128:4949–4958. doi: 10.1242/dev.128.24.4949. [DOI] [PubMed] [Google Scholar]
- 28.a) Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew. Chem. Int. Ed. 2009;48:4030–4033. doi: 10.1002/anie.200806319. Angew. Chem.2009, 121, 4090–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shieh P, Dien VT, Beahm BJ, Castellano JM, Wyss-Coray T, Bertozzi CR. J Am. Chem. Soc. 2015;137:7145–7151. doi: 10.1021/jacs.5b02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem. Int. Ed. 2011;50:8051–8056. doi: 10.1002/anie.201101817. Angew. Chem.2011, 123, 8201–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a) Rieder U, Luedtke NW. Angew. Chem. Int. Ed. 2014;53:9168–9172. doi: 10.1002/anie.201403580. Angew. Chem.2014, 126, 9322–9326. [DOI] [PubMed] [Google Scholar]; b) Thalhammer F, Wallfahrer U, Sauer J. Tetrahedron Lett. 1990;31:6851–6854. [Google Scholar]
- 31.a) Rossin R, Renart Verkerk P, van den Bosch SM, Vulders RCM, Verel I, Lub J, Robillard MS. Angew. Chem. Int. Ed. 2010;49:3375–3378. doi: 10.1002/anie.200906294. Angew. Chem.2010, 122, 3447–3450. [DOI] [PubMed] [Google Scholar]; b) Devaraj NK, Thurber GM, Keliher EJ, Marinelli B, Weissleder R. Proc. Natl. Acad. Sci. USA. 2012;109:4762–4767. doi: 10.1073/pnas.1113466109. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Koo H, Lee S, Na JH, Kim SH, Hahn SK, Choi K, Kwon IC, Jeong SY, Kim K. Angew. Chem. Int. Ed. 2012;51:11836–11840. doi: 10.1002/anie.201206703. Angew. Chem.2012, 124, 12006–12010. [DOI] [PubMed] [Google Scholar]; d) Lee SB, Kim HL, Jeong H-J, Lim ST, Sohn M-H, Kim DW. Angew. Chem. Int. Ed. 2013;52:10549–10552. doi: 10.1002/anie.201304026. Angew. Chem.2013, 125, 10743–10746. [DOI] [PubMed] [Google Scholar]; e) Emmetiere F, Irwin C, Viola-Villegas NT, Longo V, Cheal SM, Zanzonico P, Pillarsetty N, Weber WA, Lewis JS, Reiner T. Bioconjugate Chem. 2013;24:1784–1789. doi: 10.1021/bc400322h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a) Shieh P, Hangauer MJ, Bertozzi CR. J Am. Chem. Soc. 2012;134:17428–17431. doi: 10.1021/ja308203h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2014;111:5456–5461. doi: 10.1073/pnas.1322727111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Patterson DM, Jones KA, Prescher JA. Mol. BioSyst. 2014;10:1693–1697. doi: 10.1039/c4mb00092g. [DOI] [PubMed] [Google Scholar]; b) Cole CM, Yang J, Šečkutė J, Devaraj NK. ChemBioChem. 2013;14:205–208. doi: 10.1002/cbic.201200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.