Abstract
RNA interference (RNAi) is a conserved sequence-specific gene regulatory mechanism1–3 mediated by the RNA-induced silencing complex (RISC), which is composed of a single-stranded guide RNA and an Argonaute protein. The PIWI domain, a highly conserved motif within Argonaute, has been shown to adopt an RNase H fold4,5 critical for the endonuclease cleavage activity of RISC4–6. Here we report the crystal structure of Archaeoglobus fulgidus Piwi protein bound to double-stranded RNA, thereby identifying the binding pocket for guide-strand 5′-end recognition and providing insight into guide-strand-mediated messenger RNA target recognition. The phosphorylated 5′ end of the guide RNA is anchored within a highly conserved basic pocket, supplemented by the carboxy-terminal carboxylate and a bound divalent cation. The first nucleotide from the 5′ end of the guide RNA is unpaired and stacks over a conserved tyrosine residue, whereas successive nucleotides form a four-base-pair RNA duplex. Mutation of the corresponding amino acids that contact the 5′ phosphate in human Ago2 resulted in attenuated mRNA cleavage activity. Our structure of the Piwi–RNA complex, and that determined elsewhere7, provide direct support for the 5′ region of the guide RNA serving as a nucleation site for pairing with target mRNA and for a fixed distance separating the RISC-mediated mRNA cleavage site from the anchored 5′ end of the guide RNA.
Small interfering RNAs (siRNAs), the cleavage products of RNase III enzyme Dicer, consist of RNA duplexes that contain two-nucleotide 3′ overhangs and 5′-phosphorylated ends. These unique features at the termini of 19–23-base-pair duplexes, which distinguish siRNAs from other endogenous RNAs, are also required for their functional role in RNAi-mediated processes. Extensive structural and biochemical studies on PAZ (for PIWI/Argonaute/Zwille)–RNA recognition8–10, culminating in the structures of PAZ bound to single-stranded RNA11 and siRNA-like duplexes12, have established that the 3′ overhang is specifically recognized by the PAZ domain, a module conserved in both Dicer and Argonaute proteins. Biochemical studies in Drosophila melanogaster lysates have shown that the 5′ phosphate on the guide RNA is essential for its assembly into RISC13,14, whereas related studies have established that the 5′ phosphate of the partner strand is sensed in the process of RISC assembly by means of R2D2 (ref. 15). In addition, the cleavage-site position in the mRNA, paired to the guide RNA in the RISC complex, is defined by the 5′ end of the guide RNA strand16. Recent crystal structures have established that the PIWI domain, a highly conserved module in Argonaute proteins, adopts an RNase H fold4–6. The Piwi protein also contains within it a conserved basic pocket that probably constitutes the binding site for the 5′ phosphate of the guide RNA5.
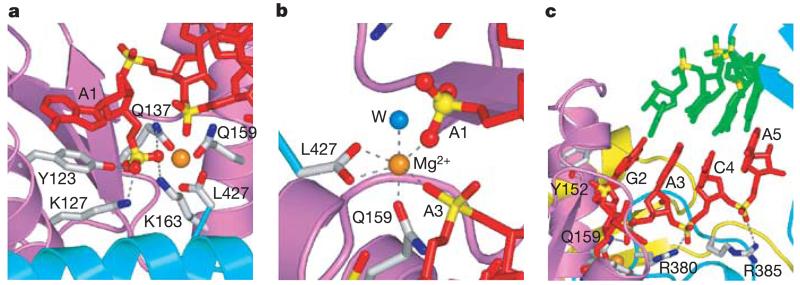
Here we describe the 2.5-Å crystal structure of the A. fulgidus Piwi protein (Fig. 1a) bound to a 5′-phosphate-containing 21-mer RNA, capable of self-complementary duplex formation (Fig. 1b). The crystallographic asymmetric unit contains two Piwi proteins, each bound to a short four-base-pair segment of duplex, positioned adjacent to the 5′ phosphate and its attached unpaired nucleotide (Fig. 1c). The structure of the A. fulgidus Piwi protein, which consists of a short N-terminal element and two domains labelled A and B (Fig. 1a), is similar (root-mean-square deviation = 0.65Å for Cα) in the free state5 and in the RNA-bound complex. The RNA, whose sequence from the 5′ end is p-A1-G2-A3-C4-A5 (Fig. 1b), forms a short A-form duplex, involving pairing of the G2-A3-C4-A5 segment with its complement towards the 3′ end of the sequence (Fig. 1c, d). The RNA duplex is positioned in a basic channel spanning the A–B inter-domain interface of the protein (Fig. 1e), whereas the 5′ phosphate is inserted into a conserved basic pocket located primarily within domain A and the C terminus of domain B (Figs 1e and 2a). A network of hydrogen bonds involving the side chains of Y123, K127, Q137 and K163 and a bound divalent cation anchor the 5′ phosphate into this pocket (Fig. 2a). The bound divalent cation, most probably Mg2+, is octahedrally coordinated to three protein-based oxygens, two nucleic-acid phosphate oxygens and a water molecule in the complex (Fig. 2b). This divalent cation, which is coordinated to the C-terminal carboxylate in an otherwise basic pocket, was also observed in the structure of the free A. fulgidus Piwi protein (identified as Cd2+)5, except that two coordinated chlorides are replaced by two phosphate oxygens on complex formation. These results are consistent with the formation of a preformed 5′-phosphate-binding pocket in Piwi, and by extension in Argonaute proteins. They also support an earlier prediction of the potential location of the 5′-phosphate-binding pocket within the Piwi scaffold5.
Figure 1.
Crystal structure of the A. fulgidus Piwi–RNA complex. a, The A. fulgidus Piwi protein consists of a yellow N-domain (1–37), a magenta domain A (38–167) and a cyan domain B (168–427). b, The sequence and pairing alignment of the 21-mer RNA. The red and green segments are observed in the crystal structure of the complex, whereas those in black are disordered. c, The relative alignments of the two Piwi proteins together with their bound RNAs within the asymmetric unit. d, e, Ribbon (d) and electrostatic (e) views of the complex. The RNA is shown in a stick representation.
Figure 2.
The 5′-phosphate-binding site in the A. fulgidus Piwi–RNA complex. a, Positioning of the 5′ phosphate in a basic pocket lined by residues of domain A (K127, Y123, Q137, Q159 and K163), domain B (C-terminal L427) and a bound divalent cation in orange. Bases A1 and G2 are splayed apart, with unpaired A1 stacked on Y123. b, The octahedral coordination of the bound divalent cation, presumably Mg2+, involving protein and RNA oxygens and a water molecule. c, Positioning of the short four-base-pair duplex segment involving the G2-A3-C4-A5 segment (red) with its complementary partner (green). Note intermolecular hydrogen bonds to the non-bridging phosphate oxygens of the A1-G2-A3-C4-A5 segment.
We propose that it is the guide RNA strand whose 5′ phosphate is anchored in the basic pocket of the Piwi protein. The A1 base at the 5′ end of the guide RNA strand is unpaired and stacks on the aromatic ring of invariant Y123 (Fig. 2a), thereby accounting for non-sequence-specific recognition of the first nucleotide within the 5′-end-binding pocket. The non-bridging oxygens of the backbone phosphates at the A1–G2, G2–A3, A3–C4 and C4–A5 steps are hydrogen-bonded to the side chains of Y152, Q159, R380 and R385, respectively (Fig. 2c). The two paired RNA strands bind in a strongly asymmetric manner, such that the 5′ end of the guide RNA strand (coloured red in Fig. 2c) interacts extensively with the protein, burying 1,500Å2 of protein surface, whereas the mRNA strand (coloured green in Fig. 2c) positioned opposite it has minimal contacts with the protein, burying 350Å2 of protein surface.
We have generated a set of single and double mutants of A. fulgidus Piwi protein residues that contact the 5′ phosphate and attached A1 residue of the bound RNA, in an effort to investigate the importance of these highly conserved residues to RNA recognition. Filter binding studies (averages of three runs) establish that the apparent binding affinity for RNA (Kd,app) decreases from threefold to sevenfold on proceeding from the wild-type protein (0.19 μM) to mutants Q137A (0.66 μM), K163A (0.73 μM) and K127A (1.46 μM), all of which should disrupt hydrogen bonding to the 5′ phosphate, and mutant Y123A (0.99 μM), which should disrupt stacking with the A1 residue (Fig. 3b). The same trend in binding affinity loss is observed for double mutants Q137A/K163A (0.75 μM), K127A/Q137A (1.15 μM) and K127A/K163A (1.56 μM) (Fig. 3c). These results indicate that 5′ phosphate stabilization might involve multiple hydrogen-bonding interactions and that the disruption of any single interaction or pair of such interactions results in only a modest decrease in binding affinity.
Figure 3.
Mutation studies of conserved 5′-phosphate-binding residues in A. fulgidus Piwi protein and human Ago2 protein. a, Sequence alignment of conserved 5′-phosphate-binding residues across various species of Piwi and Argonaute proteins. Prefix Hs, human; Dm, Drosophila; Aa, Aquifex aeolicus, Af, Archaeoglobus fulgidus. b, Representative binding curves (average of three runs) for A. fulgidus Piwi single-site mutants with 21-nucleotide RNA labelled with 5′-32P by double-filter binding assay. The deduced Kd,app values are listed below. c, The corresponding binding curves for A. fulgidus PIWI dual-site mutants. d, The cleavage activities of wild-type human Ago2 and its mutants. The black bar at the right of the image represents the region of the target RNA complementary to the siRNA used. The expression levels of the proteins used in the assays were assessed by western blotting with anti-HA antibody and are shown in the lower panel.
Because the A. fulgidus Piwi protein shares significant sequence homology with human Argonaute proteins (Fig. 3a), we mapped the 5′-end-binding pocket in human Ago2, the RISC component that mediates small RNA-guided cleavage of target mRNA6,17, and set out to investigate whether mutation of key residues lining the 5′-end-binding pocket would affect human Ago2 mRNA cleavage activity. Guided by the crystal structure of the A. fulgidus Piwi protein–RNA complex, we introduced single as well as double alanine mutations at the corresponding residues located in the putative 5′-end-binding pocket of human Ago2, namely K533, Q545 and K570 (Fig. 3a). The wild-type human Ago2 guided the cleavage of the radiolabelled target RNA efficiently (Fig. 3d, lane 4). The single alanine mutants, K533A, Q545A and K570A, showed reduced cleavage activity (Fig. 3d, lanes 5 to 7), whereas the double alanine mutants manifested more severe defects in cleavage capacity (Fig. 3d, lanes 1 to 3). These results indicate that the 5′-end-binding pocket observed in A. fulgidus Piwi protein is conserved in human Ago2 and is important for its small RNA guided cleavage activity.
We have also directly investigated the effect of 5′-phosphorylation of RNAs on binding affinity. These results, based on a double filter-binding competition assay, establish that the 5′-phosphate-containing RNA (Supplementary Fig. S2a, panels labelled B) binds the A. fulgidus Piwi protein about an order of magnitude more tightly than its non-phosphorylated counterpart (Fig. S2a, panels labelled A). It therefore seems that 5′-phosphorylation indeed provides additional affinity for guide RNA recognition of A. fulgidus Piwi protein.
There are no specific intermolecular contacts directed to the 2′-OH groups of the RNA in the crystal structure of the A. fulgidus Piwi protein–RNA complex, indicating that the Piwi protein might also be targeted by other types of nucleic acid. We have investigated the apparent binding affinities of A. fulgidus Piwi protein to both single-stranded and double-stranded 21-mer DNA and RNA oligonucleotides with the use of double filter-binding assays (Supplementary Fig. S2b). The binding is 20-fold tighter for single-stranded DNA than for ssRNA, with the binding affinity decreasing on proceeding from DNA–DNA to RNA–DNA to RNA–RNA duplexes. The preference for ssDNA and dsDNA of the archaeal Piwi protein was unexpected and will need further investigation. Nevertheless, our results are consistent with previous observations that the 2′-OH is not required for RNAi-mediated events18. It is also conceivable that other domains within Argonaute proteins and/or other RISC components could provide additional interactions that discriminate between nucleic acid types.
Our structure revealed that the stacked bases 2 to 5 at the 5′ end of the guide RNA are accessible for pairing with their complementary counterparts on the mRNA. Bioinformatics and biochemical analysis of microRNA (miRNA) genes have shown a high degree of sequence conservation restricted to positions 2–8, indicative of a significant contribution of the 5′ end to specificity and activity19–21. Our structure supports this concept, in which the mRNA would initially nucleate with the 5′ end of the protein-bound guide RNA strand19,22, followed by zippering up of the remaining segment to form the guide RNA–mRNA duplex. The observation that the first base on the guide RNA strand is not available for pairing with mRNA in the crystal structure of the complex is consistent with earlier biochemical and kinetic data showing that disruption of this pair has no effect on cleavage activity and under some conditions even favours target cleavage22.
More than half of the bound 21-mer is disordered in the crystal structure of the A. fulgidus Piwi protein–RNA complex (Fig. 1c). We have therefore constructed a model in which the trajectory of the four-base-pair duplex observed in the crystal structure of the complex was extended to form two turns of A-form helix (Fig. 4a). This duplex ends up being positioned within a basic channel5 that spans one face of both domains A and B of a monomeric Piwi protein (Fig. 4b). The proposed mRNA cleavage site is positioned opposite the putative catalytic residues associated with the RNase H fold of domain B of the Piwi protein4–6 (Fig. 4a), although the archaeal A. fulgidus Piwi protein lacks the DD components of the conserved DDE motif, normally associated with RNase H cleavage activity23 (Supplementary Fig. S1).
Figure 4.
Model of A. fulgidus Piwi protein (Af Piwi) bound to an 18-base-pair RNA duplex with 5′-phosphorylated single-nucleotide overhangs. a, The 5′-end nucleotide and four-base-pair duplex (coloured in red and green for guide strand and target strand, respectively) observed in the crystal structure of the complex were extended by 14-base-pair A-form duplex (coloured in tan and white for guide strand and target strand, respectively). The 5′ end anchoring pocket and putative catalytic site are circled, and the phosphate cleavage site is marked by a red ball. b, Electrostatic surface view of the model of the Af Piwi–dsRNA complex.
Previous studies have established that the RISC complex cleaves the mRNA strand in a site-specific manner between positions 10 and 11 as defined from the 5′ end of the guide strand16. This implies that the catalytic cleavage site is measured from the bound 5′ end of the guide strand by a fixed distance, a feature consistent with the anchoring of both the 5′ phosphate and its adjacent nucleotide of the guide strand, as observed in our structure of the A. fulgidus Piwi protein–RNA complex (Figs 2a and 4a).
Argonautes, which contain conserved PAZ and PIWI domains, seem to be the sole proteins required for RISC-mediated activities24. Active RISCs, which contain only a single guide RNA strand25, are capable of directing multiple rounds of site-specific target mRNA cleavage26. Both our structure of the A. fulgidus Piwi protein–RNA complex and that determined by others7 provides the first structural insights into 5′ end recognition of the guide RNA strand, the nucleation step associated with guide-RNA–mRNA recognition and the distance-based identification of the mRNA cleavage site. Further attempts at structural characterization of full-length Argonaute–RNA complexes, if successful, could provide structure-based information related to the catalytic cleavage mechanism, a central event of RNA interference.
Methods
Protein and RNA preparations
The full-length A. fulgidus Piwi cDNA was amplified by polymerase chain reaction (PCR) from Archeoglobus fulgidus genomic DNA (ATCC) and cloned into pET28a (Novagen) with an amino-terminal His6 tag. The protein was overexpressed in the Escherichia coli host cell BL21-Gold(DE3) (Stratagene) induced by 1 mM isopropyl β-D-thiogalactoside at 20 °C, and purified by Ni-chelating and heparin Hitrap columns. After gel-filtration chromatography on Superdex 200 (Amersham Biosciences), the pure A. fulgidus Piwi with attached His tag was concentrated and stored at 30 mg ml−1 in 10 mM Tris-HCl pH 8.0, 1 M NaCl and 1 mM dithiothreitol. Selenomethionine-labelled protein was produced by inhibiting the methionine biosynthesis pathway in the host cell. Site-directed mutations were made by a two-step PCR-based overlap extension method and confirmed by DNA sequencing. All the mutants were purified as described for the wild-type protein except that the Heparin column step was omitted. RNAs were synthesized chemically in our laboratory or purchased from Dharmacon, then deprotected and purified by denaturing polyacryamide-gel electrophoresis.
Crystallization and data collection
Crystals of native and selenomethionine-substituted A. fulgidus Piwi protein–21-mer RNA complex were grown by hanging-drop vapour diffusion at 20 °C. About 15 mg ml−1 protein–RNA complex in 0.1 M KCl, 10 mM HEPES-K pH 7.5 buffer was mixed 1:1 with a well solution containing 6% polyethylene glycol 4000, 40 mM KCl, 10 mM MgCl2 in 50 mM sodium cacodylate pH 6.5. These crystals grew to a maximum size of 0.2 mm × 0.2 mm × 0.2 mm over the course of 10 days.
For data collection, crystals were flash frozen (100 K) in the above reservoir solution supplemented with 30% glycerol with a 10% step increase. A total of 720 frames of 0.5° oscillation were collected on the 2.5Å crystals of selenomethionine-labelled A. fulgidus Piwi protein–21-mer RNA complex at 0.9776Å at National Synchrotron Light Source (NSLS) beamline X26C, Brookhaven National Laboratory. The data were processed with HKL2000 (http://www.hkl-xray.com/). The crystal belonged to space group R3, with unit cell parameters listed in Supplementary Table S1. There were two complex molecules per asymmetric unit, with about 55% solvent content.
Structure determination
The structure of the A. fulgidus Piwi protein–21-mer RNA complex was solved by using the free A. fulgidus Piwi protein structure (PDBID: 1W9H) as the search model and Molrep (CCP4 package)27 was used to locate the two copies of the A. fulgidus Piwi protein. The extra density corresponding to the 5′ terminus of the RNA and several additional base pairs could be clearly traced without ambiguities from the earliest stage. The A. fulgidus Piwi protein–RNA complex model was rebuilt with the program O28 and refined with CNS (http://cns.csb.yale.edu/v1.1/) and REFMAC (CCP4 package)27. The R-free set contained 5% of the reflections chosen at random. Refinement statistics are given in Supplementary Table S1. The protein comprises residues 2–427 and the RNA comprises the 5′ phosphate, the A1 residue and four additional base pairs involving residues at the 5′ end. Disordered regions within the A. fulgidus Piwi protein included loop segments 304–308 and 331–341 for molecule A and 332–338 for molecule B in the asymmetric unit. The middle part of the duplex RNA (nine base pairs) and three nucleotides at the 3′ end cannot be traced in our final density map. The figures were prepared with PyMOL (http://pymol.sourceforge.net/).
Double filter-binding assays
The affinity of the A. fulgidus Piwi and its mutants for various oligonucleotides was detected and quantified with a double filter-binding assay29. The reactions containing 5′-32P-labelled nucleic acids and proteins were applied to filters containing two membranes: a protein-binding Protran BA-85 nitrocellulose membrane and a nucleic-acid-binding Hybond-N+ nylon membrane. After air-drying, the filters were quantified on a PhosphorImager. The data were analysed as reported previously30; details of experimental protocols are listed in Supplementary Methods.
Cleavage assays using human Ago2 and its mutants
The Flag/haemagglutinin (HA)-tagged wild-type and mutant constructs were transiently transfected into HEK-293 cells, and Ago2 proteins were immunoprecipitated from the cell lysates with anti-Flag antibody. The expression levels of the wild-type and mutant Ago2 proteins were assessed by western blotting with anti-HA antibody (Fig. 3c, lower panel). The stringently washed beads carrying equal amounts of bound Flag/HA-tagged human Ago2 proteins were preincubated with a single-stranded siRNA to reconstitute RISC activity. After an additional washing step to remove the unbound siRNA, the beads were incubated with a 32P-cap-labelled target RNA. After incubation for 1 h the RNA was recovered from the beads and analysed by denaturing gel electrophoresis. Details of the experimental protocols are listed in Supplementary Methods
Supplementary Material
Acknowledgements
We thank A. Saxena and personnel at synchrotron beamline X26C of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, for their assistance. Use of the NSLS beamline is supported by the US Department of Energy, Basic Energy Sciences, Office of Science. D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust and the Dewitt Wallace and Maloris Foundations, and T.T. is supported by a National Institutes of Health grant.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
Coordinates have been deposited in the Protein Data Bank under accession code 1YTU.
References
- 1.Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanism of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 5.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI-domain-siRNA guide complex. Nature. doi: 10.1038/nature03462. doi:10.1038/nature03462 (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 9.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 10.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 11.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 12.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nykänen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 14.Tomari Y, et al. RISC assembly defects in the Drosophila RNAi mutant Armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 15.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNA and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 24.Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl Acad. Sci. USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 26.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 27.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. Collaborative Computational Project no. 4. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard GJ. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagar MB, Lucast L, Doudna JA. Conserved but nonessential interaction of SRP RNA with translation factor EF-G. RNA. 2004;10:772–778. doi: 10.1261/rna.5266504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.