Summary
CdiB/CdiA proteins mediate inter-bacterial competition in a process termed contact-dependent growth inhibition (CDI). Filamentous CdiA exoproteins extend from CDI+ cells and bind specific receptors to deliver toxins into susceptible target bacteria. CDI has also been implicated in auto-aggregation and biofilm formation in several species, but the contribution of CdiA-receptor interactions to these multi-cellular behaviors has not been examined. Using Escherichia coli isolate EC93 as a model, we show that cdiA and bamA receptor mutants are defective in biofilm formation, suggesting a prominent role for CdiA-BamA mediated cell-cell adhesion. However, CdiA also promotes auto-aggregation in a BamA-independent manner, indicating that the exoprotein possesses an additional adhesin activity. Cells must express CdiA in order to participate in BamA-independent aggregates, suggesting that adhesion could be mediated by homotypic CdiA-CdiA interactions. The BamA-dependent and BamA-independent interaction domains map to distinct regions within the CdiA filament. Thus, CdiA orchestrates a collective behavior that is independent of its growth-inhibition activity. This adhesion should enable “greenbeard” discrimination, in which genetically unrelated individuals cooperate with one another based on a single shared trait. This kind-selective social behavior could provide immediate fitness benefits to bacteria that acquire the systems through horizontal gene transfer.
Keywords: contact-dependent growth inhibition, kind-selection, self/non-self recognition, toxin/immunity proteins, type V secretion systems
Introduction
Although they are often regarded as isolated and autonomous unicellular organisms, bacteria possess a number of molecular systems that mediate both cooperative and antagonistic relationships with other microorganisms. Contact-dependent growth inhibition (CDI) is one important mechanism of inter-cellular competition that is commonly found in pathogenic Gram-negative bacteria (Aoki et al., 2010, Hayes et al., 2014, Ruhe et al., 2013a). CDI is a function of CdiB/CdiA two-partner secretion (TPS) proteins, which inhibit the growth of neighboring bacteria upon direct cell-to-cell contact (Hayes et al., 2014, Ruhe et al., 2013a). CdiB is an Omp85 outer-membrane β-barrel protein that exports and assembles CdiA effectors onto the cell surface (Baud et al., 2014, Jacob-Dubuisson et al., 2013). CdiA proteins are very large, ranging from 180 – 630 kDa, and are predicted to form parallel β-helical filaments that extend several hundred angstroms to bind specific receptors on susceptible target bacteria (Kajava et al., 2001, Hayes et al., 2010, Hayes et al., 2014). Upon binding its receptor, CdiA delivers its C-terminal toxin domain (CdiA-CT) into the target cell to inhibit growth. Because CDI+ strains express receptors for their own CdiA effectors, they also receive toxin domains from adjacent sibling cells (Aoki et al., 2005, Aoki et al., 2008, Aoki et al., 2010, Nikolakakis et al., 2012, Anderson et al., 2012, Arenas et al., 2013, Beck et al., 2014). Though CDI+ cells exchange toxins with one another, they are protected from growth inhibition by small CdiI immunity proteins encoded immediately downstream of cdiA. CdiI binds directly to the CdiA-CT domain and neutralizes toxin activity (Aoki et al., 2010, Beck et al., 2014, Morse et al., 2012, Nikolakakis et al., 2012). Thus, CdiA is deployed to suppress the growth of competing bacteria, but isogenic sibling cells are immune to the delivered toxins.
During the initial characterization of CDI in Escherichia coli isolate EC93, Aoki et al. found that expression of the cdiBAIEC93 gene cluster causes laboratory strains of E. coli to auto-aggregate (Aoki et al., 2005). Cell-cell adhesion activity has also been reported for CdiA proteins from several other bacterial species. Before CDI was recognized as a competition system, Collmer and colleagues found that HecA promotes aggregation in Erwinia chrysanthemii EC16 cells and that this function is required for full virulence on a plant host (Rojas et al., 2002). HecA is a CdiA homolog and its C-terminal domain has a colicin E3-like nuclease activity that is specifically blocked by the VirA immunity protein (Walkeret al., 2004, Beck et al., 2014). These observations indicate that E. chrysanthemii EC16 probably uses HecA in inter-cellular competition, but the system also coordinates multi-cellular behavior during host invasion. This may be a general feature of CDI in plant pathogens, because CdiA proteins from Xylella fastidiosa (HxfB) and Xanthomonas axonopodis (XacFhaB) have also been implicated in virulence and biofilm formation (Guilhabert & Kirkpatrick, 2005, Gottig et al., 2009). HrpA from Neisseria meningitidis is yet another CdiA protein that is required for biofilm formation (Neil & Apicella, 2009), and hrpA mutants show decreased intracellular survival and escape from infected mammalian cells (Tala et al., 2008). Together, these studies demonstrate that CDI has a broader role in bacterial biology, promoting cooperative interactions between isogenic CDI+ cells during colonization of eukaryotic hosts.
Because CDI entails specific binding interactions between bacterial cells, the adhesin activity of CdiA represents the most direct mechanism to control biofilm architecture. However, previous studiesraise the possibility that CDI influences biofilm formation in other indirect ways. Gottig et al. found that X. axonopodis mutants lacking XacFhaB are hyper-motile due to the over-production of xanthan exopolysaccharides that are required for swarming motility (Gottig et al., 2009). In this instance, excessive extracellular polysaccharide has the paradoxical effect of suppressing biofilm formation by facilitating rapid migration of cells from one another. Intriguingly, Cotter and colleagues have recently reported that CdiA-CTE264 toxin activity is required for biofilm formation in Burkholderia thailandensis E264 (Anderson et al., 2012, Garcia et al., 2013). Wild-type B. thailandensis cells are immune to the DNase activity of CdiA-CTE264 (Anderson et al., 2012, Nikolakakis et al., 2012), so this domain's role in biofilm formation must be distinct from its growth-inhibition function. Because extracellular DNA is an important component of the B. thailandensis biofilm matrix, it has been suggested that the CdiA-CTE264 toxin could modulate the physical properties of matrix DNA (Garcia et al., 2013). Similarly, the tRNase activity of CdiA-CTUPEC536 appears to influence biofilm formation in uropathogenic E. coli (Diner et al., 2012). Taken together, these observations have led to the hypothesis that CdiA-CT domains possess additional signaling functions when delivered into immune sibling cells (Diner et al., 2012). In this model, CDI functions as a quorum-sensing mechanism to monitor the proximity/density of sibling cells and use that information to modulate gene expression through unknown signal transduction pathways.
Here, we use E. coli EC93 as a model to study the role of CDI in biofilm formation. The CDIEC93 system is well characterized and CdiAEC93-receptor interactions are understood at the molecular level. CdiAEC93 exploits the outer-membrane protein BamA as its receptor (Aoki et al., 2008). BamA is an essential Omp85 protein required for the assembly of all β-barrel proteins in Gram-negative bacteria (Voulhoux et al., 2003, Wu et al., 2005, Doerrler & Raetz, 2005, Werner & Misra, 2005). Although BamA is conserved between enterobacteria, the extracellular loop regions recognized by CdiAEC93 are variable between species (Smith et al., 2007, Ruhe et al., 2013b). Therefore, CdiAEC93 does not bind to BamA proteins from other bacteria, and E. coli cells that express heterologous bamA are resistant to inhibition by the CDIEC93 system (Ruhe et al., 2013b). In accord with previous studies, we find that E. coli EC93 ΔcdiA mutants are defective in biofilm formation. However, these defects are not due to changes in biofilm-associated gene expression, nor are they dependent on the CdiA-CT toxin region of CdiAEC93. Instead, CdiAEC93 promotes biofilm formation directly by mediating inter-cellular adhesion between neighboring cells. Remarkably, CDI+ cells that express heterologous bamA genes still auto-aggregate, indicating that the CdiAEC93-BamAEco binding interaction is not strictly required for CDI-dependent inter-cellular adhesion. We provide evidence that CdiAEC93 possesses an additional adhesion domain that is distinct from the BamAEco-binding region. Together, these data demonstrate that CdiA expressing bacteria can physically interact with one another in a receptor-independent manner. The proposed CdiA-CdiA binding interactions should facilitate cooperative behaviors between CDI+ bacteria and could promote the dispersal of cdi loci through horizontal gene transfer.
Results
EC93 ΔcdiA mutants are defective for biofilm formation
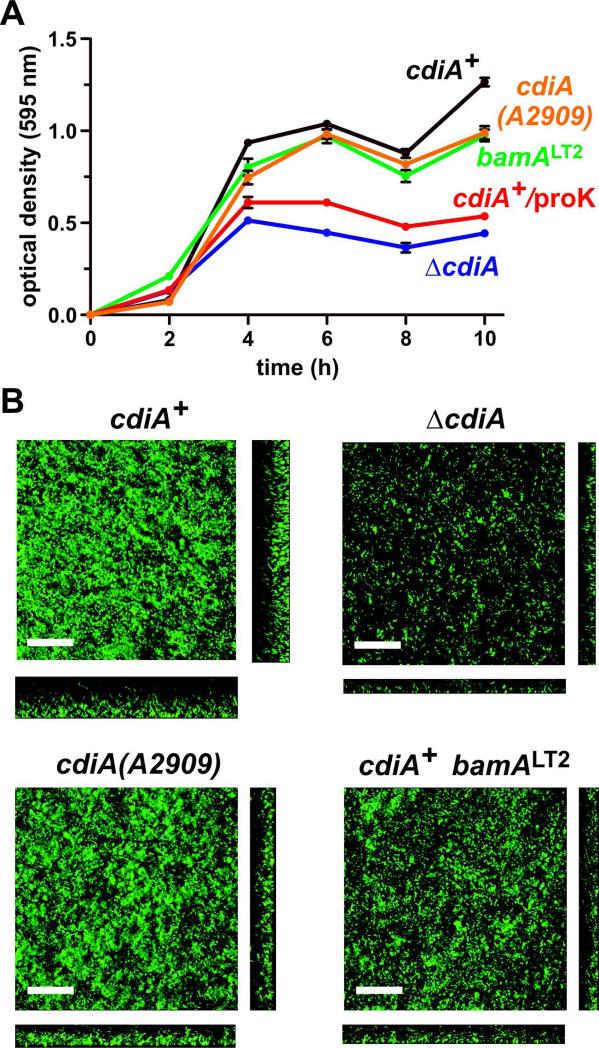
To determine whether CDI plays a role in E. coli EC93 biofilm formation, we cultured cdiA+ and ΔcdiA strains in 96-well polystyrene plates over a 10 h time period and quantified biofilm mass. Wild-type cdiA+ cells adhered to the plates within four h, then displayed a reproducible decrease in biofilm mass at 8 h, after which biomass again increased (Fig. 1A). The ΔcdiA mutant initially adhered to the plate with similar kinetics as cdiA+ cells, but generated significantly less biofilm mass over the entire time course (Fig. 1A). To examine biofilm architecture, we used confocal laser-scanning microscopy (CLSM) to visualize EC93 communities grown in static chambers. We observed a similar biofilm defect for ΔcdiA cells under these conditions, and quantification of biomass (P = 0.022) and average biofilm thickness (P < 0.0001) revealed significant differences between cdiA+ and ΔcdiA strains (Fig. 1B & Table 1). The C-terminal toxin domain of CdiA plays an important role in B. thailandensis biofilms (Garcia et al., 2013), so we tested whether deletion of the CdiA-CTEC93 region has a similar effect in E. coli EC93. We truncated CdiAEC93 after residue Ala2909, which demarcates the N-terminal boundary of the toxin domain (Aoki et al., 2010). The resulting cdiA(A2909) strain produced nearly as much biofilm mass as wild-type cells in polystyrene wells (Fig. 1A), though there were subtle decreases in biofilm mass (P = 0.09) and average thickness (P = 0.53) revealed by CLSM analysis of communities grown is static chambers (Fig. 1B & Table 1). These data are consistent with reports that cdi genes are important for biofilm formation in several other Gram-negative bacteria. However, E. coli EC93 differs from B. thailandensis E264 in that the CdiA-CTEC93 domain is not required to form a biofilm community.
Figure 1. E. coli EC93 ΔcdiA mutants are defective for biofilm formation.
A) Crystal violet staining of biofilms grown in polystyrene wells. The indicated strains of E. coli EC93 were grown in 96-well plates as described in the Methods. Planktonic cells were removed and the adherent biofilms stained with crystal violet. Staining was quantified by measuring the optical density at 595 nm (OD595). Where indicated, proteinase K was added to wild-type E. coli EC93 cultures after 2 h to determine the role of extracellular protein in biofilm formation. Each strain was tested in triplicate in two independent experiments. Presented values are the average ± SEM. B) Three-dimensional biofilm structures of E. coli EC93 strains after 24 h incubation in static biofilm chambers. Horizontal (xy) and vertical (xz) projections of biofilms are shown. Representative images are shown from one of two independent experiments. Inset scale bars equal 40 μm.
Table 1.
COMSTAT analysis of E. coli EC93 biofilmsa
| Strain genotype | Biomass (μm3 μm−2) | Average thickness (μm) | Maximum thickness (μm) | Roughness coefficient |
|---|---|---|---|---|
| cdiA+ | 3.71 (0.42) | 9.23 (0.08) | 30.54 (2.02) | 0.57 (0.03) |
| Δ cdiA | 0.92 (0.01) | 0.09 (0.00) | 18.92 (0.44) | 1.98 (0.00) |
| cdiA(A2909) | 2.38 (0.09) | 7.58 (2.22) | 27.72 (1.32) | 0.96 (0.18) |
| cdiA+ bamA LT2 | 1.48 (0.11) | 3.69 (2.38) | 23.76 (0.00) | 1.37 (0.35) |
Values represent the average ± SEM from two independent experiments
EC93 ΔcdiA mutants have no defects in growth rate, motility or biofilm gene expression
A number of factors could contribute to reduced biofilm formation in ΔcdiA mutants. We first tested whether the phenotype results from a decreased growth rate and found that cdiA+ and ΔcdiA cells grow at identical rates (Fig. S1A). Cell motility and the production of extracellular polysaccharide matrices also play key roles in biofilm formation; and previous work shows that X. axonopodis cdiA mutants are hyper-motile and have perturbed extracellular polysaccharide metabolism (Gottig et al., 2009). We examined the motility of ΔcdiA mutants on swim plates and found no significant changes from that of cdiA+ cells (P = 0.3378, Figs. S1B & S1C). Moreover, quantitative real-time PCR (qRT-PCR) analysis revealed that the levels of fliC transcripts (which encode flagellin monomers) were nearly identical in sessile populations of cdiA+ and ΔcdiA cells (Fig. S1D and data not shown). Thus, cdiA+ and ΔcdiA cells display similar flagellum synthesis and motility. We then examined the expression of genes required for the production poly-N-acetyl-D-glucosamine (PGA), cellulose and colanic acid, which are common polysaccharides used in the extracellular matrix of E. coli biofilms (Itoh et al., 2005, Wang et al., 2004, Serra et al., 2013, Danese et al., 2000, Prigent-Combaret et al., 1999). Because fliC message levels were equivalent in the cdiA+ and ΔcdiA backgrounds, we used this transcript as a reference standard for ΔΔCT analyses as described (Schmittgen & Livak, 2008). We isolated total RNA from biofilms grown on polystyrene wells for four h and quantified the relative levels of pgaA, bcsA and wcaA transcripts to monitor PGA, cellulose and colanic acid biosynthetic pathways, respectively. The pga message was essentially unaltered in ΔcdiA mutants, whereas bcsA and wcaA transcripts were elevated in ΔcdiA cells compared to wild-type (Fig. S1D). Together, these findings indicate that the ΔcdiA biofilm defect cannot be attributed to differences in growth rate, cell motility or decreased expression of extracellular matrix genes.
CdiA-BamA binding contributes to E. coli EC93 biofilm formation
CdiA (Aoki et al., 2005) and other two-partner exoproteins (Locht et al., 1993) have well-characterized adhesin properties, suggesting that CdiAEC93 might influence biofilm formation through its binding interactions with BamA. To test this model, we incubated wild-type cdiA+ cells in polystyrene wells for 2 h to allow initial surface attachment, then supplemented the culture with proteinase K (20 μg mL−1) and continued the incubation for 8 h. Our previous studies have shown that this treatment is sufficient to remove CdiAEC93 from the cell surface (Ruhe et al., 2014). Moreover, this concentration of proteinase K had no effect on E. coli EC93 cdiA+ or ΔcdiA cell growth (Fig. S2). As anticipated, the biofilm produced by proteinase-treated cells was similar to that of ΔcdiA mutants and had significantly less biomass than untreated cdiA+ cells (Fig. 1A). This result suggests that extracellular protein is important for E. coli EC93 biofilm formation, and supports a model in which CdiAEC93 functions as an adhesin and/or a proteinaceous matrix to hold cells together.
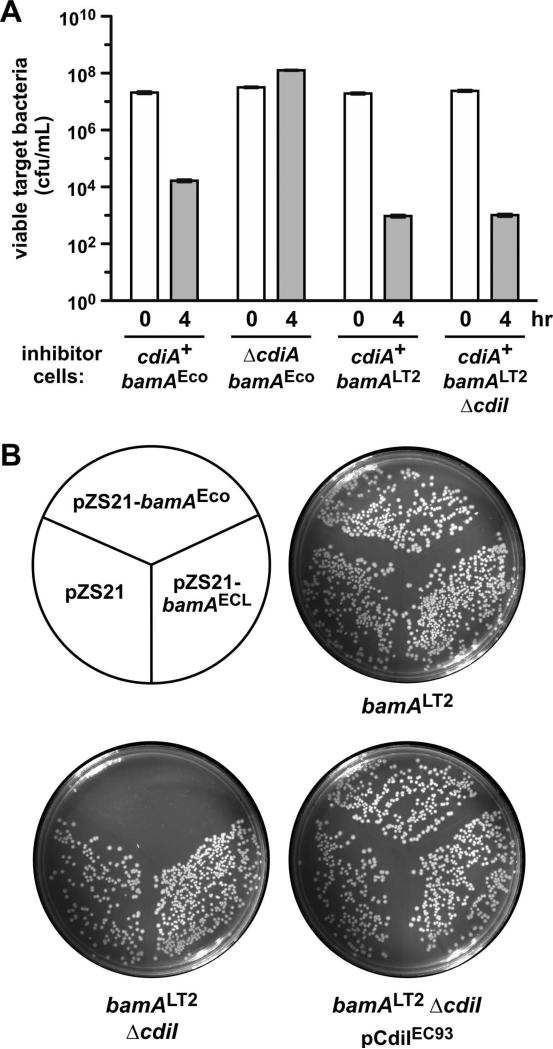
Because E. coli EC93 cells express BamAEco receptors, we reasoned that CdiAEC93-BamAEco binding interactions should contribute significantly to biofilm assembly. To test this model, we examined E. coli EC93 cells in which bamAEco is replaced with the bamALT2 gene from Salmonella Typhimurium LT2. BamALT2 is not recognized as a receptor by CdiAEC93 (Ruhe et al., 2014, Ruhe et al., 2013b), and therefore bamALT2 cells cannot form cell-cell aggregates through CdiA-receptor binding interactions. To demonstrate that EC93 bamALT2 cells lack a functional CdiAEC93 receptor, we deleted the cdiIEC93 immunity gene to generate EC93 bamALT2 ΔcdiI cells. The resulting strain retained full CDI activity in competition co-cultures with E. coli X90 target bacteria (Fig. 2A), indicating that it continues to deploy CdiAEC93 effectors. Moreover, plasmid-borne bamAEco could not be introduced into EC93 bamALT2 ΔcdiI cells unless they were first provided with a copy of the cdiIEC93 immunity gene on a compatible plasmid (Fig. 2B). Together, these experiments show that EC93 bamALT2 cells produce a functional CDIEC93 apparatus, but lack the CdiAEC93 receptor and therefore cannot be targeted by sibling cells. Surprisingly, the EC93 bamALT2 strain did not phenocopy the ΔcdiA mutant in a static biofilm assay. Instead, bamALT2 cells produced more biofilm mass in polystyrene wells than the ΔcdiA mutant and nearly as much as wild-type EC93 cells (Fig. 1A). However, EC93 bamALT2 biofilms grown in static chambers contained less average biomass (P = 0.035) and had reduced biofilm thickness (P = 0.15) compared to wild-type EC93 (Fig. 1B & Table 1). These data indicate that CdiAEC93-BamAEco binding contributes to biofilm formation, but CDI also appears to facilitate cell-cell interactions in a receptor-independent manner.
Figure 2. E. coli EC93 bamALT2 cells retain the CDI+ inhibitor cell phenotype.
A) E. coli EC93 strains of the indicated genotype were used as inhibitor cells in competition co-cultures with E. coli X90 target bacteria. Viable target bacteria were enumerated as colony forming units per mL at 0 and 4 h. The mean ± SEM is presented for two independent competition experiments. B) The cdiI immunity gene can be deleted from E. coli EC93 bamALT2 cells. Strains CH10060 (bamALT2), CH10151 (bamALT2 ΔcdiI) and CH10151 carrying pDAL741 (cdiI+) were transformed with the indicated plasmids that express bamAEco or bamAECL, followed by plating onto LB agar supplemented with kanamycin.
Receptor-independent auto-aggregation of CDI+ cells
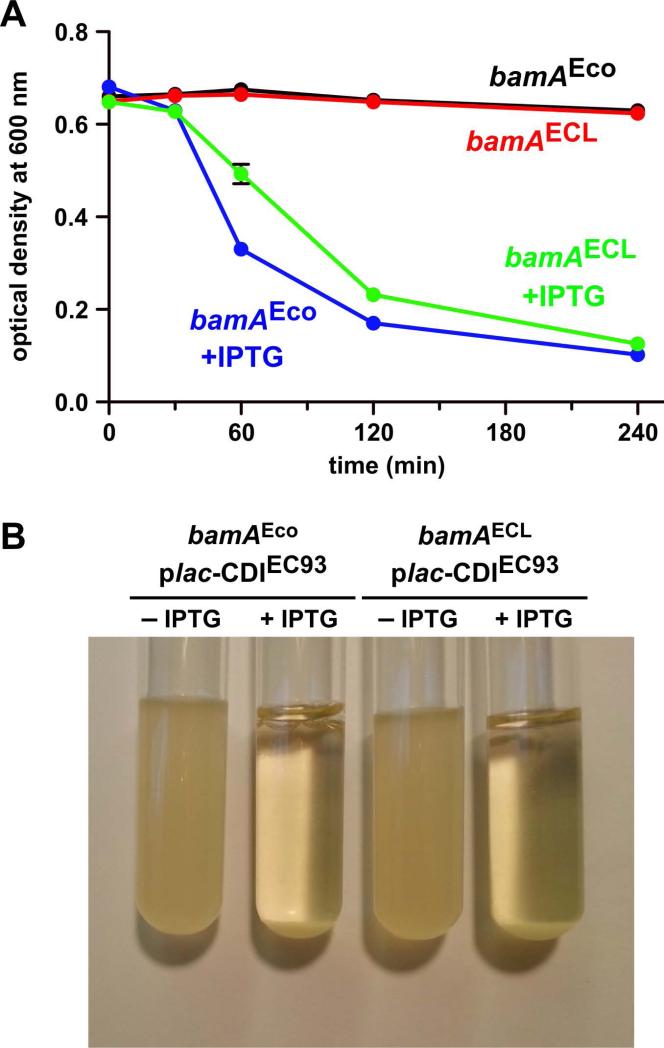
Expression of the cdiBAIEC93 gene cluster causes auto-aggregation of laboratory E. coli K-12 strains (Aoki et al., 2005). Because auto-aggregation and biofilm formation are often related phenomena, we examined whether BamAEco is strictly required for CDI-dependent aggregation. We first confirmed that cdiBAIEC93 expression is sufficient to promote auto-aggregation in E. coli EPI100 cells. The cdiBAIEC93 gene cluster was placed under the control of an IPTG-inducible promoter and auto-aggregation was monitored by measuring the optical density (at 600 nm) at the surface of standing liquid cultures as a function of time. Because aggregates sediment more rapidly than individual cells, OD600 values decline at a faster rate when cells aggregate. In the absence of IPTG, cell density at the culture surface remained nearly constant over 4 h (Fig. 3A). However, upon induction with IPTG, the culture showed significant sedimentation over the same time period (Figs. 3A & 3B). We then introduced the IPTG-inducible cdiBAIEC93 expression construct into E. coli EPI100 ΔbamA cells that express plasmid-borne bamAECL from Enterobacter cloacae ATCC 13047. Like BamALT2, the BamAECL protein is not a receptor for CdiAEC93 (Ruhe et al., 2014, Ruhe et al., 2013b). We again monitored cell aggregation through sedimentation and surprisingly found that CDI+ bamAECL cells fell out of suspension almost as rapidly as CDI+ bamAEco cells (Figs. 3A & 3B). Thus, expression of the cdiBAIEC93 gene cluster promotes auto-aggregation in a BamAEco-independent maxnner.
Figure 3. Expression of the cdiBAIEC93 gene cluster induces auto-aggregation.
A) Strains CH9604 (bamAEco) and CH9591 (bamAECL) that carry plasmid pCH12732 were grown in LB medium with and without IPTG, then adjusted to OD600 ~ 0.6 for auto-aggregation assays. B) Photograph of the cell suspensions from panel A after four h of incubation without shaking.
Cells must express CdiAEC93 to participate in cellular aggregates
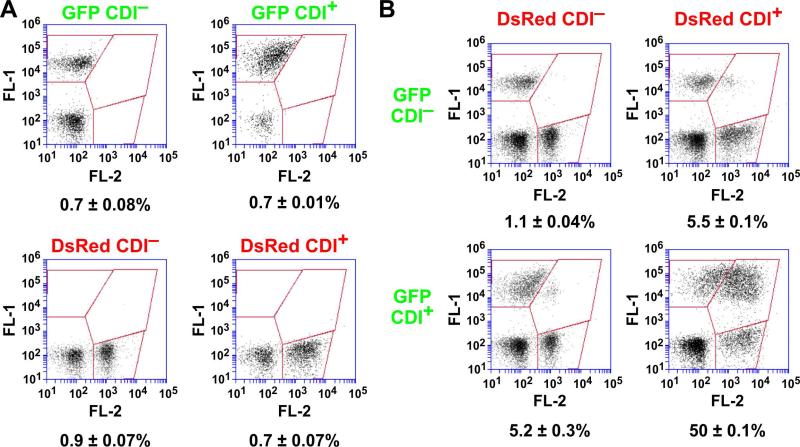
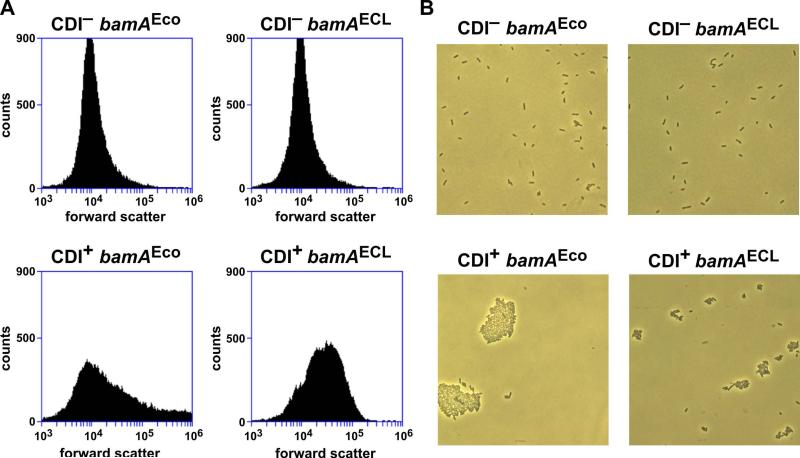
CdiAEC93 fragments are released into the growth medium (Aoki et al., 2005), raising the possibility that these peptides form a β-amyloid-like matrix that non-specifically binds bacteria together. If so, then CDI+ bacteria should be able to capture CDI− cells in the aggregates. We tested this prediction using flow cytometry to detect and quantify cell-cell binding interactions (Aoki et al., 2008, Ruhe et al., 2013b). We generated GFP- and DsRed-labeled EPI100 bamAECL cells, then provided the strains with the cdiBAIEC93 expression plasmid (CDI+) or an empty vector control (CDI−). As expected, the resulting GFP CDI−, GFP CDI+, DsRed CDI– and DsRed CDI+ strains showed very low levels of dual green/red fluorescence when analyzed individually by flow cytometry (Fig. 4A). We then mixed GFP- and DsRed-labeled cells at a 1:1 ratio and incubated with shaking to facilitate cell-cell binding. Analysis of the GFP CDI−/DsRed CDI− co-culture showed that ~1% of the flow events had both green and red fluorescence (Fig. 4B). Similarly, only about 5% of the events had dual fluorescence when CDI+ cells were mixed with a CDI− population (Fig. 4B). In contrast, approximately 50% of the events exhibited dual fluorescence when GFP-labeled CDI+ cells were mixed with DsRed-labeled CDI+ cells (Fig. 4B). This latter result indicates that bacteria must express CdiAEC93 in order to join the aggregates. Moreover, because these strains lack BamAEco receptors, the cell-cell binding interactions are presumably distinct from those that form between inhibitor and target cells during CDI. This conclusion is supported by forward light scatter analysis, which indicated that CDI+ bamAEco cell aggregates are larger and more heterogenous than the CDI+ bamAECL aggregates (Fig. 5A). Light microscopy also showed that CDI+ bamAEco cells form significantly larger aggregates than CDI+ bamAECL cells (Fig. 5B). By contrast, forward scatter analysis and light microscopy showed that most CDI− cells exist as isolated individuals, regardless of their bamA background (Figs. 5A & 5B). Thus, CdiAEC93-expressing bacteria physically interact in a BamAEco-independent manner. Because CdiAEC93 expression is required to participate in these aggregates, we hypothesized that this cell-cell adhesion is mediated by homotypic CdiA-CdiA binding interactions.
Figure 4. CDI expression is required for auto-aggregation.
A) CH9591 (bamAECL) cells were transformed with pTNC-WEB (CDI−) or pDAL660Δ1-39 (CDI+) plasmids and labeled with GFP or DsRed. Each of the four cell populations was analyzed by flow cytometry to detect background levels of dual green/red fluorescent events. B) The individual cell populations show in panel A were mixed at 1:1 ratio and incubated with shaking at 37 °C. The suspensions were then analyzed by flow cytometry using FL1 (533/30nm, GFP) and FL2 (585/40nm, DsRed) fluorophore filters.
Figure 5. Flow cytometric and microscopic analysis of CDI-dependent aggregates.
A) Strains CH9604 (bamAEco) and CH9591 (bamAECL) that carry either pTNC-WEB (CDI−) or pDAL660Δ1-39 (CDI+) were grown in LB medium and analyzed by flow cytometry for forward scatter. B) The cells from prepared for panel A were also visualized by light microscopy to determine the number of bacterial cells per aggregate.
Receptor-dependent and -independent binding regions map to different positions within CdiAEC93
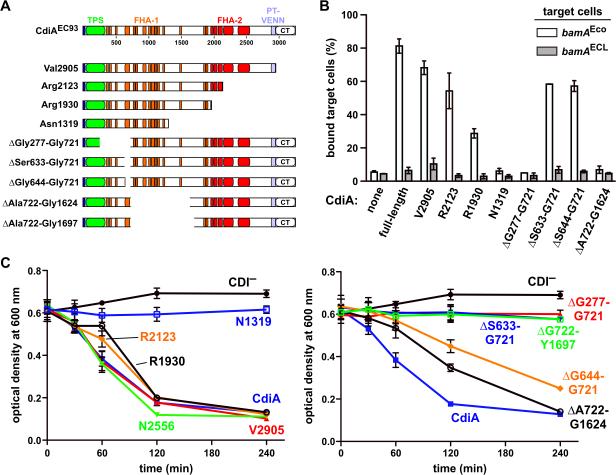
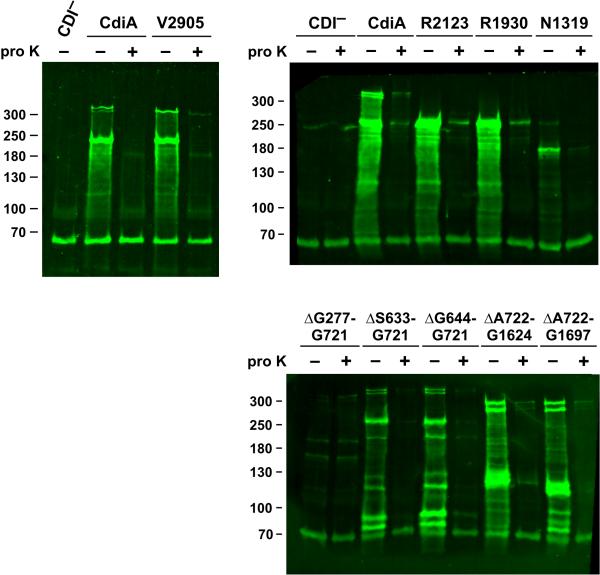
We next sought to determine whether the two adhesin activities are mediated by distinct regions of the CdiAEC93 filament. We generated a series of CdiAEC93 truncation constructs (Fig. 6A) and analyzed their BamA-binding properties using flow cytometry. Cells expressing CdiAEC93 truncated at residues Val2905 and Arg2123 still aggregated specifically with bamAEco target bacteria, but truncation to residue Arg1930 significantly attenuated cell-binding activity (Fig. 6B). Further truncation to residue Asn1319 completely abrogated the ability to bind bamAEco target cells (Fig. 6B). These data indicate that the BamAEco-binding region lies somewhere within the N-terminal 1930 residues of CdiAEC93. We then tested internal deletion constructs and found that CdiAEC93 proteins lacking residues Ser633 – Gly721 and Ser644 – Gly721 retained BamAEco-binding activity, but deletion of Ala722 – Gly1624 abrogated this activity (Figs. 6A & 6B). This latter results suggests that the BamAEco-binding region is located between residues Ala722 and Gly1624, but it is also possible that CdiA(ΔAla722-Gly1624) is not secreted or perhaps not assembled properly on the cell surface. Therefore, we analyzed each truncation and deletion variants by immunoblot using polyclonal antibodies against the N-terminal TPS transport domain of CdiAEC93. In general, each CdiAEC93 variant accumulated to approximately the same level as wild-type protein, but the CdiA(Asn1319) and CdiA(ΔGly277-Gly721) proteins were much less abundant (Fig. 7). The latter mutation deletes a portion of the TPS transport domain, which is predicted prevent export across the outer membrane. To monitor CdiAEC93 secretion, we treated intact cells with proteinase K to degrade extracellular protein and compared them to untreated cells using immunoblot analysis. With the exception of the ΔGly277-Gly721 mutant, all of the variants were sensitive to extracellular protease (Fig. 7), indicating that most of the CdiAEC93 variants were exported to the cell surface. These data suggest that CdiA(ΔAla722 – Gly1624) is processed properly and does not bind target bacteria because the BamAEco-binding region has been deleted.
Figure 6. Receptor-dependent and -independent adhesion domains map to distinct positions within CdiAEC93.
A) Architecture of CdiAEC93. The CdiAEC93 is composed of an N-terminal TPS transport domain (green) followed by two distinct regions of filamentous hemagglutinin peptide repeats (FHA-1 and FHA-2). The pretoxin-VENN (PT-VENN) domain demarcates the C-terminal toxin region. The various CdiAEC93 truncations and in-frame deletions are shown schematically. B) BamAEco-dependent target-cell binding assays. DL4259 cells that carry pTNC-WEB (none) or the indicated CDI cosmids were mixed at a 5:1 ratio with CH9604 (bamAEco) and CH9591 (bamAECL) target cells that express DsRed. The suspensions were analyzed by flow cytometry using FL1 (533/30nm, GFP) and FL2 (585/40nm, DsRed) fluorophore filters. The percentage of bound target cells was calculated as the number of dual green/red fluorescent events divided by the total number of red fluorescent events. The average ± SEM is presented for two independent cell-binding experiments. C) Auto-aggregation of CdiAEC93 mutants. Mono-cultures of E. coli CH9591 (bamAECL) that express the indicated CdiAEC93 truncation and in-frame deletion variants were suspended and allowed to sediment at 1 × g to monitor auto-aggregation.
Figure 7. Immunoblot analysis of CdiAEC93 truncation and deletion variants.
CH9591 (bamAECL) cells that produce the indicated CdiAEC93 proteins were treated with 100 μg mL−1 of extracellular proteinase K (proK) where indicated. Proteinase K digestion was quenched with 1 mM PMSF and total urea-soluble protein was extracted. Proteins were resolved on SDS 6% polyacrylamide gels and transferred to nitrocellulose for immunoblot analysis using polyclonal antibodies to the CdiAEC93 TPS domain. The migration position of molecular weight standards are indicated in kDa. The predicted mass of full-length CdiAEC93 is ~319 kDa.
We then examined CdiAEC93 truncation and deletion variants for receptor-independent auto-aggregation. CdiAEC93 proteins truncated at Val2905, Arg2123 and Arg1930 all supported auto-aggregation in the bamAECL genetic background, but cells expressing the Asn1319 truncation protein did not sediment (Fig. 6C). Forward scatter analysis also indicated that all truncations except CdiA(Asn1319) promoted auto-aggregation in the bamAECL background (Fig. S3). Sedimentation analysis of the internal deletion variants revealed that CdiA(ΔSer644-Gly721) and CdiA(ΔAla722-Gly1624) support receptor-independent aggregation, whereas the other deletion constructs do not (Fig. 6C). Together, these experiments identified internal deletion constructs that retain specific adhesion activities. CdiA(ΔAla722-Gly1624) does not bind to BamAEco, but supports receptor-independent auto-aggregation (Figs. 6B & 6C). Conversely, CdiA(ΔSer633-Gly721) specifically binds to BamAEco, yet lacks auto-aggregation activity (Figs. 6B & 6C). Collectively, these data demonstrate that the BamAEco and receptor-independent adhesion domains map to different positions on the CdiAEC93 filament.
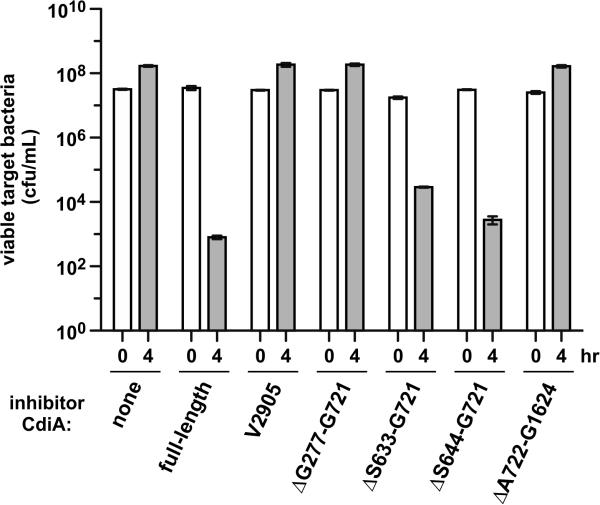
Having localized the receptor-independent adhesion domain to the N-terminal region of CdiAEC93, we asked whether this portion of the filament plays any role in CDI. We used EPI100 cells that express CdiAEC93 deletion variants as inhibitor strains in competition co-cultures with E. coli X90 target bacteria. X90 targets grew about 5-fold in mock competitions with CDI− EPI100 cells, but viable X90 cells were reduced ~40,000-fold in co-cultures with EPI100 cells that express full-length CdiAEC93 (Fig. 8). Inhibitors that express truncated CdiA(Val3105) were unable to inhibit target cells (Fig. 8), demonstrating that the C-terminal toxin domain is required for growth inhibition. The CdiA(ΔGly277-Gly721) protein also failed to inhibit target bacteria (Fig. 8), consistent with the observation that this effector is not secreted and therefore cannot bind to target cells. In contrast, CdiA(ΔSer633-Gly721) and CdiA(ΔSer644-Gly721) mutants both retained inhibition activity, reducing target-cell counts approximately 600- and 11,000-fold, respectively (Fig. 8). CdiA(ΔAla722-Gly1624), which lacks BamAEco-binding activity, was unable to inhibit target cells and phenocopied the export-defective (ΔGly277-Gly721) and toxin-deletion (Val3105) mutants (Fig. 8). Taken together, these data show that the two adhesion activities have distinct functions and locations on the CdiAEC93 filament. The BamAEco-binding domain appears to reside between residues Ala722 and Gly1624 and is essential for toxin delivery into target cells. In contrast, the receptor-independent adhesion domain maps to a region N-terminal to Ser644 and is not required for CDI-mediated growth inhibition.
Figure 8. Competition co-cultures with CdiAEC93 truncation and deletion variants.
Stain CH9591 (bamAECL) cells that express the indicated CdiAEC93 variants were used as inhibitor cells in competition co-cultures with E. coli X90 target cells. Inhibitors were mixed at a 10:1 ratio with targets and incubated with shaking at 37 °C for four h. Viable target bacteria were enumerated as colony forming units per mL and counts are presented as the average ± SEM for two independent experiments.
Discussion
TPS and auto-transporter proteins have long been implicated in bacterial biofilm formation (Rojas et al., 2002, Neil & Apicella, 2009, Gottig et al., 2009, Irie et al., 2004, Serra et al., 2011, Anderson et al., 2012). These type V secretion systems often function as adhesins, implying that the secreted exoproteins mediate attachment to the substratum and/or other bacteria during biofilm formation. In addition to directly mediating adhesion, results suggest that CDI modulates biofilm formation indirectly through changes in gene expression (Gottig et al., 2009). Additionally, Cotter and co-workers report that the enzymatic activity of the CdiA-CTE264 toxin is required for biofilm formation in B. thailandensis E264 (Garcia et al., 2013). In contrast, we find no significant alterations in cell motility or biofilm gene expression in E. coli EC93 ΔcdiA mutants; and the CdiA-CTEC93 region does not appear to play the same critical role in E. coli EC93 biofilms as it does in B. thailandensis. Presumably, these discrepancies reflect fundamental differences between CDI systems. For example, E. coli EC93 expresses its cdi genes constitutively (Aoki et al., 2005), whereas the B. thailandensis system is regulated by quorum sensing (Majerczyk et al., 2014). Moreover, homology between CdiAEC93 and CdiAE264 is largely limited to the N-terminal TPS transport domain and the effectors carry unrelated toxin domains (Fig. S4). The CdiA-CTE264 region has Mn2+-dependent DNase activity (Garcia et al., 2013, Nikolakakis et al., 2012), whereas the CdiA-CTEC93 toxin domain forms membrane pores (Aoki et al., 2009, Ruhe et al., 2014). Garcia et al. have suggested that CdiA-CTE264 activity could modify the extracellular DNA matrix and thereby promote B. thailandensis biofilm formation (Garcia et al., 2013). In contrast, our data suggest that CdiAEC93 acts primarily as an adhesin to tether bacterial cells to one another. However, CdiAEC93-expressing cells still auto-aggregate when they lack functional BamAEco receptors. The simplest explanation for these data is that CdiAEC93 possesses an additional adhesion activity. We hypothesize that CdiAEC93 may mediate cell adhesion through homotypic interactions, because bacteria must express CdiAEC93 in order to participate in receptor-independent aggregates. Homotypic interactions have been proposed previously for other type V secretion systems, including the Ag43 autotransporter from E. coli (Girard et al., 2010) and Bordetella FHA (Menozzi et al., 1994). FHA appears to bind several different ligands including mammalian integrins (Ishibashi et al., 1994, Relman et al., 1990). Notably, other CdiA proteins were originally described as adhesins that bind to eukaryotic host cells. MhaB1 and MhaB2 from Moraxella catarrhalis were found to increase bacterial association with human epithelial cells in culture (Balder et al., 2007). HrpA from N. meningitidis was reported to facilitate adhesion to epithelial cells, though the hrpA phenotype is only manifest in lipopolysaccharide-deficient mutant backgrounds (Schmitt et al., 2007). Taken together, these observations suggest that CdiA proteins often possess multiple adhesion domains.
De novo biofilm formation requires that bacteria first adhere to a surface. After this initial attachment phase, cells secrete extracellular matrix molecules, which help bind the bacteria together and allow large three-dimensional structures to be built. EC93 ΔcdiA mutants have different initial attachment phenotypes depending on the abiotic substratum. EC93 cdiA+ and ΔcdiA cells adhere to polystyrene surfaces at similar rates, but ΔcdiA mutants appear to have difficulty establishing biofilms on glass surfaces. However, the more profound defect appears to be in biofilm maturation, in accord with previous findings with N. meningitidis and B. thailandensis cdi mutants (Neil & Apicella, 2009, Anderson et al., 2012, Garcia et al., 2013). Collectively, these observations suggest that CdiA augments cell-cell interactions and reinforces the extracellular matrix. Intriguingly, the E. coli EC93 cdiBAI locus is linked to a predicted fimbria operon. Fimbriae are adhesive appendages that often mediate attachment to abiotic substrates and are therefore critical for the early stages of biofilm formation. This genetic linkage could perhaps indicate cooperation between the fimbria and CdiAEC93 adhesion systems during biofilm formation. We note a similar linkage in uropathogenic E. coli strains, where the cdi genes are carried on pathogenicity island II together with type I fimbria and the pyelonephritis associated pili (pap) genes (Dobrindt et al., 2002). These associations suggest that CDI plays an integral role in biofilm lifestyles, and that its adhesin activities contribute to bacterial virulence and pathogenicity.
CDI provides a mechanism to discriminate neighboring cells through both kin- and kind-selection. Kin are defined as individuals that are related to one another across their entire genomes and whose genetic relationships are the result of direct vertical descent (Strassmann et al., 2011). By contrast, individuals of the same kind are those that share a specific trait – often specified by a single genetic locus – but are not necessarily related at other loci (Strassmann et al., 2011). Adhesion between isogenic CDI+ cells represents a form of kin selection, which increases the relatedness of the population and provides benefits by resisting forces that would disrupt the community. But E. coli EC93 cells also have the potential to adhere to other E. coli strains, because virtually all isolates carry and express identical bamAEco receptor genes. Thus, the growth-inhibition function of CDI could prevent intrusions by other E. coli strains, or conversely allow the CDI+ strain to attach to and invade an established E. coli microcolony. In contrast, receptor-independent CdiA-CdiA interactions represent a possible kind-selective mechanism, whereby unrelated bacteria cooperate if they express similar cdiA alleles. Strains of Citrobacter rodentium, Salmonella typhi and Enterobacter cloacae all encode CdiA proteins that share significant (>50%) sequence identity with CdiAEC93 over the homotypic interaction region. In principle, these strains could participate in CdiA-mediated aggregation and cooperate with E. coli EC93 cells by co-inhabiting the same biofilm community. Thus, cdiA genes may act as so-called greenbeard alleles, which allow individuals to recognize others with the same trait and direct preferential treatment to them (Dawkins, 1976, West & Gardner, 2010, Strassmann et al., 2011). This kind-selective function could be critical for the maintenance and propagation of cdi genes, which are usually found within genomic islands and other mobile genetic elements (Poole et al., 2011). Because greenbeard discrimination promotes cooperation between unrelated individuals, it may provide selective pressure to spread cdi loci through horizontal gene transfer.
CDI-mediated growth inhibition is usually limited to closely related bacteria (Aoki et al., 2005, Aoki et al., 2010, Nikolakakis et al., 2012, Arenas et al., 2013, Anderson et al., 2012, Beck et al., 2014, Anderson et al., 2014, Koskiniemi et al., 2015). For E. coli EC93, the molecular basis of this discrimination is understood and reflects variations in cell-surface epitopes of the BamA receptor between bacterial species (Ruhe et al., 2013b). Therefore, the specificity of CdiA-receptor interactions limits the target-cell range. This feature of CDI makes biological sense if related bacteria are more likely to compete for the same environmental resources. However, these observations raise important questions about the evolution of cdi genes, particularly with respect to horizontal gene transfer. The genes encoding CDI systems are often associated with transposable elements and are sometimes encoded within genomic islands, suggesting that cdi genes are transferred horizontally between different bacteria. This hypothesis is also consistent with the observation that cdi loci are not necessarily found in all strains of a given species (Aoki et al., 2010, Poole et al., 2011). Therefore, if cdi genes are transferred between different bacterial species, it is likely that the recipient bacterium will not carry an appropriate surface receptor for the acquired CDI system. Because all characterized CDI systems recognize self receptors, CdiA-receptor interactions probably evolve after the horizontal gene transfer event. This is presumably a slow process, raising the question of what selective pressures allow retention of cdi loci in the interim. One possibility is that the toxin/immunity functions of CDI enforce retention. Although CDI toxins cannot be delivered between cells in the absence of a suitable CdiA receptor, synthesis of the CdiA-CT toxin from internal translation initiation sites could stabilize cdi genes through an immunity protein addiction mechanism similar that that proposed for type II prokaryotic toxin/antitoxin modules (Jensen & Gerdes, 1995, Wozniak & Waldor, 2009). The results presented here suggest that receptor-independent adhesion could provide an additional selective advantage to cells that have newly acquired cdi genes. TPS systems are autonomous and have the potential to function in a variety of Gram-negative bacteria. Therefore, newly acquired cdiBA genes could plausibly produce properly assembled CdiB/CdiA complexes on the cell surface, thereby enabling receptor-independent auto-aggregation. If these auto-adhesive properties provide a significant fitness benefit, then bacteria that retain the cdi genes would derive a selective advantage thereby providing the opportunity for cdiA to acquire new receptor-binding function.
Materials and Methods
Strains and growth conditions
Bacterial strains used in this study are listed in Table 2. Bacteria were routinely grown in LB broth or LB agar at 37 °C unless otherwise noted. Growth media was supplemented with antibiotics (ampicillin, 100 μg mL−1; kanamycin, 50 μg mL−1; tetracycline 12.5 μg mL−1) where indicated. E. coli EC93 ΔcdiA (CH10094) and bamALT2 (CH10053) strains have been described previously (Ruhe et al., 2013b, Ruhe et al., 2014). The toxin-encoding region of cdiA was deleted from E. coli EC93 to generate strain CH10024. Regions of upstream and downstream of the cdiA-CT sequence were amplified with primers ΔcdiA(CT)-up-Sac/ΔcdiA(CT)-up-Bam and ΔcdiA-dn-Eco/ΔcdiA-dn-Xho, respectively (Table S1). The resulting products were sequentially ligated to plasmid pKAN (Hayes et al., 2002) to generate pKAN-cdiAΔCT (Table 2). The SacI/EcoRI fragment from plasmid pKAN-cdiAΔCT was introduced into E. coli EC93 cells that express phage λ Red recombinase genes from plasmid pSIM6 as described (Thomason et al., 2007). The FRT-flanked kanamycin-resistance cassette was removed using FLP recombinase as described (Cherepanov & Wackernagel, 1995). The cdiI gene was deleted from E. coli EC93 bamALT2 cells (strain CH10060) using essentially the same approach. Upstream and downstream homology regions were amplified using primers EC93-cdiI(KO)-Sac/EC93-cdiI(KO)-Bam and EC93-cdiI(KO)-Eco/EC93-cdiI(KO)-Xho, respectively. The two products were sequentially ligated to plasmid pKAN to generate plasmid pKAN-ΔcdiI. The SacI/XhoI fragment from plasmid pKAN-ΔcdiI was introduced into E. coli CH10060 cells that express phage λ Red recombinase genes.
Table 2.
Bacterial strains
| Strains | Description a | Reference |
|---|---|---|
| X90 | F′ laclq lac′ pro′/ara Δ(lac-pro) nal1 argE(amb) rifr thi-1, RifR | (Beckwith & Signer, 1966) |
| EPI100 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacXcZΔM15 ΔlacX recA1 endA1 araD139 Δ(ara, leu)7697 galU galK ⌊− rpsL nupG, StrR | Epicentre |
| DY378 | W3110 λcI857 Δ(cro-bioA) | (Thomason et al., 2007) |
| EC93 | Escherichia coli isolate from rat feces | (Aoki et al., 2005) |
| DL4259 | E. coli MC4100 that expresses GFP-mut3 | (Morse et al., 2012) |
| CH2016 | X90 (DE3) Δrna ΔslyD::kan, RifR KanR | (Garza-Sánchez et al., 2006) |
| CH9350 | X90 ΔbamA::cat pZS21-bamA+, CmR KanR | (Ruhe et al., 2013b) |
| CH9371 | X90 ΔbamA::cat pZS21-bamAECL, CmR KanR | (Ruhe et al., 2013b) |
| CH9591 | EPI100 ΔbamA::cat pZS21-bamAECL, CmR KanR | (Ruhe et al., 2013b) |
| CH9604 | EPI100 ΔbamA::cat pZS21-bamA+, CmR KanR | (Ruhe et al., 2013b) |
| CH10024 | EC93 cdiA(A2909) | This study |
| CH10053 | EC93 bamALT2::kan, KanR | (Ruhe et al., 2014) |
| CH10060 | EC93 bamALT2 | This study |
| CH10094 | EC93 ΔcdiA | (Ruhe et al., 2014) |
| CH10131 | EC93 bamALT2 ΔcdiI::kan, KanR | This study |
| CH10151 | EC93 bamALT2 ΔcdiI | This study |
Abbreviations: AmpR, ampicillin-resistant; CmR, chloramphenicol-resistant; KanR, kanamycin-resistance; RifR, rifampicin-resistant; TetR, tetracycline-resistant
Motility was assayed on freshly prepared tryptone broth agar (0.4% agar). Overnight cultures were adjusted to OD600 = 1.0 and 10 μL of the suspension was spotted onto the center of the plate. Plates were incubated in humid chambers at 37 °C for 12 h. Bacterial competition co-cultures were conducted in LB media without antibiotics. Inhibitors and target bacteria (E. coli X90) were grown individually to mid-log phase at 37 °C, then mixed at a 10:1 ratio in fresh LB at a final OD600 of 0.33. Samples were plated onto selective media at 0 h to enumerate colony forming units (CFU) per mL for each population. The co-cultures were incubated at 37 °C with shaking for 4 h, then samples plated onto selective media to quantify viable target cell counts as CFU mL−1.
Plasmid constructions
Super-folding gfp was amplified with primers gfp(mut3)-Bam-for/sfGFP-rev-Bam ligated into the BamHI site of plasmid pTrc99a. The gfp fragment was then subcloned into plasmid pCH450 using EcoRI and PstI restriction sites. The coding sequence for DsRed was amplified from plasmid DsRedExpress2 using primers dsRed-Eco-for/dsRed-Xho-rev and ligated into plasmid pCH450 using EcoRI and XhoI restriction sites. The coding sequence for the TPS transport domain of CdiAEC93 was amplified using primers EC93cdiA-V33-Spe-for/EC93cdiA-G285-Xho-rev and ligated to plasmid pSH21P using SpeI and XhoI restriction sites. The resulting plasmid was used to over-produce N-terminal His6-tagged TPS transport domain for purification and generation of polyclonal antibodies. Truncation and deletion mutations within cosmid-borne cdiA were generated using a series of plasmid pDAL660Δ1-39 derivatives that carry random transprimer insertions as described (Aoki et al., 2005). Plasmids that carry in-frame stop codons after CdiA residues Val2905, Asn2556, Arg2123, Arg1930 and Arg1319 were used directly for cell-cell binding assays. In-frame deletions were created by digesting selected transprimer-containing plasmids with PmeI and SfiI, followed by re-ligation and transformation into EPI100 cells.
The IPTG-inducible cdiBAI expression plasmid was built in several steps. The rrnB transcription terminators from excised from plasmid pUC18T-miniTn7T-Kan (Choi et al., 2008) with SacI/NheI and ligated to SacI/SpeI digested plasmid pKAN. The lac promoter was then amplified with primers lac-Sacfor/lac-OE-rev and the 5′-end of cdiB amplified with cdiB-OE-for/cdiB-Sac-rev. The two PCR products were combined using over-lapping end PCR (Aiyar et al., 1996) with primers lac-Sac-for/cdiB-Sac-rev, and the fused product ligated to pKAN-rrnB using the SacI restriction site. The lacIq gene was then amplified with primers lacI-Eco-for/lacI-Xho-rev and ligated using EcoRI and XhoI restriction sites. Finally, a fragment upstream of cdiB was amplified with primers CDI-up-Kpn-for/CDI-up-Xho-rev and ligated using XhoI and KpnI restriction sites. The final plasmid was amplified with primers CDI-up-Kpn-for/cdiB-Sac-rev and the resulting product was recombineered into plasmid pDAL660Δ1-39 to generate plasmid pCH12732.
Biofilm assays
Overnight cultures of E. coli EC93 and derivatives were inoculated into fresh tryptone broth supplemented with 0.5% glycerol at a final OD600 of 0.05. The cell suspension (200 μL) was added to the interior well(s) of a 96-well flat-bottom polystyrene plate and incubated at 37 °C for the indicated times. For proteinase K treated samples, 2 μL of 1 mg mL−1 proteinase K freshly dissolved in 1 × PBS was added to wells after 2 h of incubation. Suspended cells were removed by inversion of the plate followed by gentle tapping onto a clean paper towel. 200 μL of 0.1% crystal violet solution was added to sample wells and blank wells and incubated for 15 min at ambient temperature. The stain was removed by inversion of the plate and tapping onto a fresh paper towel. Wells were gently washed three times with 1 × PBS. Wells were destained with 0.2 mL of 95% ethanol followed by vigorous pipetting. Staining was quantified by measuring absorbance of the ethanol solution at 595 nm.
For confocal laser-scanning microscopic (CLSM) analysis of biofilms, overnight cultures of E. coli EC93 were diluted to OD600 of 0.02 in LB medium and 3 mL of the suspension inoculated into glass chambers (Lab-Tek) for incubation at 30 °C under static conditions. After 24 h, planktonic bacteria were removed by inverting the chambers and gentle washing with LB medium. Adherent cells were then stained with 1 μM SYTO-9 (Life Technologies) in the dark for 30 min. Biofilms were visualized with an LSM 5 Pascal laser-scanning microscope (Zeiss). Three-dimensional images were reconstructed using Imaris 7.6 and analyzed using COMSTAT (Heydorn et al., 2000). Each experiment included 2 independent biological replicates, with three images taken for each replicate.
Gene expression analysis
E. coli EC93 biofilms were produced as described above. After incubation on polystyrene plates for 4 h, planktonic cells were gently removed with a pipette and the volume immediately replaced with 200 μL of Qiagen RNA Protect Bacterial Reagent. Biofilms were harvested from the plate with rigorous pipeting and RNA extracted from the suspension using the Qiagen RNEasy kit with on-column DNase I digestion according to manufacturer's instructions. Total cDNA was prepared using the SuperScript III First Strand Synthesis kit. The relative abundance of pgaA, wcaA, bcsA, and fliC transcripts was determined by quantitative real-time PCR using primer pairs pgaA-for/pgaA-rev, wcaA-for/wcaA-rev, bcsA-for/bcsA-rev and fliC-for/fliC-rev, respectively with the SYBR Green Real-Time PCR Master Mix according to manufacturer's instructions. Comparative CT analysis was performed as described (Schmittgen & Livak, 2008) using pgaA and fliC transcript levels as references.
Auto-aggregation assay
Strains CH9604 (bamAEco) and CH9591 (bamAECL) were transformed with IPTG-inducible CDIEC93 system plasmid and tested for auto-aggregation upon induction. Cells were grown to OD600 ~1.0 in TB supplemented with 0.5% glycerol with or without 1.5 mM IPTG. Cells were harvested and resuspended in 20 mL of spent media at OD600 ~ 0.6 – 0.7. The resuspended cells were transferred to a 13 cm glass culture tube and incubated at 37 °C without agitation. The OD600 was measured at the top of the cell suspension at 30, 60, 120 and 240 min. BamAEco independent auto-aggregation was determined in the same manner for CdiAEC93 truncation and deletion variants expressed in strain CH9591 (bamAECL), except no IPTG induction was required to express these proteins.
Flow cytometry
BamAEco receptor binding studies were conducted using strain DL4259, which expresses gfp-mut3 from the papIB promoter (Webb et al., 2013). DL4259 was transformed with cosmids that express CdiAEC93 truncation and deletion variants, then grown in LB medium supplemented with ampicillin in baffled flasks at 37 °C until the cells became fluorescent. Fluorescent DL4259 cells were mixed at a 5:1 ratio with DsRed-expressing CH9604 (bamAEco) or CH9591 (bamAECL) cells at a final OD600 of 0.2. the cell suspension were shaken at 30 °C for 20 min, diluted into 1 × PBS, vortexed briefly, then analyzed on an Accuri C6 flow cytometer using FL1 (533/30nm, GFP) and FL2 (585/40nm, DsRed) fluorophore filters. Percent target-cell binding was calculated as the number of dual green/red fluorescent events divided by the total number of red fluorescent events (Ruhe et al., 2013b).
For receptor-independent binding assays, CH9591 (bamAECL) cells harboring pWEB-TNC (CDI−) or pDAL660Δ1-39 (CDI+) were labeled with GFP (pCH450::sfGFP) or DsRed (pCH450::DsRed), and the green- and red-labeled populations were mixed at a 1:1 ratio in 0.55 LB medium supplemented with ampicillin, tetracycline and 0.2% arabinose. Cell mixtures were grown with shaking at 37 °C until the cells became fluorescent, then the cultures were diluted into 1 × PBS, vortexed briefly and analyzed by flow cytometry. Forward-scatter analyses were performed on strains CH9604 (bamAEco) and CH9591 (bamAECL) that carry CDI expression cosmids. Overnight cultures were diluted 1:100 in fresh LB media supplemented with ampicillin and grown with shaking for 4 h at 37 °C. Cells were diluted into 1 × PBS, mixed gently, and analyzed by flow cytometry for forward scatter and by light microscopy to visualize cell aggregates.
CdiAEC93 TPS transport domain purification
The His6-tagged TPS transport domain (CdiAEC93 residues Val33 – Gly285) was overproduced in E. coli strain CH2016 harboring plasmid pSH21P::cdiA(V33-G285). An overnight culture was diluted 1:100 in fresh LB supplemented with ampicillin and grown to mid-log phase at 37 °C in a baffled flask. Expression was then induced with the addition of 1.5 mM IPTG for 90 min. Cells were harvested by centrifugation and the pellet was frozen at −80°C. Cells were broken by freeze-thaw in urea lysis buffer (8 M urea, 150 mM NaCl, 0.05% Triton X-100, and 20 mM imidazole) as described previously (Garza-Sánchez et al., 2006). The CdiAEC93 TPS domain was purified via Ni2+-affinity chromatography under denaturing conditions in urea lysis buffer, followed by elution in urea lysis buffer supplemented with 25 mM EDTA. The purified TPS domain was judged to be >95% pure by SDS-PAGE analysis and was used to generate rabbit polyclonal antibodies (Cocalico Biologicals Inc., Reamstown, PA).
Proteinase K treatment and immunoblot analysis
Overnight cultures of strain CH9591 (bamAECL) carrying CDI cosmids were diluted 1:100 into 2 mL fresh LB without antibiotics and grown at 37 °C to OD600 ~1.0. Cells were harvested, resuspended in 2 mL of 1 × PBS, and split into two 1 mL aliquots. 100 μg mL−1 proteinase K (final concentration) or buffer was added to each cell aliquot followed by incubation at 37°C for 1 h. Cells were harvested and resuspended in 1 mL 1 × PBS, 1 mM freshly prepared phenylmethylsulfonyl fluoride (PMSF). Cells were harvested by centrifugation and washed three times with 1 mL 1 × PBS supplemented with 1 mM PMSF. Washed cells were broken by freeze-thaw lysis in 75 μL of urea lysis buffer, 1 mM PMSF. Urea lysates (50 μg total protein) were run on SDS-6% polyacrylamide gels and blotted onto nitrocellulose for immunoblot analysis using polyclonal antibodies to the N-terminal TPS transport domain of CdiAEC93. Blots were visualized using IRDye® 680 (LI-COR) labeled anti-rabbit secondary antibodies and an Odyssey® infrared imager.
Supplementary Material
Table 3.
Plasmids
| Plasmid | Description a | Reference |
|---|---|---|
| pTrc99a | IPTG-inducible expression plasmid, AmpR | GE Healthcare |
| pCH450 | pACYC184 derivative with E. coli araBAD promoter for arabinose-inducible expression, TetR | (Hayes & Sauer, 2003) |
| pSIM6 | Expresses phage λ Red proteins from heat-shock inducible promoter, AmpR | (Datta et al., 2006) |
| pKAN | pBluescript with FRT-flanked kanamycin-resistance cassette ligated into Smal restriction site, AmpR KanR | (Hayes & Sauer, 2003) |
| pKAN-BamA(KO) | pKAN containing regions upstream and downstream of bamAEco, AmpR KanR | This study |
| pWEB-TNC | Cosmid cloning vector, AmpR CmR | Epicentre |
| pDsRedExpress2 | Constitutive expression of DsRed, AmpR | Clontech |
| pDAL741 | pBR322 derivative that expresses cdiIEC93 immunity gene, AmpR | (Aoki et al., 2005) |
| pCP20 | Heat-inducible expression of FLP recombinase, CmR AmpR | (Cherepanov & Wackernagel, 1995) |
| pSH21P | pET21b-derived expression plasmid, appends an N-terminal His6 epitope tag, AmpR | (Koskiniemi et al., 2014) |
| pCH450-DsRed | L-arabinose inducible expression of DsRed, TetR | This study |
| pTrc99A-sfGFP | IPTG inducible expression of superfolding GFP, AmpR | This study |
| pCH450-GFP | L-arabinose inducible expression of superfolding GFP, TetR | This study |
| pKAN-cdiAΔCT | Construct to delete the cdiA-CT region from E. coli EC93, AmpR KanR | This study |
| pKAN-ΔcdiI | Construct to delete the cdiI immunity gene from E. coli EC93, AmpR KanR | This study |
| pZS21 | pSC101-derived plasmid vector, KanR | (Lutz & Bujard, 1997) |
| pZS21-bamA+ | pZS21 derivative that expresses bamAEco, KanR | (Kim et al., 2007) |
| pZS21-bamAECL | Expresses bamAECL from Enterobacter cloacae ATCC 13047, KanR | (Ruhe et al., 2013b) |
| PDAL660Δ1-39 | Constitutive expression of the cdiBAlEC93 gene cluster, CmR AmpR | (Aoki et al., 2005) |
| pDAL878 | Derivative of pDAL660Δ1-39 in which the cdiA-CT/cdiI region has been deleted. Produces truncated CdiA(V2905stop) protein, CmR AmpR | (Webb et al., 2013) |
| pCH12699 | Derivative of pDAL660Δ1-39 that produces CdiA(ΔG722-G1624), CmR AmpR | This study |
| pCH12700 | Derivative of pDAL660Δ1-39 that produces CdiA(ΔG722-Y1697), CmR AmpR | This study |
| pCH12702 | Derivative of pDAL660Δ1-39 that produces CdiA(ΔG277-G721), CmR AmpR | This study |
| pCH12703 | Derivative of pDAL660Δ1-39 that produces CdiA(ΔS633-G721), CmR AmpR | This study |
| pCH12704 | Derivative of pDAL660Δ1-39 that produces CdiA(ΔS644-G721), CmR AmpR | This study |
| pCH12705 | Derivative of pDAL660Δ1-39 that produces truncated CdiA(R2123stop), CmR AmpR | (Aoki et al., 2005) |
| pCH12706 | Derivative of pDAL660Δ1-39 that produces truncated CdiA(R1930stop), CmR AmpR | (Aoki et al., 2005) |
| pCH12707 | Derivative of pDAL660Δ1-39 that produces truncated CdiA(R1319stop), CmR AmpR | (Aoki et al., 2005) |
| pCH12732 | IPTG-inducible expression of cdiBAIEC93 gene cluster, CmR AmpR | This study |
| pCH12736 | Derivative of pDAL660Δ1-39 that produces truncated CdiA(N2556stop), CmR AmpR | (Aoki et al., 2005) |
Abbreviations: AmpR, ampicillin-resistant; CmR, chloramphenicol-resistant; KanR, kanamycin-resistance; RifR, rifampicin-resistant; TetR, tetracycline-resistant
Acknowledgments
Z.C.R. was supported by the Tri-Counties Blood Bank Postdoctoral Fellowship. This work was supported by grants AI114261 (F.H.Y.) and GM102318 (D.A.L & C.S.H.) from the National Institutes of Health.
References
- Aiyar A, Xiang Y, Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol. 1996;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 2012;8:e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 2014;10:e1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact- dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol. 2009;191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J, Schipper K, van Ulsen P, van der Ende A, Tommassen J. Domain exchange at the 3′ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics. 2013;14:622. doi: 10.1186/1471-2164-14-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balder R, Hassel J, Lipski S, Lafontaine ER. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect Immun. 2007;75:2765–2775. doi: 10.1128/IAI.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud C, Guerin J, Petit E, Lesne E, Dupre E, Locht C, Jacob-Dubuisson F. Translocation path of a substrate protein through its Omp85 transporter. Nat Commun. 2014;5:5271. doi: 10.1038/ncomms6271. [DOI] [PubMed] [Google Scholar]
- Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith JR, Signer ER. Transposition of the lac region of Escherichia coli. I. Inversion of the lac operon and transduction of lac by phi80. J Mol Biol. 1966;19:254–265. doi: 10.1016/s0022-2836(66)80003-7. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol. 2008;74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Dawkins R. The Selfish Gene. Oxford Univ. Press; Oxford: 1976. p. 224. [Google Scholar]
- Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev. 2012;26:515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrler WT, Raetz CR. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem. 2005;280:27679–27687. doi: 10.1074/jbc.M504796200. [DOI] [PubMed] [Google Scholar]
- Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol. 2013 doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard V, Cote JP, Charbonneau ME, Campos M, Berthiaume F, Hancock MA, Siddiqui N, Mourez M. Conformation change in a self-recognizing autotransporter modulates bacterial cell-cell interaction. J Biol Chem. 2010;285:10616–10626. doi: 10.1074/jbc.M109.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4:e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhabert MR, Kirkpatrick BC. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005;18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Irie Y, Mattoo S, Yuk MH. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J Bacteriol. 2004;186:5692–5698. doi: 10.1128/JB.186.17.5692-5698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi Y, Claus S, Relman DA. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte CR3 (CD11b/CD18). J Exp Med. 1994;180:1225–1233. doi: 10.1084/jem.180.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. Depolymerization of beta- 1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Guerin J, Baelen S, Clantin B. Two-partner secretion: as simple as it sounds? Res Microbiol. 2013;164:583–595. doi: 10.1016/j.resmic.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Garza-Sanchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, Hayes CS, Low DA. Genetic analysis of the CDI Pathway from Burkholderia pseudomallei 1026b. PLoS One. 2015;10:e0120265. doi: 10.1371/journal.pone.0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Garza-Sanchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, Andersson DI, Hayes CS, Low DA. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet. 2014;10:e1004255. doi: 10.1371/journal.pgen.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C, Bertin P, Menozzi FD, Renauld G. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol Microbiol. 1993;9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi FD, Boucher PE, Riveau G, Gantiez C, Locht C. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun. 1994;62:4261–4269. doi: 10.1128/iai.62.10.4261-4269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil RB, Apicella MA. Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect Immun. 2009;77:2285–2293. doi: 10.1128/IAI.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm- dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D, Tuomanen E, Falkow S, Golenbock DT, Saukkonen K, Wright SD. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell. 1990;61:1375–1382. doi: 10.1016/0092-8674(90)90701-f. [DOI] [PubMed] [Google Scholar]
- Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A. 2002;99:13142–13147. doi: 10.1073/pnas.202358699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013a;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Nguyen JY, Beck CM, Low DA, Hayes CS. The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol. 2014;94:466–481. doi: 10.1111/mmi.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact- dependent growth inhibition to members of the same species. mBio. 2013b;4:e00480–00413. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, Turner D, Boesl M, Abele M, Frosch M, Kurzai O. A functional two-partner secretion system contributes to adhesion of Neisseria meningitidis to epithelial cells. J Bacteriol. 2007;189:7968–7976. doi: 10.1128/JB.00851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, Yantorno OM, Deora R. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One. 2011;6:e28811. doi: 10.1371/journal.pone.0028811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, Allison HE. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol. 2007;189:7223–7233. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Tala A, Progida C, De Stefano M, Cogli L, Spinosa MR, Bucci C, Alifano P. The HrpB HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol. 2008;10:2461–2482. doi: 10.1111/j.1462-5822.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. Recombineering: genetic engineering in bacteria using homologous recombination. Current protocols in molecular biology / edited by Frederick M. Ausubel. 2007 doi: 10.1002/0471142727.mb0116s78. [et al Chapter 1: Unit 1 16. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Walker D, Lancaster L, James R, Kleanthous C. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 2004;13:1603–1611. doi: 10.1110/ps.04658504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS ONE. 2013;8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- West SA, Gardner A. Altruism, spite, and greenbeards. Science. 2010;327:1341–1344. doi: 10.1126/science.1178332. [DOI] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.