Abstract
The structure and dynamics of the stem-loop transactivation response element (TAR) RNA from the human immunodeficiency virus type-1 (HIV-1) bound to the ligand argininamide (ARG) has been characterized using a combination of a large number of residual dipolar couplings (RDCs) and trans-hydrogen bond NMR methodology. Binding of ARG to TAR changes the average inter-helical angle between the two stems from ~47° in the free state to ~11° in the bound state, and leads to the arrest of large amplitude (±46°) inter-helical motions observed previously in the free state. While the global structural dynamics of TAR–ARG is similar to that previously reported for TAR bound to Mg2+, there are substantial differences in the hydrogen bond alignment of bulge and neighboring residues. Based on a novel H5(C5)NN experiment for probing hydrogen-mediated 2hJ(N,N) scalar couplings as well as measured RDCs, the TAR–ARG complex is stabilized by a U38-A27·U23 base-triple involving an A27·U23 reverse Hoogsteen hydrogen bond alignment as well as by a A22-U40 Watson–Crick base-pair at the junction of stem I. These hydrogen bond alignments are not observed in either the free or Mg2+ bound forms of TAR. The combined conformational analysis of TAR under three states reveals that ligands and divalent ions can stabilize similar RNA global conformations through distinct interactions involving different hydrogen bond alignments in the RNA.
Keywords: recognition, adaptation, collective motions, NMR, residual dipolar couplings
There is now ample evidence indicating that many RNAs do not fold into a single well-defined structure, but rather exist as an ensemble of interconverting conformations.1-4 The distribution of RNA conformations is often governed by the hierarchical nature of secondary and tertiary contacts in this highly modular biomolecule.5 Thus, while autonomously folding secondary structural elements such as helices are often formed, weak, scarce, and often metal ion-dependent tertiary contacts can render their relative alignment and hence RNA global conformation more subject to equilibrium fluctuations. This RNA global plasticity is often invoked to explain heterogeneous reaction trajectories that are frequently observed during catalysis6-8 and protein recognition.2,4,9 An emerging theme is that the functionally active conformation may not always be the most populated in solution. Rather, transiently populated conformational sub-states may be captured during protein recognition,2,4,9 and stabilized by binding to divalent ions during catalysis.6-8
To date, structural insight into the dynamical behavior of RNA global conformation has been hindered in part by limitations in techniques for probing rigid-body motions. Such collective motions generally occur at time-scales (microseconds–milliseconds) that are inaccessible to traditional techniques such as molecular dynamics simulations10 and NMR spin relaxation.11,12 Although NMR relaxation dispersion can be used to probe slow motional fluctuations,13 these methods provide limited information about motional amplitudes and directions, rendering it difficult to determine the structural characteristics of conformational sub-states. A similar problem is encountered with fluorescence resonance energy transfer (FRET),14 which although can be used to probe changes in RNA global conformation,15,16 provides limited structural resolution regarding the conformational sub-states.
Recent developments in techniques for partially aligning biomolecules under solution conditions have enabled measurements of NMR residual dipolar couplings (RDCs) as a new probe of biomolecular structure and dynamics.17,18 By providing long-range constraints on bond vector orientation,19,20 the measurement of RDCs has tremendously enhanced the accuracy and precision with which extended structures such as nucleic acids can be determined by NMR.21-23 By being uniquely sensitive to motional averaging over a wide window of time scales (picoseconds–milliseconds), the measurement of RDCs has also emerged as a powerful approach for probing the amplitudes and directions of collective motions in biomolecules.24-27
We recently employed RDCs to investigate the conformational dynamics of human immunodefiency virus type 1 (HIV-1) transactivation response element (TAR) RNA (Figure 1(a)).28,29 The TAR domain has been the subject of numerous investigations because its interaction with the transactivator protein (Tat) is critical for HIV-1 viral replication,30 rendering it a potential target for therapeutic development.31 Previous structural studies had established that binding to peptide mimics of Tat, including the ligand argininamide (ARG), induces a change in the TAR global conformation32-37 from a bent to coaxial interhelical alignment. However little information was available regarding global motions in TAR and its potential role in recognition. Our RDC-NMR study of TAR in the divalent ion free state (TAR-FREE)28 provided evidence that the two helices undergo large amplitude (±46°) rigid-body collective motions about an average inter-helical angle of 47°. Because the coaxially aligned bound TAR conformations appeared to be dynamically accessible in the free state, these results were interpreted as evidence that TAR complexation may proceed via tertiary capture of transiently accessible conformations. More recently, we demonstrated using RDC NMR methodology that binding of Mg2+ to TAR (TAR-Mg) leads to coaxial alignment of the two stems and a solution conformation that is similar to its X-ray structure counterpart determined previously in the presence of divalent ions,38 and the complete quenching of inter-helical motions.29
Figure 1.
(a) Secondary structure of the HIV-1 TAR analogue used here. The six-residue hairpin loop in wt-TAR (CUGGGA) has been replaced with the more stable tetraloop (UUCG). Yellow boxes indicate residues having the largest ARG-induced 1H chemical shift perturbations [(Δδ(ppm) δTAR(Mg) − δTAR(free)) >0.1 ppm]. (b) and (c) Comparison of C–H and N–H splittings measured using two frequency-based experiments in TAR–ARG in the absence (b) and presence (c) of 25 mg/ml Pf1 phage ordering media.42,43 The two experiments yield splittings in either the direct (1H) or indirect (13C) dimensions with the exception of C5–H5 for which independent splitting measurements were performed using J-scaled experiments in the indirect dimension (13C). (d) and (e) The global conformation and dynamics of HIV-1 TAR–ARG derived from order matrix analysis of RDCs44,45 and comparison with previous results for TAR-FREE28 and TAR-Mg.29 (d) The average RDC-derived inter-helical conformation is shown for TAR–ARG, TAR-FREE28 and TAR-Mg.29 Stems I and II are colored red and blue, respectively, with the helix axes depicted using arrows and the interhelical angles indicated on the structures. Stem II is superimposed in all structures to highlight differences in the global conformation. (e) Comparison of the internal generalized degree of order (ϑint = ϑstemI/ϑstemII) providing a measure of inter-helical motions in TAR–ARG, TAR-FREE28 and TAR-Mg.29 Uniformly 15N/13C-labeled HIV-1 TAR RNA was prepared using in vitro T7 RNA polymerase transcription reactions. NMR samples contained ~1.0 mM 15N/13C-labeled TAR, 15 mM sodium phosphate (pH 6.0–6.2), 25 mM sodium sulfate, 0.1 mM EDTA and 5 mM l-argininamide dihydrochloride (ARG) at pH 6.0–6.2. A second identical NMR sample was prepared which also contained ~25 mg/ml Pf1 phage42,43 yielding a deuterium line splitting of ~25 Hz. NMR experiments were recorded on Varian Inova 600 MHz instruments at 25 °C. One-bond C2–H2, C6–H6, C8–H8, C1′–H1′ and N–H splittings were measured using a modified, real-time version of the S3E and S3CT TROSY IPAP pulse sequences50 in both the direct (ω2, 1H) and indirect (ω1, 13C) dimension. Two independent measurements for C5–H5 splittings were obtained in the indirect (13C) dimension using J-scaled methodology, in which two spectra were recorded: one containing the narrow TROSY component of the C5–H5 doublet, and one containing the same component, but with the C5–H5 splitting enhanced by a factor of 0.5. Duplicate measurements were obtained for 90% of the total measurable peaks. Splittings between C4–C5, C5–C6 and C1′–C2′ nuclei were measured using 13C–13C homonuclear IPAP type of experiments (unpublished results). In all cases, RDCs were calculated as the difference between splittings measured in the presence (J + D) and absence (J) of phage. Stem-specific order tensor parameters were calculated by independently subjecting RDCs measured in stems I and II to an order matrix analysis using the program ORDERTEN_SVD.45 Idealized A-form geometries were used as input coordinates for two stems. Residues (G17 and C45) in stem I were excluded from this analysis to avoid potential complications arising from local fluctuations at the terminus. Results and statistics for the order tensor analysis are summarized in Table 1.
To further explore how the global structure and dynamics of TAR changes in response to recognition, we employed RDCs along with trans-hydrogen bond NMR methodology39-41 to investigate the conformation of TAR bound to ARG. Binding of TAR to ARG was first examined by recording 2D 13C–1H correlation spectra of uniformly 13C-/15N- labeled TAR with increasing ARG concentration. Monotonical changes in chemical shifts along both the direct (1H) and indirect (13C) dimensions were observed, indicating rapid exchange (106–103 s−1) between free and ARG-bound TAR states. The largest chemical shift perturbations (>0.1 ppm in the 1H dimension) were observed for residues A22, U23, C24, U25, G26, G28, C39, U40, and C41, indicating that ARG binds to the bulge and neighboring residues of TAR (Figure 1(a), shown in yellow), in agreement with previous NMR studies of HIV-132,34,37 and HIV-236 TAR–ARG complexes.
To examine how ARG binding affects the global structural dynamics of TAR, RDCs were measured between directly bonded C–H (1DC–H), N–H (1DN–H), and C–C (1DC–C) nuclei using 25 mg/ml of Pf1 phage as an ordering medium.42,43 To estimate the uncertainty in RDC measurements, splittings between C–H and N–H nuclei were measured using two frequency-based approaches. As shown in Figure 1(b) and (c), the agreement between the two sets of measurements is excellent, both in the absence (Figure 1(b)) and presence of phage (Figure 1(c)). The root-mean-square-deviation (RMSD) is substantially smaller than the magnitude of measured RDCs (range between −38.4 Hz and 40.7 Hz) and correlation coefficients R2 are close to ideal (Figure 1(b) and (c)). The average RDC values derived independently from the two experiments were used in subsequent analysis whenever possible, and the RMSD between the two sets of RDC measurements (2.6 Hz) was used as an estimate of the uncertainty in RDCs. A total of 116 RDCs were measured in TAR–ARG, including 18 1DC1′–H1′, 3 1DC2–H2, 16 1DC5–H5, 14 1DC6–H6, 11 1DC8–H8, 16 1DC4–C5, 15 1DC5–C6, 15 1DC1′–C2′, and 8 1DN–H RDCs.
To elucidate the global structure and dynamics of TAR–ARG, RDCs measured in stems I and II were independently subjected to an order matrix analysis44,45 using idealized A-form geometries as input coordinates for the two stems as previously described for TAR-FREE28 and TAR-Mg.29 As shown in Table 1, the measured RDCs are in very good agreement with the local geometry of idealized A-form helices. The RMSD between measured RDCs and values calculated using best-fit order tensors for stems I (3.9 Hz) and stem II (3.5 Hz) are only slightly larger than the uncertainty in RDC measurements (2.6 Hz). This argues that like in the case of TAR-FREE28 and TAR-Mg,29 the local conformation of the two stems in TAR–ARG are accurately modeled using idealized A-form helices.
Table 1.
Order tensor analysis of RDCs measured in stems I and II in the HIV-1 TAR–ARG complex
| N | CN | RMSD (Hz) | R | η | ϑ × 10−3 | ϑ int | |
|---|---|---|---|---|---|---|---|
| Stem I | 32 | 3.7 | 3.9 | 0.98 | 0.33 ± 0.05 | 1.26 ± 0.04 | 0.93 ± 0.07 |
| Stem II | 32 | 3.5 | 3.5 | 0.94 | 0.35 ± 0.03 | 1.36 ± 0.03 |
Shown are the total number of RDCs used in the order tensor calculations (N), the condition number defined as the ratio of largest to smallest singular value in singular value decomposition (CN), the root-mean-square-deviation (RMSD) and correlation coefficient (R) between measured RDCs and values calculated using the best-fit order tensors, the asymmetry parameter (η = |(Syy−Sxx)/Szz|), the generalized degree of order (), and internal generalized degree of order (ϑint = ϑi/ϑj; ϑi > ϑj). Parameters are reported using idealized A-form helices as input coordinates for the two stems.
Shown in Figure 1(d) is the relative orientation of stems I (in red) and II (in blue) in TAR–ARG determined by superimposing stem-centered order tensor frames derived from the order tensor analysis. For comparison, inter-helical conformation of TAR-FREE28 and TAR-Mg29 derived using an identical RDC-based procedure is also shown, with stem II superimposed in all three structures. The average inter-helical angle in TAR–ARG is 11.2°, and based on the uncertainly in the orientational solutions, can range between ~8° and ~14°. Despite a small departure from perfect coaxial alignment, the relative twisting between the two helices is as would be expected from a continuous A-form helix. The global TAR–ARG conformation determined here using RDCs is in good agreement with previous NMR structures of HIV-1 and HIV-2 TAR bound to ARG and other peptide mimics of Tat.32,34,36,37
Our results, however, argue that ARG binding also affects the global flexibility of the TAR conformation. The order tensor analysis yields two additional stem-specific principal order parameters describing the degree (ϑ)25 and asymmetry (η) of alignment. While these parameters should be identical for rigidly linked helices, inter-helical motions can lead to variations dependent on motional amplitudes and directions.25 In the previous study of TAR-FREE,28 the ϑ value for stem I was significantly smaller than values measured in stem II (ϑint = ϑI/ϑII = 0.59(±0.06)), indicating that the two stems undergo substantial motions relative to one another (Figure 1(e)). We have recently obtained independent support for such large amplitude global motions in TAR-FREE using direct magnetic field magnetic-induced RDCs.46 Remarkably, the stem-specific ϑ values (Table 1) determined here for TAR–ARG are in far better agreement (ϑstemI = (1.26 ± 0.04) × 10−3 and ϑstemII = (1.36 ± 0.03) × 10−3). The resulting ϑint value of 0.93(± 0.07) (Table 1) approaches unity, consistent with a rigid inter-helical conformation (Figure 1(e)). Hence, as is the case for Mg2+ (Figure 1(e)), ARG binding dramatically stabilizes the global conformation of TAR.
The observed ARG-induced modulation in the global conformational dynamics of TAR must in part originate from rearrangements in the structure and dynamics of bulge and neighboring residues connecting the two stems. Although the original NMR structure of the HIV-1 TAR–ARG complex argued for formation of a U38-A27·U23 base-triple involving a A27·U23 reverse Hoogsteen hydrogen bond alignment32 (Figure 1(a)), this has been challenged by a more recent NMR structure of the HIV-1 TAR bound to ARG and Tat-derived peptides in which remote placement of residues U23 and A27 precludes reverse Hoogsteen hydrogen bond alignment.34 While base-triple formation was demonstrated decisively in an HIV-2 TAR–ARG complex47 (compared to HIV-1 TAR, HIV-2 TAR has a two-nucleotide bulge and lacks residue C24) using direct trans-hydrogen bond NMR methodology,39-41 similar evidence has yet to be established for the HIV-1 TAR–ARG complex.
To determine whether the U38-A27·U23 base-triple forms in the HIV-1 TAR–ARG complex, we employed trans-hydrogen bond NMR methodology.39-41 Direct evidence for the U38-A27·U23 base-triple could in principle be obtained using the traditional 2JHN HNN-COSY experiment.39,40 However, in none of the TAR systems studied thus far has the imino H3 proton of U23 been observable. Here, we have employed a novel H5(C5)NN experiment which correlates the non-exchangeable H5 protons of the donor uridine with the nitrogen of the acceptor base (N1 or N7 of adenine) (Figure 2(a)). The magnetization transfer pathway is depicted in Figure 2(a) for the base-triple. Compared to other experiments for indirect means of hydrogen bond detection, including an adaptation of the J(N,N)-HNN correlated spectroscopy (COSY) experiment used for detecting base-triple formation in HIV-2–ARG complex,47 the H5(C5)NN experiment yields spectra that are simpler in appearance, consisting exclusively of (H5,N3)(U) and (H5,NA) cross-peaks. Details of the H5(C5)NN experiment are provided in the legend to Figure 2(a).
Figure 2.
Shown in Figure 2(b) are regions of the H5(C5)N3N spectrum recorded on TAR–ARG, TAR-FREE and TAR-Mg. In all cases, cross-peaks are observed between H5(U42, stem I)-N1(A20, stem I) and H5(U38, stem II)-N1(A27, stem II) corresponding to the A20-U42 and A27-U38 Watson–Crick hydrogen bond alignment in stems I and II, respectively. However, two additional cross-peaks are observed in TAR–ARG that are not observed in either of TAR-FREE or TAR-Mg. One of the cross-peaks is between H5(U23, bulge) and N7(A27, stem II), and corresponds to an A27·U23 reverse Hoogsteen hydrogen bond alignment. This constitutes direct evidence for U38-A27·U23 base-triple formation in the HIV-1 TAR–ARG complex. The second cross-peak is anomalously weaker in intensity and corresponds to the A22-U40 Watson–Crick base-pair at the junction of stem I (Figure 2(b), in red). Previous NMR studies have reported nuclear Overhauser effect (NOE) evidence for transient A22-U40 hydrogen bonding in HIV-1 TAR complexes,34,35,37 though unambiguous interpretation was difficult based on these indirect lines of evidence. Our results indicate that the A22-U40 hydrogen bond is indeed formed in TAR–ARG, but the weaker intensity of the trans-hydrogen bond cross-peak argues that it may either be transiently disrupted and/or statically departing from an idealized Watson–Crick alignment. Intriguingly, even though both Mg2+ and ARG stabilize similar coaxially aligned TAR global conformations, the hydrogen bond alignment of residues at the interface of the two helices are substantially different in the two cases.
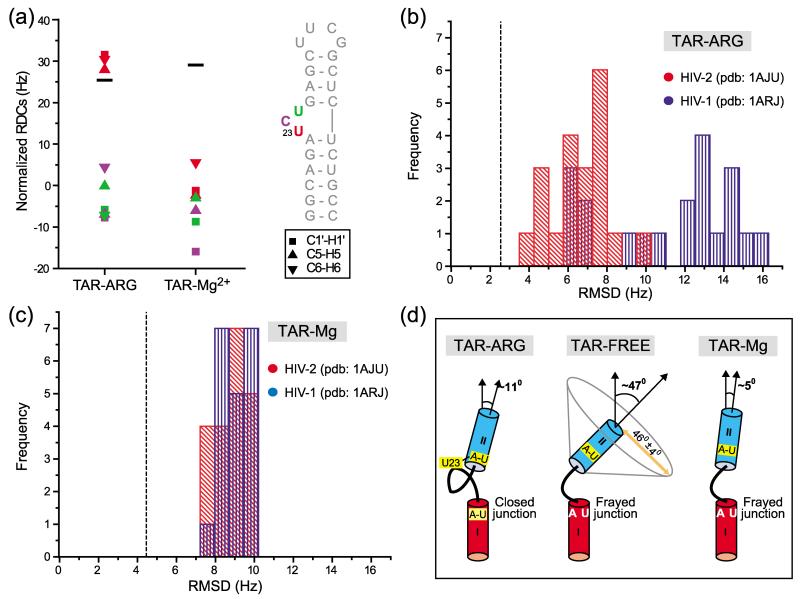
That U23 adopts distinct conformations in TAR–ARG and TAR-Mg could be directly visualized by comparing RDCs measured in this residue. Direct comparison of RDCs is possible in this case because the orientation and asymmetry of the order tensor is very similar in TAR–ARG and TAR-Mg. As shown in Figure 3(a), RDCs previously measured for U23 in TAR-Mg29 are significantly attenuated relative to the average values measured in the base moieties of stem II (Figure 3(a)). This indicates that U23 departs either statically and/or dynamically from an alignment that is coaxial with respect to stem II. In sharp contrast, the RDCs measured in TAR–ARG are substantially larger and similar to counterparts measured in stem II, consistent with a rigid alignment of U23 that is close to coaxial with respect to stem II, as would be expected from an A27(stem II)·U23 (bulge) reverse Hoogsteen hydrogen bond alignment. Smaller variations in RDCs measured in C24 and U25 are also observed, potentially indicating differences in conformation and/or dynamics of these residues in TAR-Mg and TAR–ARG.
Figure 3.
The conformation of the base-triple and neighboring residues probed using RDCs. (a) Comparison of RDCs measured for residues U23, C24 and U25 in TAR–ARG and TAR-Mg. To account for differences in the degree of alignment of TAR–ARG and TAR-Mg, RDCs measured in TAR-Mg were normalized by scaling the values by a factor of ϑstemII(TAR–ARG)/ϑstemII(TAR–Mg). Filled black bars indicate the average normalized base 1DC–H values measured in stem II for each of the two states. (b) and (c) RDC-based structure validation of 40 NMR models for residues U23, A27 and U38 derived from previous NMR structures of HIV-1 TAR–ARG and HIV-236 TAR–ARG. Shown are histogram plots of RMSDs between RDCs measured in (b) HIV-1 TAR–ARG and (c) HIV-1 TAR-Mg and values calculated using the best-fit order tensor. Results for HIV-1 TAR–ARG (PDB code 1ARJ)34 and HIV-2 TAR–ARG36 (PDB code 1AJU) are shown in blue and red, respectively. The vertical broken line corresponds to the estimated uncertainty in RDCs. A total of 17 and 13 RDCs measured in residues U23, A27, U38 in TAR–ARG and TAR-Mg, respectively, were independently subjected to order matrix analysis using the program ORDERTEN_SVD using 40 previous NOE-based NMR structures of HIV-1 (1ARJ) and HIV-2 (1AJU)36 TAR–ARG complex as input coordinates.
We were able to gain additional structural support for having a base-triple in TAR–ARG and not in TAR-Mg using an RDC-based structure validation approach. Here, coordinates for residues U23, A27 and U38 derived from NMR structures of HIV-2 TAR–ARG36 (1AJU; U23 and A27 within hydrogen bonding distance) and HIV-1 TAR–ARG34 (1ARJ; large distance between U23 and A27 precludes hydrogen bonding) were challenged for agreement with 17 RDCs measured for these residues in TAR–ARG. As shown in Figure 3(b), the RMSDs between measured RDCs and values calculated using the best-fit order tensor are substantially smaller for input coordinates derived from 1AJU (Figure 3(b), red solutions, average RMSD = 6.5 Hz) compared to 1ARJ (Figure 3(b), blue solutions, average RMSD = 11.3 Hz). Significantly, the ϑ value of (1.33 ± 0.16) × 10−3 determined for the best-fit base-triple conformation (1AJU model 11) is in excellent agreement with values determined for stems I ((1.26 ± 0.04) × 10−3) and II ((1.36 ± 0.03) × 10−3). This argues that the base-triple including bulge residue U23 is rigid in TAR–ARG, in agreement with a recent NMR spin relaxation study of HIV-2 TAR–ARG complex48 and electron paramagnetic resonance (EPR) studies of HIV-1 TAR.49 The above analysis was repeated using 13 RDCs previously measured in TAR-Mg.29 As shown in Figure 3(c), despite the smaller number of RDCs, large RMSDs are observed for both 1AJU (Figure 3(c), red solutions, average RMSD = 9.1) and 1ARJ (Figure 3(c), blue solutions, average RMSD = 8.9 Hz). Hence, while residues U23, A27 and U38 adopt a conformation that favors base-triple formation in TAR–ARG, this is not the case for TAR-Mg.
We have demonstrated that ARG binding to TAR leads to the arrest of global inter-helical motions and stabilization of a single global inter-helical alignment that falls within the envelope of conformations that appear to be dynamically accessible in TAR-FREE (Figure 3(d)). This result supports the notion that TAR–ARG complex formation may proceed via tertiary capture of a transiently accessible state having preformed recognition elements. The comparison of TAR–ARG with TAR-Mg29 is also illuminating. Although the global structural dynamics of TAR–ARG and TAR-Mg are very similar, there are substantial differences in the local conformation of bulge and neighboring residues. Besides the U38-A27·U23 base-triple in TAR–ARG, which is not observed in either TAR-Mg or TAR-FREE, an additional A22-U40 Watson–Crick hydrogen bond is uniquely observed in TAR–ARG. Based on previous NOE structures of the TAR–ARG complex,32,34,36 the guanadinium moiety of ARG is sandwiched between residues U23 and A22. Such stacking interactions involving residues at the interface of the two helices likely promote formation of the A27·U23 and A22-U40 hydrogen bond alignments and also provide a basis for stabilizing the TAR global conformation. In contrast, based on a previous X-ray structure of TAR bound to divalent ions,38 stabilization of TAR’s global conformation by Mg2+ likely occurs in part through a network of inner- and outer-sphere interactions between divalent ions and RNA ligands located throughout the inter-helical interface. Hence, ligands and divalent ions can stabilize similar RNA global conformations through distinct mechanisms that give rise to different local hydrogen bond alignments in the RNA.
Supplementary Material
Acknowledgements
This research was funded by CA46778 to D.J.P. H.M.A. acknowledges start-up funds from the University of Michigan.
Abbreviations used
- HIV
human immunodeficiency virus
- TAR
transactivation response element
- ARG
argininamide
- RDC
residual dipolar coupling
- RMSD
root-mean-square deviation
Footnotes
Supplementary data associated with this article can be found at doi: 10.1016/j.jmb.2004.02.031
Supplementary Material comprising one Figure is available on Science Direct
References
- 1.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson JR. Induced fit in RNA-protein recognition. Nature Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 3.Woodson SA. Compact but disordered states of RNA. Nature Struct. Biol. 2000;7:349–352. doi: 10.1038/75106. [DOI] [PubMed] [Google Scholar]
- 4.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 5.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 6.Scott WG. RNA structure, metal ions, and catalysis. Curr. Opin. Chem. Biol. 1999;3:705–709. doi: 10.1016/s1367-5931(99)00029-0. [DOI] [PubMed] [Google Scholar]
- 7.Hanna R, Doudna JA. Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol. 2000;4:166–170. doi: 10.1016/s1367-5931(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 8.DeRose VJ. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]
- 9.Patel DJ. Adaptive recognition in RNA complexes with peptides and protein modules. Curr. Opin. Struct. Biol. 1999;9:74–87. doi: 10.1016/s0959-440x(99)80010-4. [DOI] [PubMed] [Google Scholar]
- 10.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nature Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 11.Kay LE. Protein dynamics from NMR. Nature Struct. Biol. 1998;5:513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]
- 12.Ishima R, Torchia DA. Protein dynamics from NMR. Nature Struct. Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 13.Palmer AG, III, Kroenke CD, Loria LP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 14.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 15.Lilley DM, Wilson TJ. Fluorescence resonance energy transfer as a structural tool for nucleic acids. Curr. Opin. Chem. Biol. 2000;4:507–517. doi: 10.1016/s1367-5931(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 16.Walter NG, Harris DA, Pereira MJ, Rueda D. In the fluorescent spotlight: global and local conformational changes of small catalytic RNAs. Biopolymers. 2001;61:224–242. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 17.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins—information for structure determination in solution. Proc. Natl Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 19.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Quart. Rev. Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 20.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 21.Mollova ET, Pardi A. NMR solution structure determination of RNAs. Curr. Opin. Struct. Biol. 2000;10:298–302. doi: 10.1016/s0959-440x(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald D, Lu P. Residual dipolar couplings in nucleic acid structure determination. Curr. Opin. Struct. Biol. 2002;12:337–343. doi: 10.1016/s0959-440x(02)00328-7. [DOI] [PubMed] [Google Scholar]
- 23.Al-Hashimi HM, Gorin A. Application of NMR residual dipolar couplings in studies of RNA: towards high throughput investigations. Appl. Genom. Proteomic. 2003;2:3–16. [Google Scholar]
- 24.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nature Struct. Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 25.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J. Am. Chem. Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 26.Meiler J, Peti W, Griesinger C. Dipolar couplings in multiple alignments suggest alpha helical motion in ubiquitin. J. Am. Chem. Soc. 2003;125:8072–8073. doi: 10.1021/ja029816l. [DOI] [PubMed] [Google Scholar]
- 27.Tolman JR, Al-Hashimi HM. NMR studies of biomolecular dynamics and structural plasticity using residual dipolar couplings. Annu. Rep. NMR Specrosc. 2003;51:105–166. [Google Scholar]
- 28.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 29.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen BR. Transactivation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 31.Froeyen M, Herdewijn P. RNA as a target for drug design, the example of Tat-TAR interaction. Curr. Top. Med. Chem. 2002;2:1123–1145. doi: 10.2174/1568026023393200. [DOI] [PubMed] [Google Scholar]
- 32.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 33.Zacharias M, Hagerman PJ. The bend in RNA created by the transactivation response element bulge of human-immunodeficiency-virus is straightened by arginine and by Tat-derived peptide. Proc. Natl Acad. Sci. USA. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aboul-ela F, Karn J, Varani G. The structure of the human-immunodeficiency-virus type-1 Tar RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 35.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucl. Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodsky AS, Williamson JR. Solution structure of the HIV-2 TAR-argininamide complex. J. Mol. Biol. 1997;267:624–639. doi: 10.1006/jmbi.1996.0879. [DOI] [PubMed] [Google Scholar]
- 37.Long KS, Crothers DM. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry. 1999;38:10059–10069. doi: 10.1021/bi990590h. [DOI] [PubMed] [Google Scholar]
- 38.Ippolito JA, Steitz TA. A 1.3-angstrom resolution crystal structure of the HIV-1 transactivation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl Acad. Sci. USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 40.Pervushin K, Ono A, Fernandez C, Szyperski T, Kainosho M, Wuthrich K. NMR scaler couplings across Watson–Crick base pair hydrogen bonds in DNA observed by transverse relaxation optimized spectroscopy. Proc. Natl Acad. Sci. USA. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumdar A, Patel DJ. Identifying hydrogen bond alignments in multistranded DNA architectures by NMR. Accts Chem. Res. 2002;35:1–11. doi: 10.1021/ar010097+. [DOI] [PubMed] [Google Scholar]
- 42.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 43.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc. 1998;120:10571–10572. [Google Scholar]
- 44.Saupe A. Recent results in the field of liquid crystals. Angew. Chem., Int. Ed. Engl. 1968;7:97–112. [Google Scholar]
- 45.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J. Magn. Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. Probing motions between equivalent RNA domains using magnetic field induced residual dipolar couplings: accounting for correlations between motions and alignment. J. Am. Chem. Soc. 2003;125:10530–10531. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]
- 47.Hennig M, Williamson JR. Detection of N–H…N hydrogen bonding in RNA via scalar couplings in the absence of observable imino proton resonances. Nucl. Acids Res. 2000;28:1585–1593. doi: 10.1093/nar/28.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayie KT, Brodsky AS, Williamson JR. Base Flexibility in HIV-2 TAR RNA Mapped by Solution (15)N, (13)C NMR Relaxation. J. Mol. Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 49.Edwards TE, Okonogi TM, Sigurdsson ST. Investigation of RNA-protein and RNA-metal ion interactions by electron paramagnetic resonance spectroscopy. The HIV TAR-Tat motif. Chem. Biol. 2002;9:699–706. doi: 10.1016/s1074-5521(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 50.Meissner A, Sorensen OW. The role of coherence transfer efficiency in design of TROSY-type multidimensional NMR experiments. J. Magn. Reson. 1999;139:439–442. doi: 10.1006/jmre.1999.1788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.