Abstract
Using the components of a particularly well-studied plant virus, cowpea chlorotic mottle virus (CCMV), we demonstrate the synthesis of virus-like particles (VLPs) with one end of the packaged RNA extending out of the capsid and into the surrounding solution. This construct breaks the otherwise perfect symmetry of the capsid and provides a straightforward route for monofunctionalizing VLPs using the principles of DNA nanotechnology. It also allows physical manipulation of the packaged RNA, a previously inaccessible part of the viral architecture. Our synthesis does not involve covalent chemistry of any kind; rather, we trigger capsid assembly on a scaffold of viral RNA that is hybridized at one end to a complementary DNA strand. Interaction of CCMV capsid protein with this RNA-DNA template leads to selective packaging of the RNA portion into a well-formed capsid, but leaves the hybridized portion poking out of the capsid through a small hole. We show that the RNA-DNA protruding from the capsid is capable of binding DNA and DNA-functionalized colloidal particles. Additionally, we show that the RNA-DNA scaffold can be used to nucleate virus formation on a DNA-functionalized surface. We believe this self-assembly strategy can be adapted to viruses other than CCMV.
Graphical abstract
Small RNA viruses consist entirely of genomic RNA packaged inside a one-molecule-thick protective protein capsid (Fig.1). In addition to making up a large fraction of the world’s viral pathogens, small RNA viruses are helping to define new fields of applied science through their use as functional nanoparticles1. For example, they have been exploited as contrast agents for biomedical imaging1–1, as vectors for the delivery of small molecules and genes to cells6–11, and as nanoscale building blocks for the formation of superstructures with unique material, optical and dynamic properties12–17.
Figure 1.
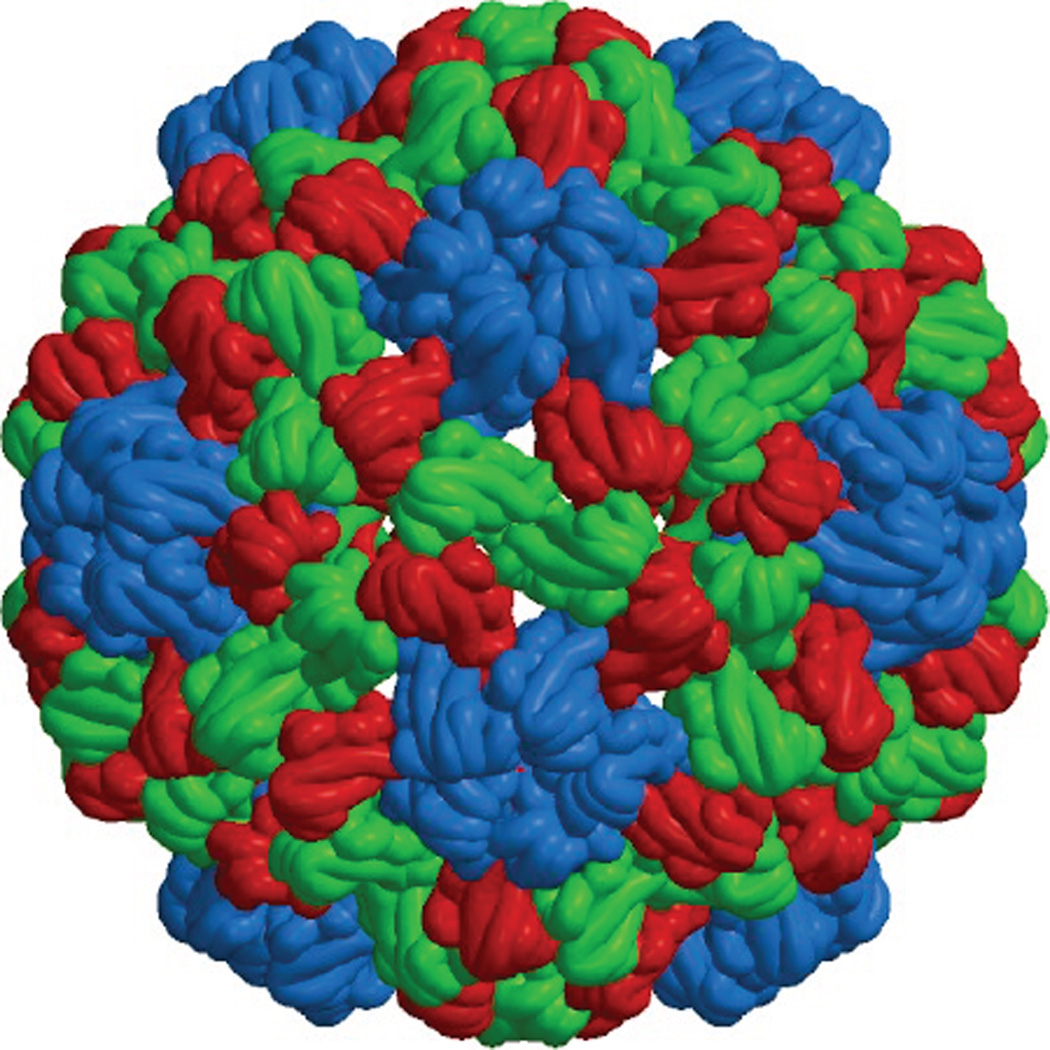
The capsid of cowpea chlorotic mottle virus (CCMV), like many small RNA viruses, has icosahedral symmetry and consists of 180 copies of its capsid protein. Diameter is 28 nm. Taken from VIPERdb18.
Much of the utility of small RNA viruses derives from their symmetric capsids18 which can be engineered to display a high density of functional moieties. Indeed, an arsenal of functionalization strategies20–31 have been developed that combine molecular biology (cloning) and/or selective covalent chemistries to isotropically label the various polyvalent surfaces (exterior, interior, and interfacial1) of the capsid. However, in situations where a high degree of labeling is not desired, monofunctional particles that display only a single copy (or a specific limited arrangement) of a particular functional group are needed. Monofunctional virus particles are difficult to produce32–34 in a controlled way due to the inherent symmetry of the capsid and its abundance of equivalent binding sites.
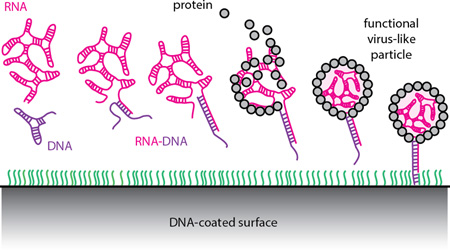
Lying just beneath the capsid, the viral RNA contains a wealth of inequivalent binding sites that could in principle be selectively targeted using the methods of DNA nanotechnology35–40. Unfortunately, the capsid is impermeable these techniques – the main evolutionary purpose of the capsid is to protect the RNA from unfavorable interactions with macromolecules from the outside world. Our work bypasses this inaccessibility through the synthesis of virus-like particles (VLPs) with a portion of one end of the RNA extending outside of the capsid (Fig. 2). With the symmetry of the particle broken by the exposed RNA, we generate robust monofunctionalization through the conjugation of desired moieties using only Watson-Crick basepairing.
Figure 2.
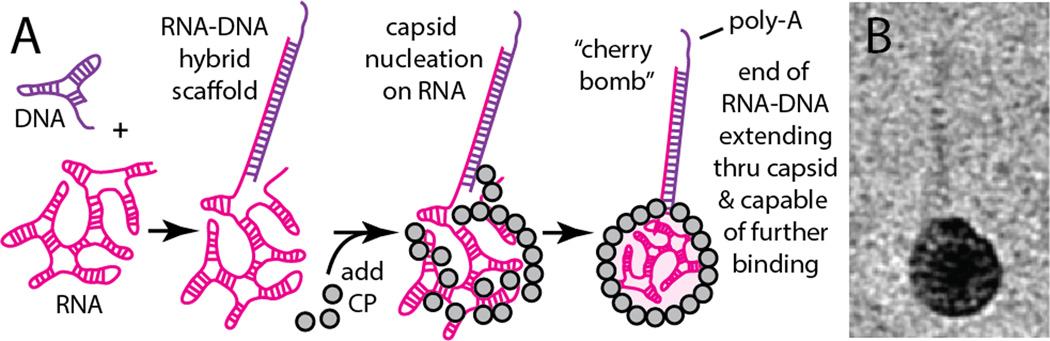
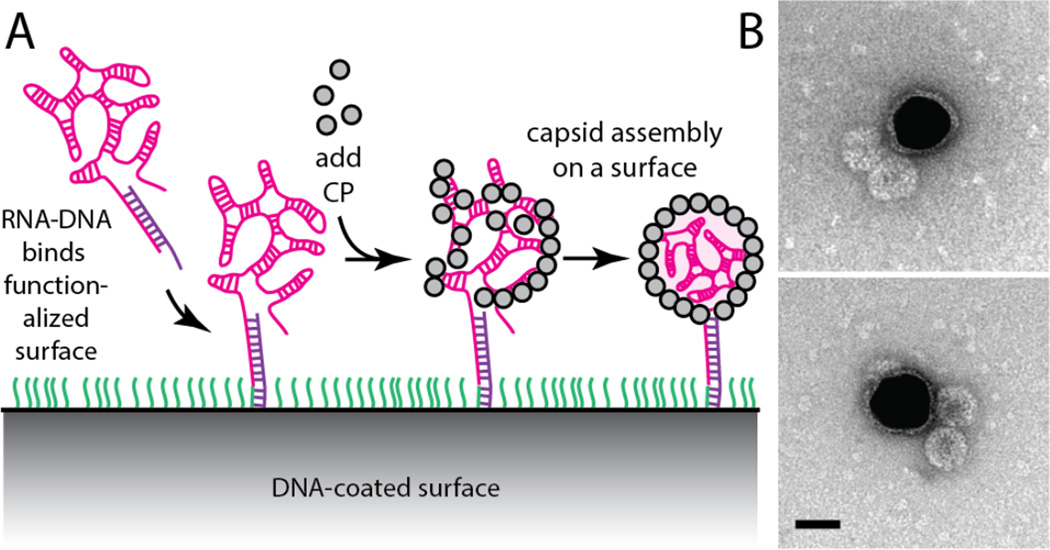
(A) Schematic illustration of assembly of the “cherry bomb”. (B) Positive-stain transmission electron micrograph (TEM) of a cherry bomb capsid (dark sphere measuring 26 nm) and its RNA-DNA appendage (lighter strand extending upward).
Our synthesis (Fig. 2A) does not require genetic modification nor covalent chemistry, but instead relies on the ability of a particularly well-studied small RNA virus, CCMV, to be disassembled and reconstituted by self-assembly in vitro41. Additionally, we exploit the qualitative structural differences between single-stranded (ss) and double-stranded (ds) nucleic acid to reshape the viral RNA that templates the assembly. Due to extensive intramolecular base pairing, ss-RNA of the length naturally packaged by CCMV (about 3 kb) is a highly branched, flexible, compact object that has physical dimensions comparable to the capsid interior42 (22-nm-internal diameter). In contrast, the same length (3 kbp) of ds-DNA occupies a much larger volume due to its increased stiffness (~50 nm persistence length) and lack of branching. As a result, ds-DNA longer than about 75 bp cannot be accommodated within the interior volume of the CCMV capsid and does not function as a template for normal capsid assembly43,44.
By hybridizing the first 185 bases at the 5′-end of a 3.2 kb ss-RNA with a complementary ss-DNA strand, we dramatically stiffen the 5′-end of the RNA. [We use the 5′-end of the 3234-base RNA molecule (“B1”) of the tripartite genome of brome mosaic virus (BMV), although either end of any similar-length sequence will likely do.] Here, the DNA strand (see SI) acts as a molecular splint. The resulting RNA-DNA hybrid can be expected to behave as a compact, flexible, branched 3-kb ss-RNA connected to a rigid, linear, 185-base ds-RNA-DNA appendage (see second-from-left cartoon in Figure 2A). The physical length of the ds portion is about 50 nm. It is very stable (85 °C melting temperature) owing to its perfect sequence complementarity. The in vitro packaging of this RNA-DNA hybrid by CCMV capsid protein (CP), using the same protocol developed earlier for pure RNA,45–47 results in the selective encapsidation of the ss-RNA portion and leaves the ds-RNA-DNA appendage poking out of the capsid and into solution. We refer to this final construct as a “cherry bomb” because of its structural resemblance (Figure 2B, additional images shown in SI, Figure S1) to the well-known explosive firework.48
While the structure of the hole that passes the ds-RNA-DNA through the capsid is not known, previous in vitro packaging studies have shown ss and ds nucleic acid traversing the capsids of CCMV and the closely related BMV. We previously observed49 that ss-RNA molecules significantly longer than wild-type are packaged by multiple CCMV capsids (“multiplets”) that each share a portion of the overlong RNA (Figure 3). And a separate study44 found that long ds-DNA is packaged by a contiguous string of many BMV capsids if ss-RNA fragments are also present. In both cases, nucleic acid is shared by connected capsids, passing through one or more holes in each capsid that are too small to be seen by negative-stain transmission electron micrograph (TEM). It is likely that the ds-RNA-DNA appendage of the cherry bomb exits the capsid through a similar hole.
Figure 3.
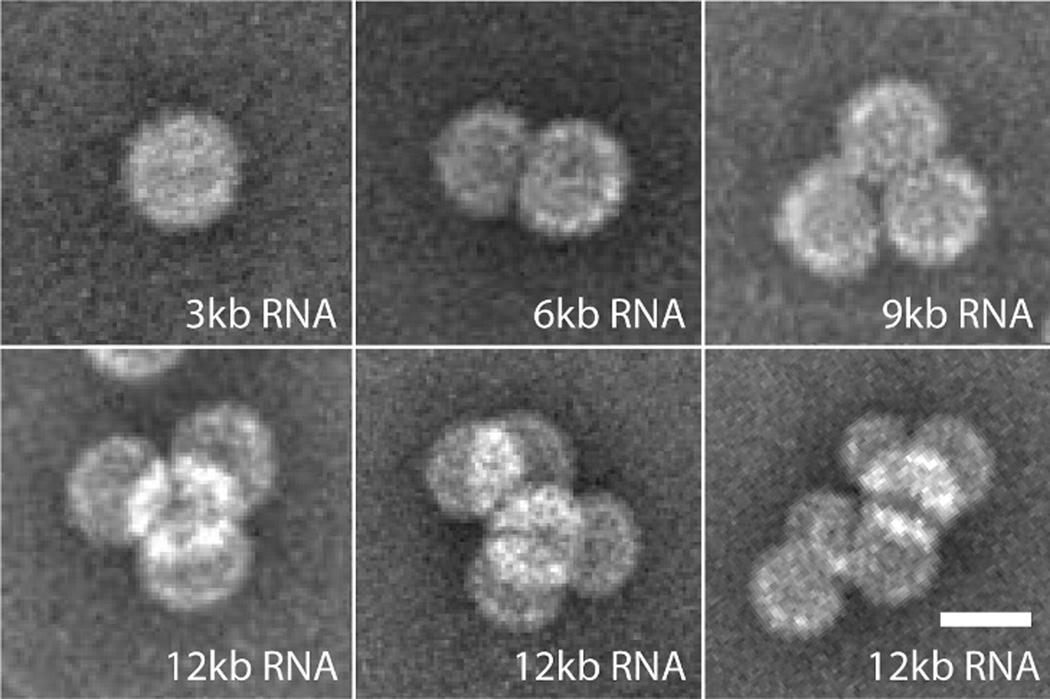
Single ss-RNA molecules progressively longer than wild-type (3 kb) are shared by two or more CCMV capsids; multiplets. Scale bar shows 25 nm. Adapted from J. Virol. 2012, 86, 3322, doi: 10.1128/ JVI.06566-11. Copyright American Society for Microbiology.
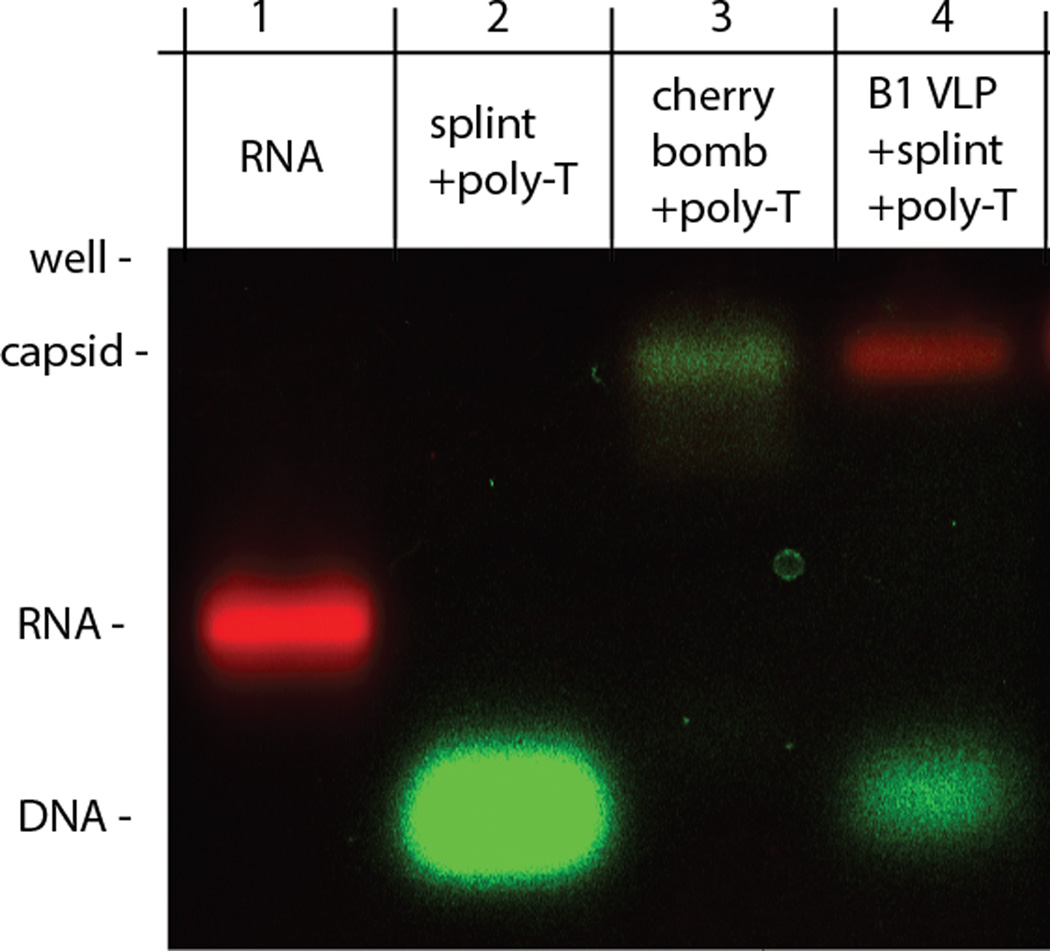
To test whether the exposed RNA-DNA appendage can be used to bind nucleic acids in solution, we designed the DNA splint with a 3’ poly-A15 overhang in addition to the 185 bases that complement the 5’-end of the RNA (see right-most cartoon in Fig. 2A). Once formed, cherry bombs were combined with fluorescent (green) poly-T15 DNA strands and analyzed by native agarose gel electrophoresis (Fig. 4). The capsids co-migrated with the fluorescent poly-T15 (Fig. 4, lane 3), confirming that the poly-A15 sticky end of the RNA-DNA appendage binds its complementary strand in solution. Control experiments in which already-assembled VLPs containing only 3.2 kb RNA (B1) were added to the splint DNA and fluorescent poly-T15 showed no nonspecific binding (Fig. 4, lane 4).
Figure 4.
Native agarose gel electrophoresis shows cherry bombs selectively bind fluorescently labeled poly-T15 DNA strands (green). Fluorescently labeled RNA shown in red.
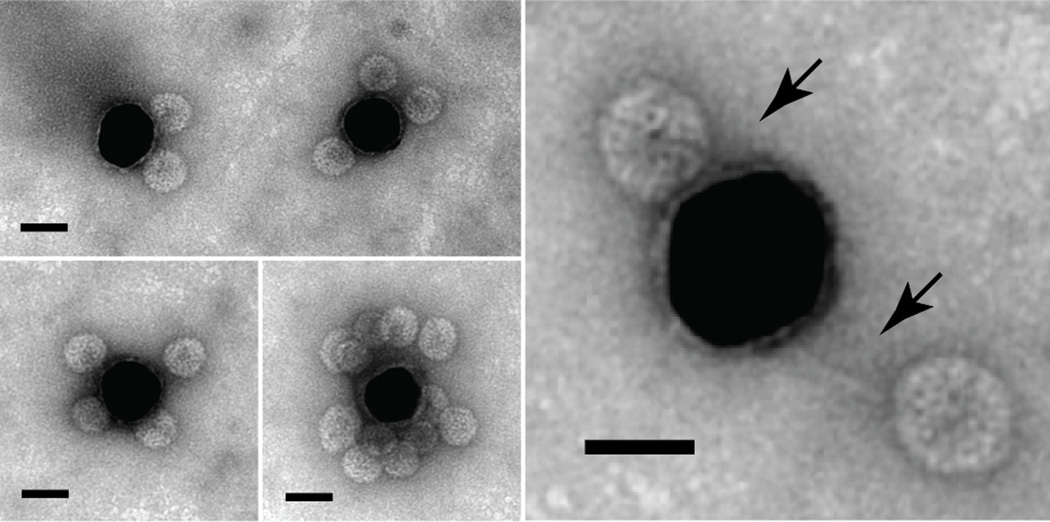
The ability of the cherry bomb to bind a functionalized surface was demonstrated by direct imaging of a mixture of these capsids with 30-nm gold nanoparticles (AuNPs) that had been previously decorated with a high density of poly-T25 DNA strands50 (Fig. S2). Negative-stain TEM (Fig. 5) shows a high concentration of capsids at the AuNP surface, and an exceptionally well-stained image reveals the RNA-DNA appendage linking the capsids to the gold surface (Fig. 5, arrows). Control experiments between B1 VLPs and functionalized AuNPs showed no non-specific binding (Fig. S3).
Figure 5.
Cherry bomb capsids bind the surface of DNA-functionalized 30-nm AuNPs. A zoomed-in image resolves the RNA-DNA duplex (arrows) linking the capsids (light spheres) and the AuNPs (dark sphere). Scale bars show 25 nm.
Separately, we tested whether capsids could be assembled around RNAs that were already tethered to a functionalized surface (Fig. 6A). Here, the hybrid RNA-DNA scaffold was prepared and equilibrated with 30-nm poly-T25-coated AuNPs at a molar ratio of 10:1 (RNA:AuNP). After hybridization of the RNA to the AuNPs, CP was added at a mass ratio of 10:1 (CP:RNA), equilibrated for 5 min on ice, and imaged by negative-stain TEM (Fig. 6B). The presence of well-defined capsids at the Au particle surface demonstrated assembly of cherry bomb capsids around the immobilized RNA-DNA. Some aggregation of CP in the presence of AuNPs was also observed (Fig. S4).
Figure 6.
(A) Schematic showing the assembly of VLPs on a DNA-functionalized (green strands) surface. (B) Cherry bomb capsids (light spheres) were grown on 30-nm AuNPs (dark spheres) as shown in (A). Scale bar shows 25 nm.
While several elegant methods have recently been described for the monofunctionalization of tobacco mosaic virus particles,51–53 they most likely cannot be applied to other viruses; they require either controlled disassembly of one end of the rod-like capsid or self-assembly of capsids on a substrate bound RNA that contains a specific packaging sequence. The homogeneous assembly pathway described here provides a general strategy for monofunctionalizing icosahedral particles. Here we note that the ability to form cherry bomb structures is probably not limited to the plant virus CCMV; multiplet capsids have been observed in the packaging of overlong RNAs by the CP of the bacterial virus fr54 and of the mammalian virus SV40,55 indicating that they too have the potential to form cherry bombs.
In addition to offering a single, highly specific binding modality for building functional viral-based materials, our method for (i) physically binding and manipulating one end of the packaged genome and (ii) nucleating capsid assembly at a surface will enable new single-particle measurements that might reveal how RNA gets into and out of viral capsids during infection. Examples of such measurements include time-resolved studies of capsid assembly and force-pulling experiments56,57 that measure the work required to pull viral RNA out of its capsid.
Supplementary Material
ACKNOWLEDGMENT
WMG and CMK acknowledge support from the NSF in the form of grant CHE 1051507. TEM images were obtained in the California NanoSystems Institute (CNSI) Electron Imaging Center for Nano-Machines, supported by NIH (1S10RR23057). Additional support provided by an NIH training grant for USPHS National Research Service Award 5T32GM008496.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Procedures, data from control experiments, and examples of CP-AuNP aggregation are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Douglas T, Young M. Science. 2006;312(5775):873. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 2.Liepold L, Anderson S, Willits D, Oltrogge L, Frank JA, Douglas T, Young M. Magn. Reson. Med. 2007;58(5):871. doi: 10.1002/mrm.21307. [DOI] [PubMed] [Google Scholar]
- 3.Malyutin AG, Easterday R, Lozovyy Y, Spilotros A, Cheng H, Sanchez-Felix OR, Stein BD, Morgan DG, Svergun DI, Dragnea B, Bronstein LM. Chem. Mater. 2014 [Google Scholar]
- 4.Prasuhn DE, Yeh RM, Obenaus A, Manchester M, Finn MG. Chem. Commun. Camb. Engl. 2007;12:1269. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 5.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. J. Am. Chem. Soc. 2008;130(8):2546. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 6.Aljabali AA, Shukla S, Lomonossoff GP, Steinmetz NF, Evans DJ. Mol. Pharm. 2012;10(1):3. doi: 10.1021/mp3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azizgolshani O, Garmann RF, Cadena-Nava R, Knobler CM, Gelbart WM. Virology. 2013;441(1):12. doi: 10.1016/j.virol.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Chem. Biol. 2007;14(10):1152. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown WL, Mastico RA, Wu M, Heal KG, Adams CJ, Murray JB, Simpson JC, Lord JM, Taylor-Robinson AW, Stockley PG. Intervirology. 2003;45(4-6):371. doi: 10.1159/000067930. [DOI] [PubMed] [Google Scholar]
- 10.Wu M, Brown WL, Stockley PG. Bioconjug. Chem. 1995;6(5):587. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]
- 11.Wu M, Sherwin T, Brown WL, Stockley PG. Nanomedicine Nanotechnol. Biol. Med. 2005;1(1):67. doi: 10.1016/j.nano.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Daniel M-C, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nano Lett. 2006;6(4):611. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 13.Maye MM. Nat. Nanotechnol. 2013;8(1):5. doi: 10.1038/nnano.2012.244. [DOI] [PubMed] [Google Scholar]
- 14.Kostiainen MA, Kasyutich O, Cornelissen JJ, Nolte RJ. Nat. Chem. 2010;2(5):394. doi: 10.1038/nchem.592. [DOI] [PubMed] [Google Scholar]
- 15.Dixit SK, Goicochea NL, Daniel M-C, Murali A, Bronstein L, De M, Stein B, Rotello VM, Kao CC, Dragnea B. Nano Lett. 2006;6(9):1993. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Lin T, Johnson JE, Finn MG. Chem. Biol. 2002;9(7):813. doi: 10.1016/s1074-5521(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 17.Cadena-Nava RD, Hu Y, Garmann RF, Ng B, Zelikin AN, Knobler CM, Gelbart WM. J. Phys. Chem. B. 2011;115(10):2386. doi: 10.1021/jp1094118. [DOI] [PubMed] [Google Scholar]
- 18.Virosphere. http://viperdb.scripps.edu/ [Google Scholar]
- 19.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL, Reddy VS. Nucleic Acids Res. 2009;37(suppl 1):D436. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillitzer E, Willits D, Young M, Douglas T. Chem Commun. 2002;20:2390. doi: 10.1039/b207853h. [DOI] [PubMed] [Google Scholar]
- 21.Blum AS, Soto CM, Wilson CD, Cole JD, Kim M, Gnade B, Chatterji A, Ochoa WF, Lin T, Johnson JE, et al. Nano Lett. 2004;4(5):867. [Google Scholar]
- 22.Strable E, Finn MG. Viruses and Nanotechnology. Springer; 2009. pp. 1–21. [Google Scholar]
- 23.Steinmetz NF, Lomonossoff GP, Evans DJ. Langmuir. 2006;22(8):3488. doi: 10.1021/la060078e. [DOI] [PubMed] [Google Scholar]
- 24.Smith JC, Lee K-B, Wang Q, Finn MG, Johnson JE, Mrksich M, Mirkin CA. Nano Lett. 2003;3(7):883. [Google Scholar]
- 25.Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Biomacromolecules. 2003;4(3):472. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 26.Gupta SS, Raja KS, Kaltgrad E, Strable E, Finn MG. Chem. Commun. 2005;34:4315. doi: 10.1039/b502444g. [DOI] [PubMed] [Google Scholar]
- 27.Pokorski JK, Breitenkamp K, Finn MG. J. Am. Chem. Soc. 2011;133(24):9242. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pokorski JK, Steinmetz NF. Mol. Pharm. 2011;8(1):29. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel KG, Swartz JR. Bioconjug. Chem. 2011;22(3):376. doi: 10.1021/bc100367u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinmetz NF, Lin T, Lomonossoff GP, Johnson JE. Viruses and Nanotechnology. Springer; 2009. pp. 23–58. [DOI] [PubMed] [Google Scholar]
- 31.Young M, Debbie W, Uchida M, Douglas T. Annu Rev Phytopathol. 2008;46:361. doi: 10.1146/annurev.phyto.032508.131939. [DOI] [PubMed] [Google Scholar]
- 32.Klem MT, Willits D, Young M, Douglas T. J. Am. Chem. Soc. 2003;125(36):10806. doi: 10.1021/ja0363718. [DOI] [PubMed] [Google Scholar]
- 33.Suci PA, Kang S, Young M, Douglas T. J. Am. Chem. Soc. 2009;131(26):9164. doi: 10.1021/ja9035187. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Chen Y, Chen H, He W, Zhang Z-P, Zhang X-E, Wang Q. J. Am. Chem. Soc. 2011;133(50):20040. doi: 10.1021/ja207276g. [DOI] [PubMed] [Google Scholar]
- 35.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382(6592):607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 36.Rothemund PWK. Nature. 2006;440(7082):297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 37.Strable E, Johnson JE, Finn MG. Nano Lett. 2004;4(8):1385. [Google Scholar]
- 38.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Nano Lett. 2010;10(7):2714. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Capehart SL, Pal S, Liu M, Zhang L, Schuck PJ, Liu Y, Yan H, Francis MB, De Yoreo JJ. ACS Nano. 2014;8(8):7896. doi: 10.1021/nn5015819. [DOI] [PubMed] [Google Scholar]
- 40.Rogers WB, Manoharan VN. Science. 2015;347(6222):639. doi: 10.1126/science.1259762. [DOI] [PubMed] [Google Scholar]
- 41.Bancroft JB, Hiebert E. Virology. 1967;32(2):354. doi: 10.1016/0042-6822(67)90284-x. [DOI] [PubMed] [Google Scholar]
- 42.Gopal A, Zhou ZH, Knobler CM, Gelbart WM. Rna. 2012;18(2):284. doi: 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee S, Pfeifer CM, Johnson JM, Liu J, Zlotnick A. J. Am. Chem. Soc. 2006;128(8):2538. doi: 10.1021/ja056656f. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer P, Herzog M, Hirth L. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;276(943):99. doi: 10.1098/rstb.1976.0100. [DOI] [PubMed] [Google Scholar]
- 45.Garmann RF, Comas-Garcia M, Gopal A, Knobler CM, Gelbart WM. J. Mol. Biol. 2014;426(5):1050. doi: 10.1016/j.jmb.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garmann RF, Comas-Garcia M, Koay MS, Cornelissen JJ, Knobler CM, Gelbart WM. J. Virol. 2014;88(18):10472. doi: 10.1128/JVI.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comas-Garcia M, Garmann RF, Singaram SW, Ben-Shaul A, Knobler CM, Gelbart WM. J. Phys. Chem. B. 2014;118(27):7510. doi: 10.1021/jp503050z. [DOI] [PubMed] [Google Scholar]
- 48. http://en.wikipedia.org/wiki/Cherry_bomb. [Google Scholar]
- 49.Cadena-Nava RD, Comas-Garcia M, Garmann RF, Rao ALN, Knobler CM, Gelbart WM. J. Virol. 2012;86(6):3318. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurst SJ, Lytton-Jean AK, Mirkin CA. Anal. Chem. 2006;78(24):8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi H, Rubloff GW, Culver JN. Langmuir. 2007;23(5):2663. doi: 10.1021/la062493c. [DOI] [PubMed] [Google Scholar]
- 52.Eber FJ, Eiben S, Jeske H, Wege C. Angew. Chem. Int. Ed. 2013;52(28):7203. doi: 10.1002/anie.201300834. [DOI] [PubMed] [Google Scholar]
- 53.Jung S, Yi H. Langmuir. 2014;30(26):7762. doi: 10.1021/la501772t. [DOI] [PubMed] [Google Scholar]
- 54.Hohn T. J. Mol. Biol. 1969;43(1):191. doi: 10.1016/0022-2836(69)90088-6. [DOI] [PubMed] [Google Scholar]
- 55.Kler S, Wang JC-Y, Dhason M, Oppenheim A, Zlotnick A. ACS Chem. Biol. 2013;8(12):2753. doi: 10.1021/cb4005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413(6857):748. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 57.Liu N, Peng B, Lin Y, Su Z, Niu Z, Wang Q, Zhang W, Li H, Shen J. J. Am. Chem. Soc. 2010;132(32):11036. doi: 10.1021/ja1052544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.