Abstract
Schistosomiasis affects millions of people in developing countries and is responsible for more than 200,000 deaths annually. Because of toxicity and limited spectrum of activity of alternatives, there is effectively only one drug, praziquantel, available for its treatment. Recent data suggest that drug resistance could soon be a problem. There is therefore the need to identify new drug targets and develop drugs for the treatment of schistosomiasis. Analysis of the Schistosoma mansoni genome sequence for proteins involved in detoxification processes found that it encodes a single cytochrome P450 (CYP450) gene. Here we report that the 1452 bp open reading frame has a characteristic heme-binding region in its catalytic domain with a conserved heme ligating cysteine, a hydrophobic leader sequence present as the membrane interacting region, and overall structural conservation. The highest sequence identity to human CYP450s is 22%. Double stranded RNA (dsRNA) silencing of S. mansoni (Sm)CYP450 in schistosomula results in worm death. Treating larval or adult worms with antifungal azole CYP450 inhibitors results in worm death at low micromolar concentrations. In addition, combinations of SmCYP450-specific dsRNA and miconazole show additive schistosomicidal effects supporting the hypothesis that SmCYP450 is the target of miconazole. Treatment of developing S. mansoni eggs with miconazole results in a dose dependent arrest in embryonic development. Our results indicate that SmCYP450 is essential for worm survival and egg development and validates it as a novel drug target. Preliminary structure-activity relationship suggests that the 1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethan-1-ol moiety of miconazole is necessary for activity and that miconazole activity and selectivity could be improved by rational drug design.
Author Summary
Over 600 million people in endemic countries are at risk of contracting schistosomiasis, which results in over 200,000 deaths each year and significant illness to most people that are infected. There are concerns that the drug widely used for the treatment of schistosomiasis, praziquantel, may be losing efficacy due to evolution of drug resistant worms. Since the disease mainly affects the poor in developing countries, pharmaceutical companies have little interest in developing new drugs and none are currently being tested. In this paper we focus on a novel parasite protein, cytochrome P450, which we propose to be a new drug target. Worms are unusual in having only one cytochrome P450 gene; humans have 57 cytochrome P450 genes. By using reverse genetic and chemical approaches we found that the schistosome cytochrome P450 is essential for worm survival and egg development and, therefore, is an essential and druggable target. Drugs that target fungal cytochrome P450s and are already in use for treating several human diseases were identified as potential hits for further development for schistosomiasis treatment.
Introduction
Schistosomiasis is a helminthiasis caused by trematode worms of three main schistosome species, Schistosoma mansoni, S. haematobium, and S. japonicum. The disease is responsible for approximately 280,000 deaths annually and significant morbidity in more than 200 million people [1,2]. Schistosomiasis belongs to a class of neglected tropical diseases whose control has been given limited attention by the pharmaceutic industry because they affect poor people in developing nations. Currently, praziquantel (PZQ) is the only treatment for schistosomiasis [3]. However, studies indicate that PZQ-resistant laboratory strains can be isolated and clinical isolates with increased PZQ resistance have been reported [4]. Therefore, it is a matter of time before resistance fully evolves. In addition, PZQ is much less active against juvenile worms and often results in incomplete cures [5–8] and its mechanism of action, including its biotransformation are not fully understood [3].
Biotransformation pathways play vital roles in providing essential molecules for cell survival and to modify harmful molecules in order to facilitate their elimination. Xenobiotic biotransformation occurs in three phases. Phase I metabolism involves the oxidative, reductive, or hydrolytic transformations of xenobiotics, of which the most important are catalyzed by CYP450 enzymes. In phase II transformation, metabolites undergo conjugation reactions with endogenous compounds such as glutathione, glucuronic acid, amino acids, and sulfate in reactions mainly catalyzed by glutathione S-transferases (GSTs), UDP-glucuronosyltransferases, N-acetyltransferases, methyltransferases and sulfotransferases. Phase III transformations utilize membrane-bound transport proteins, which carry modified molecules across membranes for excretion [9]. There has been an extensive study of phase II metabolizing enzymes including the glutathione S-transferase family in schistosomes. For example, the main GSTs identified in S. mansoni have been shown to bind to several commercially available anthelmintics [10] and are currently important vaccine candidates [11]. Recently, a sulfotransferase was implicated in the mechanism and selectivity of action of oxamniquine in schistosomes [12]. In addition, Phase III biotransformation proteins, including the ATP-binding cassette (ABC) transporters, have been identified and their role in praziquantel susceptibility, immunoregulation within the host, parasite egg development and maturation, and translocation of important signaling molecules such as glyco- and phospholipids is being studied [13]. However, very little is known about phase I metabolizing CYP450 enzymes in schistosomes.
CYP450s are heme-containing monooxygenases. In concert with NADPH CYP450 reductases, the heme group of CYP450s serves as a terminal oxidase, i.e., a source of electrons to split molecular oxygen, with one oxygen atom added to the substrate and the other atom accepting reducing equivalents from NADPH to form water [14]. Characterized CYP450 reductase proteins are well conserved and occur as single copy genes in individual organisms. However, the CYP450 proteins are quite diverse, with most organisms having multiple CYP450 genes (Table 1) [9,15]. Analysis of the S. mansoni genome database has identified only one potential CYP450 gene [16]. In a previous study, extracts of adult S. mansoni and S. haematobium were shown to metabolize some typical CYP450 substrates and immunoblotting experiments with an anti-rat CYP450 antibody had cross-reactivity with both S. mansoni and S. haematobium homogenates with a specific band at ~50 kDa, well within typical CYP450 molecular weight range [17].
Table 1. Comparison of the number of CYP450 and CYP450 reductase genes from different species compiled from Nelson et al. [15].
| Organism | CYP450 | CYP450 Reductase |
|---|---|---|
| Schistosoma mansoni | 1 | 1 |
| Schmidtea mediterranea | 39 | 1 |
| Human | 57 | 1 |
| Mus musculus | 103 | 1 |
| Gallus gallus | 41 | 1 |
| Danio rerio | 81 | 1 |
| Drosophila melanogaster | 90 | 1 |
| Caenorhabditis elegans | 81 | 1 |
In addition to biotransformation activities, CYP450 proteins are involved in the metabolism of many essential endobiotic compounds. Synthesis of membrane sterols, cholesterol and ergosterol depends on CYP450s as does synthesis and degradation of steroid hormones [18,19]. Cellular levels of retinoic acid, the active metabolite of vitamin A, which is essential for embryonic development, postnatal survival, and germ cell development, are regulated and metabolized by several CYP450 proteins [18]. Other CYP450s are involved in the metabolism of prostaglandins, prostacyclins, and leukotrienes [19], all derivatives of fatty acids and important for cell signaling and immune response. In Caenorhabditis elegans CYP450 proteins are thought to be involved in meiosis, egg polarization, and egg shell development [20].
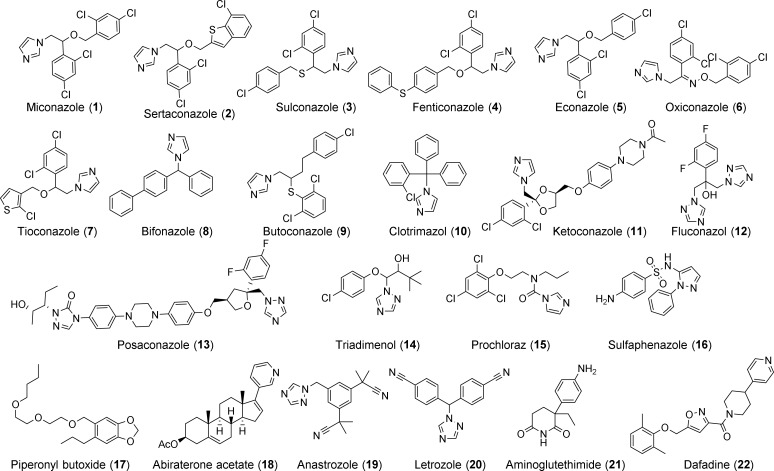
In this study, we hypothesize that the single CYP450 gene present in schistosomes is essential for worm survival and that blocking its function would lead to worm death and/or interference in parasite development. We used both genetic and pharmacological approaches to test this hypothesis. Treating larval parasites with SmCYP450-specific double-stranded RNA led to significant decreases in CYP450 mRNA and resulted in worm death. Screening a collection of CYP450 inhibitors (Fig 1) we found that low micromolar concentrations of imidazole antifungal CYP450 inhibitors had schistosomicidal activity against adult and larval worms and blocked embryonic development in the egg. We conclude that SmCYP450 is essential for parasite survival and egg development, and it is proposed as a novel target for antischistosomal drug development, with miconazole analogs as starting points in drug discovery.
Fig 1. Chemical structures of CYP450 inhibitors used in this study.
Materials and Methods
Ethics statement
In all of the experiments involving the use of animals, maintenance and use of these animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Rush University Medical Center (IACUC number 14–080; DHHS animal welfare assurance number A3120-01). Animals were euthanized with a lethal dose of Nembutal.
Chemicals and reagents
CYP450 inhibitors (Fig 1) were purchased from Sigma Aldrich (miconazole, clotrimazole, ketoconazole, posaconazole, triadimenol, sertaconazole, bifoconazole, econazole, butoconazole, dafadine, fluconazole), Santa Cruz Biotechnology (piperonyl butoxide, tioconazole, fenticonazole, prochloraz, sulconazole, oxiconazole, anastrozole, letrozole, aminoglutethimide), and Cayman Chemical Company (abiraterone acetate). Sulfaphenazole was synthesized according to published procedures [21,22].
Experimental organisms
A Puerto Rican strain of S. mansoni maintained in Biomphalaria glabrata snails and the same strain of S. mansoni maintained in NIH Swiss mice was supplied by the Biomedical Research Institute (Rockville, Maryland, USA). All adult worms, schistosomula, and egg cultures were incubated in Basch’s Media 169 [23]. Basal Medium Eagle was from Life Technologies; glucose and fungizone were from Fisher Scientific; hypoxanthine, serotonin, insulin, hydrocortisone, triiodothyronine were from Sigma Aldrich; MEM vitamins, Schneider’s Drosophila Medium, and gentamicin were from Gibco; HEPES buffer from Mediatech, Inc.; penicillin/streptomycin from Cellgro; and fetal bovine serum was from HyClone Laboratories, Inc.
Cercariae were shed from infected Biomphalaria glabrata snails and mechanically transformed to schistosomula as described [24]. To collect liver-stage, juvenile parasites mice were perfused 23 days post infection and to collect adult worms mice were perfused 6–7 weeks after infection with Dulbecco’s modified Eagle’s medium (Gibco) using methods described previously [24]. Live worms were washed thoroughly with DMEM. Eggs were obtained from the livers of the mice 7 weeks post infection. Livers were placed in ice-cold PBS and stored at 4°C overnight and processed the following day as described [24]. Parasite material was stored at -80°C for later use in stage specific SmCYP450 mRNA quantitation.
Analysis of the SmCYP450 sequence and investigation of sequence variation
The CYP450 open reading frame was amplified from adult mixed cDNA using P450_5' and P450_3' (all primers listed in Table 2) and GoTaq Flexi DNA Polymerase (Promega). PCR product was cloned into pCRII (Invitrogen) and plasmids were purified (Plasmid Mini Kit (QIAGEN) and sequenced at the University of Illinois-Chicago Core Sequencing Center (UIC). Alignment of the obtained open reading frame with the genome sequence was done using the Needleman-Wunsch Global Sequence Alignment Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Prediction of the molecular weight of the encoded protein was done at the Swiss Institute of Bioinformatics Resource Portal (http://web.expasy.org/compute_pi/).
Table 2. List of primers used for PCR, RT-PCR and qRT-PCR.
| Primer | Sequence |
|---|---|
| T3 | GCTCGAAATTAACCCTCACTAAAGGG |
| SK | CGCTCTAGAACTAGTGGATC |
| SL | AACCGTCACGGTTTTACTCTTGTGATTTGTTGCATG |
| P450_5' | ATGGATACCTTTGAATTTTATG |
| P450_3' | TTACTTCCATACATCGGTACG |
| CYP450-R1 | CACCATAAGTTGCAACAACG |
| CYP450-R3 | GTTGAGAAGCAGACACATCC |
| CYP450-revComF2 | CTGACTATTGTGTACAGCATA |
| SmCYP450-For | GTGGACAATTCTGTTGTCTA |
| SmCYP450-Rev | CTCCAAACTGTACATCTCATC |
| T7-T3 | TAATACGACTCACTATAGGGATTAACCCTCACTAAAGGGA |
| qPCR-p450-F | TGCTGGTACTGACACCACGTCTTT |
| qPCR-p450-R | GGAACTACACTAGCCCAACGATGA |
| qPCR-tubulin-F | CACGAGCAGTTAAGCGTTGCAGAA |
| qPCR-tubulin-R | TATTTGCCGTGACGAGGGTCACAT |
| CYP450-interF2 | TATGCTGTACACAATAGTCAG |
| ORF_CYP450 Reverse | TTAAATACTTGTTCTTCTATTTCC |
| GAPDH_S.mansoni FWD | ATGTTCGTTGTTGGTGTGAATG |
| GAPDH_S.mansoni REV | TTCCGTTTATGTCTGGAATGA |
Internal Coordinate Mechanics (ICM) homology modeling tool (http://www.molsoft.com) [25,26] was used to generate a CYP3050A1 model based on the CYP2C5 (PDB ID 1nr6) template and the structure-superimposition-guided sequence alignments performed using the iterative dynamic programming and superimposition steps implemented in the ICM Homology Modeling module [27]. Alignments were further adjusted manually to preserve integrity of the a-helices and b-sheets, patterns of positive (blue) and negative (red) charges, aromatic (purple) and hydrophobic (green) functionalities, and finally, proline (ochre) and cysteine (yellow) side chains. Global optimization was performed using the Biased Probability Monte Carlo (BPMC) conformational search combined with the electrostatic energy term [28]. Loop search and side chain refinement was conducted for up to 100,000 iterations, which included full energy minimization at each step, to result in a model with satisfactory local strain parameters [29].
To determine if a subset of CYP450 mRNAs was trans-spliced, the S. mansoni trans-spliced leader sequence was used in PCR with either CYP450-R1 or CYP450-R3 specific internal primers (Table 2). A modified 5’ rapid amplification of cDNA ends (RACE) with Q5 DNA polymerase (New England Biolabs) was done in a nested PCR using an adult cDNA library (kindly provided by Dr. Philip LoVerde) as the template and vector primer T3 + gene-specific CYP450-revComF2 in the first stage and the vector primer SK + gene-specific SmCYP450-Rev (Table 2) for the second stage. The product of the second PCR was cloned into pCR4 (Invitrogen). To determine if the SmCYP450 mRNA is alternatively spliced, the complete ORF was amplified using Q5 DNA polymerase from adult male, adult female, and egg cDNA (synthesized as described below) with P450_5' and P450_3'. PCR products were cloned into pCR4. Plasmid DNAs were isolated (GeneJET Plasmid Miniprep Kit, Thermo Scientific) and sequenced at the UIC sequencing core.
RNA interference (RNAi)
Plasmid Construction
A 566 bp SmCYP450 sequence close to the N-terminal region of the SmCYP450 gene was amplified using SmCYP450-For and SmCYP450-Rev and cloned into a pCRII vector (Invitrogen) according to the manufacturer’s protocol. The sequence was verified through Sanger sequencing at the UIC sequencing core. A new primer (T7-T3) was designed flanking the 566 bp sequence so that it included the full T7 promoter primer followed by part of the T3 primer sequence. PCR was carried out using T7 and T7-T3 primers with Taq DNA polymerase (Thermo Scientific) at 96°C 2 min, followed by 40 cycles at 94°C, 1 min; 48°C, 2 min; 72°C, 1.5 min; then 72°C, 7 min. The PCR product was run on a 1% agarose gel containing ethidium bromide to verify the insert size. The PCR product was cut out from the gel and cleaned with Gel Extraction kit (Qiagen) and the concentration determined.
CYP450 dsRNA Synthesis
Both a published method [30] using T7 RNA polymerase (New England BioLabs) and the MEGAclear kit (Life Technologies) were used to synthesize SmCYP450 dsRNA. In the first method, synthesis was carried out in a 100 μl reaction mix using 100 μg/ml BSA (NEB), 500 μM each of rNTPs (NEB), 1 x RNA Pol reaction buffer (40 mM Tris-Cl, 6 mM MgCl2, 10 mM dithiothreitol), and 800 units/ml RNase inhibitor (NEB) at 40°C for 4.5 hours. The resultant product was treated with RNAase free DNase I (NEB) at 37°C for 10 min and cleaned using Zymogen DNA-free RNA kit and eluted with DNase/RNase free water, or the DNase I treated samples were precipitated in 75% DEPC treated ethanol and 4 M LiCl and re-suspended in DEPC treated water. The concentration of the cleaned ssRNA was determined using a Nanodrop spectrophotometer. Synthesis using the MEGAclear kit followed manufacturer’s protocols with RNA products cleaned as described above. The RNAs were annealed to form dsRNA by incubating at 75°C, 50°C, and 37°C for 3 min each, and the concentration was determined by Nanodrop spectrophotometry. A negative control dsRNA was synthesized as described above from the ccdB and camR- bacterial gene insert of pJC53.2 plasmid obtained from Dr. James Collins (UIUC) [30].
RNAi Cultures
Freshly prepared schistosomula (300–400) were placed in each well containing 1 ml Basch’s media in a 24-well plate and incubated overnight in a 37°C and 5% CO2. The following day SmCYP450 dsRNA or control irrelevant dsRNA was added to each well to a final concentration of 10 or 30 μg/ml. Treatments were done in duplicate. Over several days worms were observed for dead (dark, granular appearance and non-motile) or alive (translucent and motile) as described [31].
Inhibitor treatment
To determine the activity of CYP450 inhibitors, 10 worm pairs in 5 ml Basch’s media per well in 6-well plates were cultured overnight at 37°C and 5% CO2 and the following day CYP450 inhibitors (Fig 1) were added to each well. The media were replenished every 48 hr with fresh media and inhibitors. Dead worms were identified as those that showed no motility when observed for several minutes. For larval worms, 300–400 freshly prepared schistosomula were placed in each well in a 24-well plate containing 1 ml Basch’s Media and incubated overnight at 37°C and 5% CO2. The following day compounds were added to each well and the parasites observed for several days without changing the media or adding fresh compounds. Live and dead parasites were classified as before.
To monitor the effects of miconazole on egg development we followed a recently published method [32]. Freshly perfused adult worm pairs were incubated in Basch’s media overnight. The following day worms were removed and miconazole (5 or 10 μM) or an equal volume of DMSO was added to the eggs produced. Eggs were further incubated a total of 72 hr in the presence of miconazole. Each group of treated eggs was then collected and centrifuged (500 x g, 5 min) and the supernatant discarded. The egg pellets were each washed in excess PBS and centrifuged. The eggs were then fixed in 100% methanol at room temperature for 10 min. After removing the methanol the eggs were incubated in DAPI (4’6-diamidino-2-phenylindole) Fluoromount-G (SouthernBiotech) overnight at 4°C for nuclear staining. Images were captured using Zeiss Axiovert Z1 imaging microscope and analyzed with AxioVision software LE (release 4.8.2 SP3, 2013).
Combined inhibitor and RNA interference
To see if their activities had additive effects, schistosomula were treated with dsCYP450 RNA at a concentration that alone did not kill schistosomula (10 μg/mL) and miconazole at concentrations that resulted in minimal killing (2.5 or 5 μM) or each alone. Schistosomula cultures were set up as described above. A control experiment was set up with irrelevant dsRNA with and without 5 μM miconazole. Parasites were observed as described above.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from frozen worm and egg samples using the TRIzol Reagent (Life Technologies) per the manufacturer’s recommendation in a 2 ml Lysis Matrix Tubes (MP Biomedicals) containing 500 μl TRIzol reagent. Tubes were shaken three times for 20 seconds each using a tissue homogenizer (FastPrep-24 5G Instrument, MP Biomedicals). The samples were incubated on ice for 5 minutes in between each lysis process. After lysis, another 500 μl TRIzol Reagent was added to each sample, mixed and incubated at room temperature for 5 min. The resultant sample was spun at 13,000 x g for 1 min to pellet cellular debris. Following centrifugation, supernatants were transferred to a new 1.5 ml microfuge tube and extracted with chloroform/isopropanol according to the manufacturer’s instructions. The gelatinous, white RNA precipitate obtained after the chloroform/isopropanol extraction was resuspended in DEPC treated water in 75% ethanol and spun at 6500 x g for 5 min at 4°C. After centrifugation the supernatant was removed and the RNA pellet briefly air-dried and re-suspended in DEPC-treated water, heated briefly at 55°C quantified on a Nanodrop spectrophotometer. Total RNA was used for cDNA synthesis (iScript, BIO-RAD) per the manufacturer’s recommendation. The synthesized cDNA for each sample was quantified by Nanodrop spectrophotometry and stored at -20°C.
Quantitative RT-PCR (qPCR) and semi-quantitative RT-PCR
Primers used for qPCR are shown in Table 2. α–tubulin (GenBank accession M80214) was used to normalize the results. The reactions were each carried out in a 20 μl reaction using ROX Passive Reference Dye (Bio-Rad) according to the manufacturer’s protocol. The amplification was monitored in a 7900HT Fast Real-Time PCR Machine (Applied Biosystems) under the following cycle conditions: (stage 1, 95°C 30 sec, stage 2, 95°C 5 sec, 60 C 30 sec) x 50, plus a one cycle dissociation curve. Fold differences were calculated using the 2-ΔΔCT as described [33] with α–tubulin transcript levels serving as the internal standard. Reactions were done in triplicate. Semi-quantitative RT-PCR was used to assess the relative abundance of SmCYP450 mRNA after RNAi silencing using Platinum Taq DNA polymerase (Life Technologies. Glyceraldehyde 3-phosphate dehydrogenase (GenBank accession M92359) was used as a control gene (primers GAPDH_S.mansoni FWD and GAPDH_S.mansoni REV) and SmCYP450 cDNA was amplified with primers CYP450-interF2 and ORF_CYP450 Reverse.
Results
The SmCYP450 coding sequence is similar to CYP450 proteins in other organisms
Cloning and sequence analysis shows that the SmCYP450 coding sequence is 1452 base pairs encoding a protein of 483 amino acids with a predicted molecular weight of 55.28 kDa. The family assignment as CYP3050A1 was made by Dr. David R. Nelson according to the CYP450 nomenclature [34,35]. The sequence was found to be longer than the sequence reported in GeneBank (Smp_156400, 1245 base pairs,) due to a miscalled junction of the 5th intron/6th exon during genome annotation. The sequence obtained was submitted to GenBank with the accession number KT072747. The gene is composed of 7 exons and 6 introns spanning 15,378 base pairs (not including 5’ and 3’ noncoding sequences). Sequence analysis shows it to be comparable to CYP450 proteins from other organisms. The signature heme-binding motif [14,36], [FW]-[SGNH]-x-[GD]-{F}-[RKHPT]-{P}-C-[LIVMFAP]-[GAD], is present (the bold, underlined residues are present in SmCYP450) (Fig 2). The ‘P450-signature’ sequence, [AG]-G-X-[DE]-T-[TS], which forms a channel for electron transfer [36], is also present in the SmCYP450 peptide. The protein has an N-terminal membrane spanning region followed by the poly-proline domain, which is important for protein folding and structural integrity [37]. The turns by the poly-proline region provide a junction between the transmembrane region and the main catalytic domain typical for most CYP450 proteins [37]. The organization of the predicted secondary structure of the SmCYP450 protein sequence follows other CYP450 proteins, beginning from helix A in the N-terminal region of the protein sequence and ending with helix L, which contains the heme-binding sequence (Fig 2). Likewise, with the exception of the absence of the J and J’ helices, the tertiary structure of SmCYP450 protein is predicted to be similar to known CYP450 proteins (Fig 3).
Fig 2. Comparison of Schistosoma mansoni CYP450 protein (Sman) with CYP450 proteins from other species.
Multiple alignment of CYP450 proteins from S. mansoni (csm305A); rabbit CYP450 2C5 (1nr6_a); human CYP450 2C9 (1r9o_a); human CYP450 2C19 (4gqs_a); human CYP450 1A1 (4i8v_a); and human CYP450 2b6 (4rrt_a). The residues are shown in one letter code and colored by type: red- negatively charged, blue—positively charged, yellow—Cys, green—hydrophobic, cyan—Gly, ochre—Pro, purple—aromatic. The residues are shown in brighter colors for conserved positions. The ‘P450-signature’ sequence, which forms a channel for electron transfer, and the CYP450 consensus motif responsible for heme-binding and interaction with molecular oxygen and the relevant substrates are boxed. Predicted helices in the secondary structure based on homology modelling of SmCYP450 are indicated by the bold letters A-L based on rabbit CYP450 2C5 [38].
Fig 3. Structural modeling of S. mansoni CYP450 (CYP3050A1) and comparison to the structure determined for rabbit CYP450 2C5 (1nr6_a) [38].
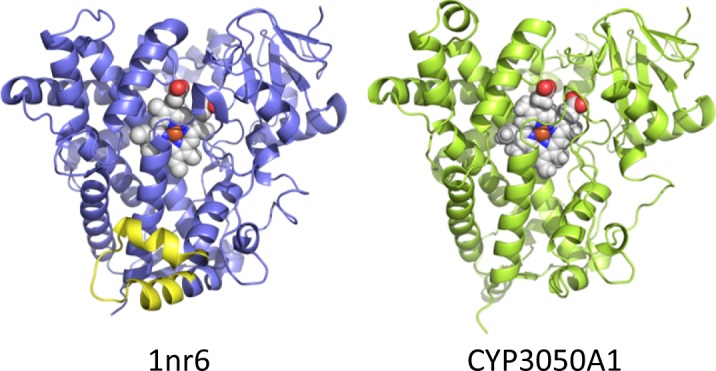
The heme is shown is each model as a space-filling projection. The J and J’ helices in rabbit CYP450 2C5, which are absent in S. mansoni CYP450, are highlighted in yellow.
There is a single CYP450 protein in S. mansoni
It is possible that a diversity of CYP450 proteins or alternative subcellular targeting of SmCYP450 results from alternative splicing or post-transcriptional modifications of the mRNA produced from the single S. mansoni CYP450 gene. This was addressed by analyzing cDNAs from a variety of developmental stages and by using modified 5’RACE and PCR with the schistosome spliced leader sequence to search for multiplicity of CYP450 mRNAs. We analyzed 33 clones from adult female worm cDNA, 23 clones from adult male worm cDNA, 11 clones from egg cDNA, and 28 clones generated by 5’ RACE and all sequences were identical. Therefore, we found no evidence for alternative splicing or other sequence variations. PCR with the spliced leader sequence and two different internal CYP450-specific primers resulted in no PCR products; therefore, the SmCYP450 mRNA does not appear to be trans-spliced. Therefore, it appears that the SmCYP450 gene encodes a single CYP450 protein.
S. mansoni CYP450 is differentially expressed during parasite development in the mammalian host
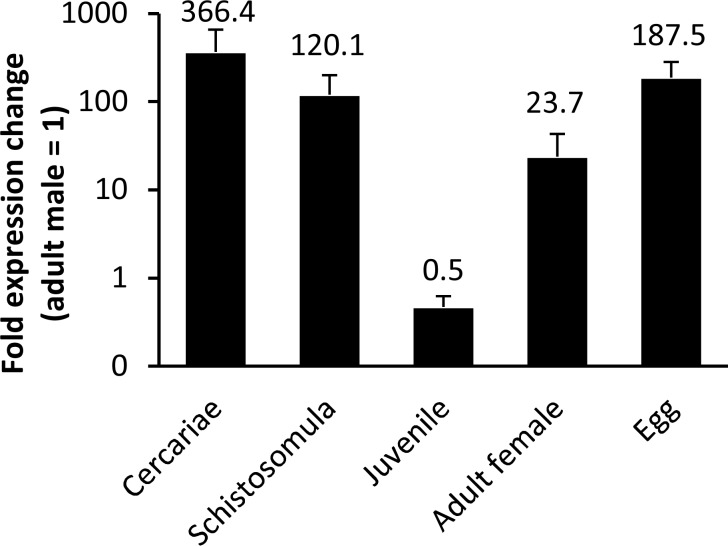
Using qRT-PCR we found that SmCYP450 mRNA was present at all developmental stages investigated and that it is differentially present during development (Fig 4). Eggs, the larval stages of development (cercariae and schistosomula) and adult female worms had higher mRNA levels than adult male worms. Liver stage parasites had the lowest SmCYP450 mRNA expression levels, about 50% that of adult males.
Fig 4. CYP450 messenger RNA abundance during the lifecycle of Schistosoma mansoni.
Whole RNA was extracted from different stages of S. mansoni (cercariae, 1-day old schistosomula; juvenile liver worms (23 days post infection), adult males (49 days post infection), adult females (49 days post infection) and eggs) using TRIzol reagent and chloroform/ethanol extraction protocol. cDNA was synthesized from whole RNA and used for qRT-PCR, with reactions done in triplicate. Adult males (= 1) were used as calibrator stage and mRNA abundance was normalized to α-tubulin. Error bars indicate standard error of the mean with n ≥ 3 biological replicates. Numbers indicate fold change relative to adult males and all values are significantly different from adult males; p < 0.05; student t-test. The results indicate that S. mansoni CYP450 is expressed in all stages investigated and that its expression is developmentally regulated.
S. mansoni CYP450 dsRNA treatment leads to schistosomula death
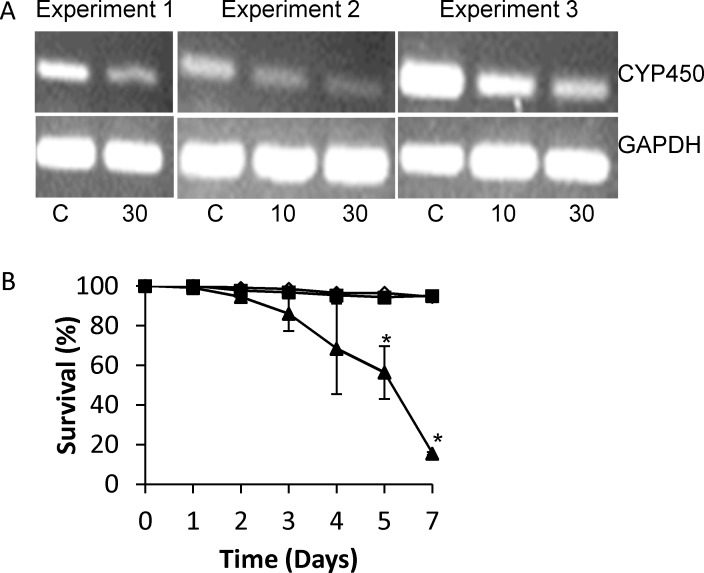
To determine if SmCYP450 is essential for schistosomula survival we used RNAi to silence SmCYP450 expression. Treating worms with 10 μg/mL or 30 μg/mL SmCYP450 specific dsRNA for two or three days resulted in a dose-dependent reduction in SmCYP450 message (Fig 5A). No change was seen in SmCYP450 mRNA after treatment with 30 μg/mL irrelevant dsRNA or in GAPDH mRNA abundance after treatment with either dsRNA (Fig 5A). Treatment with 30 μg/mL SmCYP450 specific dsRNA resulted in 80% schistosomula survival by day 3, 40% survival by day 5, and 15% survival by day 7. In contrast, 95% and 94.5% of schistosomula were alive on day 7 after treatment with 30 μg/mL irrelevant dsRNA or 10 μg/mL SmCYP450 specific dsRNA, respectively (Fig 5B).
Fig 5. Effect of silencing Schistosoma mansoni CYP450 in cultured larval worms.
Freshly prepared schistosomula (300–400) were placed in each well containing 1 ml Basch’s Media in a 24-well plate and overnight in a 37°C with 5% CO2. The following day schistosomula were treated with 10 or 30 μg/ml S. mansoni CYP450 dsRNA or 30 μg/ml negative control dsRNA. Over several days worms were observed for dead (dark, granular appearance) or alive (translucent). (A) mRNA expression patterns in schistosomula treated with S. mansoni CYP450 specific dsRNA or negative control dsRNA control after 3 days of treatment (Experiments 1 and 2) or 2 days treatment (Experiment 3). The control gene for cDNA input is S. mansoni glyceraldehyde 3-phosphate dehydrogenase (GAPDH). C, schistosomula treated with 30 μg/mL irrelevant dsRNA; 10, schistosomula treated with 10 μg/mL SmCYP450 dsRNA; 30, schistosomula treated with 30 μg/mL SmCYP450 dsRNA. (B) Effect of S. mansoni CYP450 dsRNA on schistosomula survival in cultures with 30 μg/mL negative control dsRNA (black square), 10 μg/mL S. mansoni CYP450-specific ds RNA (open triangle), and 30 μg/mL S. mansoni CYP450-specific ds RNA (black triangle). Treatments were done in triplicate and repeated 3 times. Error bars indicate standard error of the mean; *, p < 0.05; student t-test.
The imidazole subgroup of azole antifungal CYP450 inhibitors is active against S. mansoni
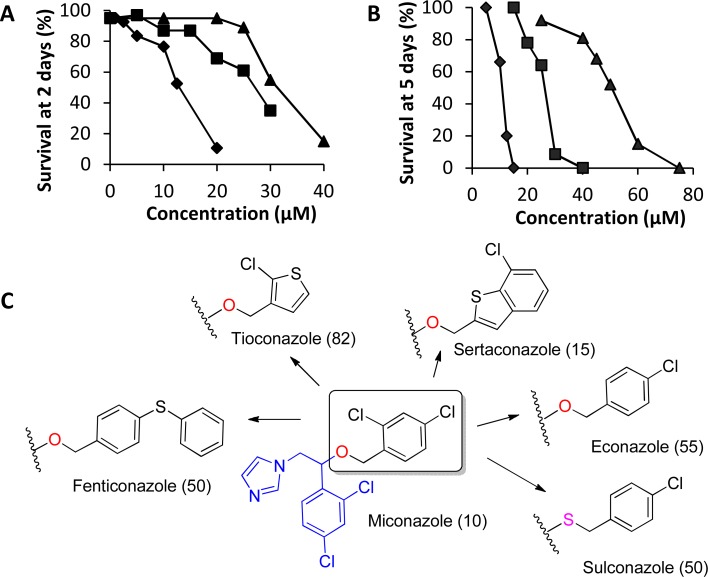
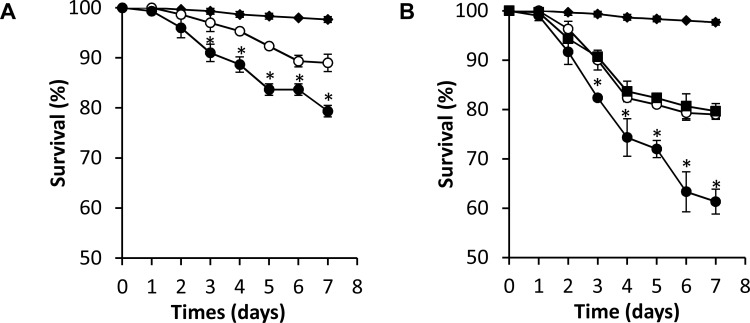
CYP450 enzymes are inhibited by numerous anti-infective and anticancer agents. We next asked if clinically relevant CYP450 inhibitors (Fig 1) affected parasite survival. Several antifungal imidazoles (miconazole, clotrimazole, ketoconazole) but not closely related triazole antifungals (fluconazole, posaconazole and triadimenol) were active against both larval and adult worms (Fig 6A and 6B and Table 3). Miconazole, clotrimazole, and ketoconazole had ED50 (Effective Dose producing 50% worm death) values of 10 μM, 20 μM, and 40 μM, after 5 day treatments against adult worms and 12.5 μM, 27.5 μM, and 30 μM after 2 day treatments against schistosomula, respectively. Other CYP450 inhibitors, such as prochloraz, sulfaphenazole, piperonyl butoxide, dafadine, letrozole, aminoglutethimide, abiraterone acetate, and anastrozole had no significant schistosomicidal activity against either larval or adult worms (Table 3). Expansion of the anti-fungal imidazole series was done to generate preliminary structure activity relationships of this compound series. Our studies revealed that imidazoles that retained the 1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethan-1-ol moiety of miconazole had significant schistosomicidal activity against both larval and adult worms, while those which lacked this moiety had much reduced or no schistosomicidal activity (Table 3, Fig 6C).
Fig 6. Activity of anti-fungal imidazole CYP450 inhibitors on larval and adult Schistosoma mansoni worms.
Survival of schistosomula (A) after 2 d culture and adult worms (B) after 5 d culture for miconazole (black diamond), clotrimazole (black square), and ketoconazole (black triangle). (C) In house SAR on known miconazole analogs against adult worms. Numbers in the parenthesis are survival (%) of adult worms on day 7 in 10 μM of respective compound.
Table 3. Results with selected cytochrome P450 inhibitors used in this study.
| Entry | Compound | Survival at 7 days (%) | Function and CYP450 class inhibited | |||
|---|---|---|---|---|---|---|
| Adult | Schistosomula | |||||
| 5 μM | 10 μM | 5 μM | 10 μM | |||
| 1 | Miconazole | 57 | 10 | 72 | 22 | antifungal CYP51 |
| 2 | Sertaconazole | 95 | 15 | 65 | 6 | |
| 3 | Sulconazole | 100 | 50 | 90 | 56 | |
| 4 | Fenticonazole | 75 | 50 | 92 | 79 | |
| 5 | Econazole | 100 | 55 | 87 | 66 | |
| 6 | Oxiconazole | 100 | 60 | 89 | 71 | |
| 7 | Tioconazole | 100 | 85 | 86 | 82 | |
| 8 | Bifoconazole | 100 | 100 | 80 | 79 | |
| 9 | Butoconazole | 100 | 80 | 77 | 60 | |
| 10 | Clotrimazole | n.d. 1 | 100 | 97 | 90 | |
| 11 | Ketoconazole | n.d. | 100 | 98 | 97 | |
| 12 | Fluconazole | 100 | 100 | 98 | 94 | |
| 13 | Posaconazole | 100 | 100 | 94 | 88 | |
| 14 | Triadimenol | 100 | 100 | 99 | 99 | |
| 15 | Prochloraz | 100 | 100 | 94 | 82 | |
| 16 | Sulfaphenazole | 100 | 100 | 99 | 93 | antibacterial CYP2C9 |
| 17 | Piperonyl butoxide | 100 | 100 | 100 | 100 | pesticide CYP6D1 |
| 18 | Abiraterone acetate | 100 | 100 | 100 | 100 | prostate cancer CYP17A1 |
| 19 | Anastrozole | 100 | 100 | 100 | 100 | breast cancer CYP19A1 |
| 20 | Letrozole | 100 | 100 | 100 | 100 | |
| 21 | Aminoglutethimide | 100 | 100 | 100 | 100 | |
| 22 | Dafadine | 100 | 100 | 100 | 100 | CYP27A1 |
1n.d., not determined.
Miconazole targets SmCYP450
Does the potent schistosomicidal activity of miconazole act through inhibition of worm CYP450 or does it have other targets in the worm? To address this question we tested low doses of miconazole against worms treated with 10 μg/mL dsRNA CYP450, which caused no significant worm death itself. While 5 μM miconazole alone resulted in 80% survival after 6 days, combinations of 5 μM miconazole and 10 μg/mL SmCYP450-specific dsRNA resulted in 60% survival (p = 0.0042). Combining 2.5 μM miconazole (90% survival alone) and 10 μg/mL SmCYP450-specific dsRNA resulted in 75% survival (p = 0.007) (Fig 7A). Addition of 30 μg/mL irrelevant dsRNA treatment had no effect on killing by 5 μM miconazole (Fig 7B). These results strongly suggest that miconazole schistosomicidal activity is specific for SmCYP450.
Fig 7. Combinations of miconazole and RNAi have increased killing activity, suggesting that they function through inhibition of the same target.
(A) Schistosomula cultured with 10 μg/ml S. mansoni CYP450 dsRNA (black diamond); 2.5 μM miconazole (open circle); or 10 μg/ml S. mansoni CYP450 dsRNA and 2.5 μM miconazole (black circle). (B) Schistosomula cultured with 10 μg/ml S. mansoni CYP450 dsRNA (black diamond); 5 μM miconazole (black square); 5 μM miconazole plus 30 μg/ml irrelevant dsRNA and 5 μM miconazole (open circle); or 5 μM miconazole plus 10 μg/ml S. mansoni CYP450 dsRNA and 5 μM miconazole (black circle). All experiments were done in triplicate. Error bars indicate standard error of mean; *, p < 0.05; student t-test).
Miconazole treatment results in impaired schistosome egg development
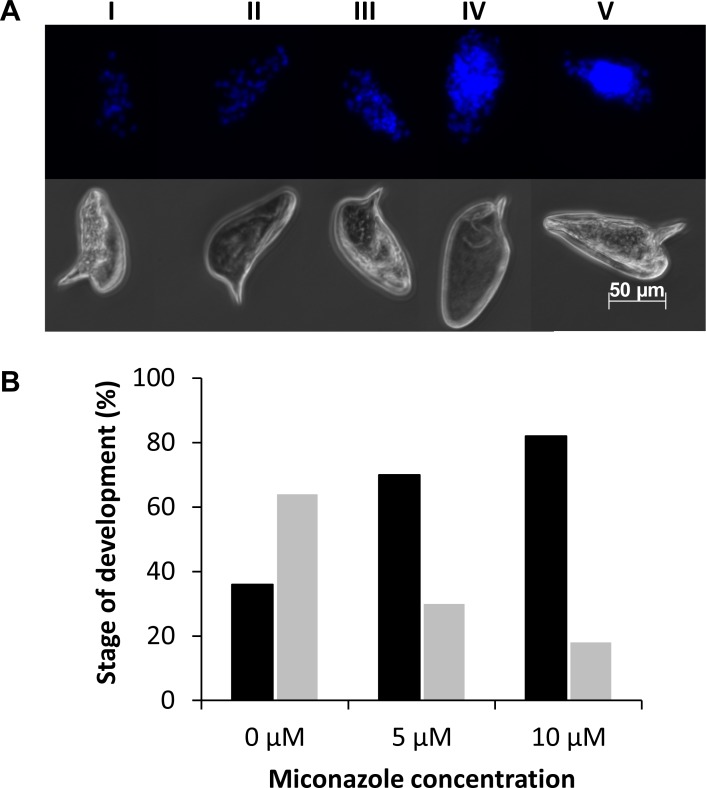
To determine if miconazole interferes with egg development and maturation we treated eggs deposited by freshly perfused adult worm pairs with miconazole and monitored embryo development using a recently described method [32,39]. Egg development was scored based on the number and arrangement of cell nuclei (Fig 8A). Our results indicate that there is a general interference of egg development and accumulation of early embryonic stages (I, II and III) and decrease in late stage embryos (IV and V) in the miconazole treatments compared to the DMSO controls. Only 30% (18/62) of eggs treated with 5 μM miconazole and 18% (10/56) treated with 10 μM miconazole reached the latter stages of egg development (stages IV and V) compared to 64% (35/55) in DMSO control (Fig 8B). These results indicate that miconazole affects embryonic development.
Fig 8. Effect of miconazole on egg development.
(A) Example of egg development scoring scheme. Upper panel shows fluorescent images of eggs representative of each developmental stage scored; the bottom panel shows brightfield images of the same eggs. (B) Scoring of egg development in cultured eggs treated with 0, 5, or 10 μM miconazole. The percentage of eggs scored at developmental stages I-III (black bars) and eggs scored at developmental stages IV-V (gray bars) are indicated. For 0 μM miconazole, n = 55 eggs scored; for 5 μM miconazole, n = 56 eggs scored; for 10 μM miconazole, n = 62 eggs scored.
Discussion
Because schistosomiasis control relies on a single drug and there is field evidence for the evolution of drug resistance [3,4], there is an urgent need to identify new, druggable worm targets. In this study we present the first detailed characterization of the CYP450 from S. mansoni and provide strong evidence that it is an essential and druggable target in the worm.
The SmCYP450 exists as a single copy gene in the S. mansoni genome [16]. This is in stark contrast to humans, which have 57 genes and alternative splicing and genetic variations that can lead to the production of many more distinct protein species [40,41], and to the free-living flatworm Schmidtea mediterranea, which has at least 39 CYP450 genes [15]. The loss of CYP450 family members in parasitic helminths has been noted previously [42]. However, the fact that parasitic flatworms have retained one CYP450 signifies that it plays an important and perhaps essential function. We add here that in S. mansoni there appears to be no post-transcriptional modifications (alternative or trans-splicing, RNA editing) to the mRNA. Therefore, it is likely that a single protein product is produced from the SmCYP450 gene. Since there was no evidence for alternative splicing to insert different leader sequences at the N-terminus, the protein product is likely only targeted to the endoplasmic reticulum.
The predicted protein has generally low sequence identity with the other CYP450s; the highest identity to human CYP450 proteins is 22% to CYP2C9. Importantly, the CYP450 consensus motif responsible for heme-binding and interaction with molecular oxygen and the relevant substrates and the ‘P450-signature’ sequence are conserved in the SmCYP450 protein sequence. Curiously, SmCYP450 lacks a number of motifs found in many characterized CYP450. The majority of CYP450s contain an ‘EXXR motif’ in helix K. The glutamic acid and arginine residues form a charge pair with a third amino acid more distant in the meander region. This is frequently an arginine in the so-called ‘PERF motif’. Putative functions of the EXXR motif and PERF motif may be to associate heme with the newly synthesized CYP450 polypeptide and/or to maintain the CYP450 tertiary architecture [43]. This is key to the structural fold of CYP450s and previous studies in which mutagenesis directed at the side-chains of glutamic acid or arginine in the EXXR motif or at the invariant cysteine in the L-helix resulted in completely inactive and misfolded proteins [44]. However, these motifs are not present in CYP450s from parasitic Trematodes (e.g., Schistosoma, Clonorchis sinensis) and Cestodes (e.g., Echinococcus multilocularis) [42]. Their absence is not without precedent as the EXXR motif is also absent in most members of a CYP157 subfamily in Streptomyces spp [45]. The Trematode CYP450 proteins also lack the J and J’ helices, which occur to the N-terminal side of and include the EXXR motif. How these differences affect protein structure and function remains to be determined.
CYP450s function in an electron transport chain in which electrons are passed from NADPH through a flavoenzyme either directly to the CYP450 heme or indirectly through cytochrome b5 or ferredoxin. In the endoplasmic reticulum, the flavoenzyme is NADPH CPY450 reductase. Additional partners of CYP450s in the endoplasmic reticulum include cytochrome b5 and cytochrome b5 reductase. In mammals, ferredoxin reductase and ferredoxins (also known as adrenodoxin reductase and adrenodoxins) are found in the mitochondria and are involved in steroid hormone synthesis mediated by CYP450s. The S. mansoni genome contains one CYP450 reductase, two cytochrome b5s, two cytochrome b5 reductases, one ferredoxin reductase, and two ferredoxins with potential to support SmCYP450 activity. Previous studies in schistosomes have found that ferredoxin reductase is mitochondrial and likely functions there in redox defenses [46,47]. Since we currently have no evidence for mitochondrial targeting of SmCYP450 protein, it is not likely that it functions in concert with ferredoxin reductase/ferredoxins.
Unlike the only previously characterized trematode CYP450, which showed highest expression in adult hermaphrodites [42], SmCYP450 is expressed at the highest levels in larval and egg stages. It is important to note that the developmental cycles and tissue locations of these organisms are significantly different. After active host localization and penetration, S. mansoni has extensive interactions with host skin, lungs, liver, and vascular epithelia, while Opisthorchis worms reside in the bilary ducts after excysting from metacercariae in the duodenum. As sequence identity between S. mansoni and O. felineus CYP450 proteins is only 37% it is quite possible that CYP450s have different functions in the worms.
The function of the SmCYP450 is not yet known. Different development stages may require different CYP450 metabolites and/or experience different immunological stresses. For instance larval parasites penetrate the skin of human host and begin migration through the skin and other tissue and may encounter different stress and immunological responses than adult worms in the mesenteric system. Larval schistosomes synthesize and secrete eicosanoids [48–53], which are signaling molecules derived from arachidonic acid, some of which are produced by CYP450s. The eicosanoids produced by schistosomes may down modulate host immune function [54,55]. Eicosanoids produced by adult worms may control other functions such as vasodilator activity, and/or vasoconstrictive action [55].
Other potential functions of SmCYP450 are in the metabolism of cholesterol and steroid hormones. Adult worms have been shown to convert cholesterol into several metabolites including pregnenolone, the first committed metabolite in steroid hormone biosynthesis [56,57]. Male worms transfer cholesterol and uncharacterized cholesterol metabolites to female worms [56] and synthetic steroids have been shown to affect worm egg production in vivo [56]. More recently, a catechol-estrogen conjugate (downstream products of CYP450 metabolism of estradiol and estrone), which has anti-estrogen affects, was identified in schistosome worm extracts and in the serum of infected humans [58]. Retinoic acid is essential for embryonic development in all metazoan organisms investigated, including free-living flatworms [59]. Retinoic acid activity is controlled through its tightly regulated synthesis from vitamin A (all-trans retinol) in a 2-step process by retinol dehydrogenases to all-trans retinal and by retinaldehyde dehydrogenases to all-trans-retinoic acid and is terminated via its breakdown by CYP450s [18,60]. Although retinoic acid signaling or metabolism in schistosomes is largely unknown, they have enzymes involved in retinoic acid metabolism (10 retinol dehydrogenases and 2 retinaldehyde dehydrogenases) and nuclear receptors related to retinoic acid receptors [61–64]. Ecdysteroids are hormones involved in insect molting and development and CYP450s are involved in their synthesis and transformation from farnesyl diphosphate and cholesterol. Ecdysteroids have been detected in schistosomes and their levels shown to vary during development [65,66]. S. mansoni synthesizes ecdysone and 20-OH ecdysone, which were shown to be potent stimulators of growth and vitellogenesis [67]. β-Ecdysterone was found to be effective in stimulating host location activities in S. mansoni miracidia [68]. Worms have two nuclear receptors related to insect ecdysone receptors, but their function in ecdysteroid signaling has not been determined [69,70]. Identification of the function of SmCYP450 will be targeted in future studies.
Our findings indicate S. mansoni has a single CYP450 protein, with highest sequence identity to human CYP450s CYP2C9 and CYP1A1. In order to compare the differences between S. mansoni CYP450 and human CYP450s we tested several different classes of CYP450 inhibitors. Although miconazole and structurally related imidazoles had schistosomicidal activity against adult and larval worms, other CYP450 inhibitors did not. These observations gave rise to an early exploration to investigate the structure-activity-relationships (SAR) of imidazole class of compounds, especially miconazole analogs (Fig 6C). Miconazole analogs were obtained by substituting the (2,4-dichlorophenyl)methanol moiety with different aryl groups. Sertaconazole, which results from substitution with a (7-chlorobenzo[b]thiophen-2-yl) methanol group, was equipotent to miconazole against adult and larval worms. Replacement with (4-chlorophenyl) methanol group results in econazole. Replacement of the oxygen by a sulfur in the econazole led to sulconazole. Modification of the econazole by substitution of a phenylthio group for the 4-chloro led to fenticonazole. Replacement with an oxime moiety into the miconazole gave oxiconazole. Econazole, sulconazole, fenticonazole and oxiconazole were less potent than miconazole. Substitution with (2-chlorothiophen-3-yl) methanol moiety results tioconazole, which is much less active. Our results indicate that miconazole constitutes a promising scaffold for targeting schistosome worms. Evidence that schistosomicidal activity of miconazole and analogs resides in the 1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethan-1-ol moiety of miconazole suggests routes to improved activity by rational drug design in future studies.
Miconazole had previously been included in a medium throughput phenotypic screen against schistosomula in an effort to repurpose approved drugs [71]. In, this study, compounds were screened at 1 μM against schistosomula and miconazole was found to be inactive, which is consistent with our results. However, for our screening purposes we tested compounds at higher concentrations and therefore, identified the schistosomicidal activity of this class of compounds. Although the concentrations required for worm killing activity in vitro may not be attained in vivo due to low biological availability, improved pharmacological properties can be incorporated into miconazole analogs to overcome these limitations. Our results indicate that the schistosomicidal activity of miconazole is due to inhibition of SmCYP450. Low concentrations of miconazole alone resulted in low schistosomicidal activity and partial reduction of SmCYP450 mRNA alone resulted in no larval worm death. However, combination treatments produced more than an additive response: 10% death in 2.5 μM miconazole alone increased to 20% with partial mRNA silencing and 20% death in 5 μM miconazole alone increased to 40% with partial mRNA silencing. The simplest explanation for this effect is that partial mRNA silencing results in decreases in SmCYP450 protein, which although it is not lethal to the worms itself, results in increased activity of miconazole due to a reduction in its protein target abundance. This strongly suggests that both SmCYP450 dsRNA and miconazole target the same pathway.
In schistosomes, egg development is a multi-stage process. Within the host mesentery and vasculature, a mature female releases approximately 300 encapsulated embryos (pre-mature eggs) per day [72,73]. Prior to that and within the mature female the early development of eggs occurs in several pre-zygotic and post zygotic stages [74]. Using methods recently developed to facilitate monitoring egg development [32,38] we investigated the effect of miconazole on egg development and maturation. Treatment with miconazole resulted in a dose-dependent impairment of ex vivo egg development, with most miconazole-treated eggs remaining at the early stages of embryonic development (Stages I-III) compared to control treatments, in which most eggs reached later stages of embryonic development (stages IV and V). CYP450 proteins are known to be involved in egg development in C. elegans, with CYP31A2 and CYP31A3 essential for the production of lipids required for egg shell development [20]. In addition, retinoic acid is essential for embryonic development in all metazoan organisms investigated, including free-living flatworms, and as indicated above, retinoic acid metabolism is mediated by CYP450 proteins. Inhibition of retinoic metabolism by miconazole could interfere with embryogenesis and egg development. There has not been a direct identification of SmCYP450 protein in eggs [75]. The newly oviposited egg is not fully formed and undergoes embryonic and subshell envelope development [76]. It is not known if SmCYP450 functions in embryonic or subshell envelope development or both, but our work shows for the first time that miconazole can block egg development.
Schistosomiasis remains a challenging disease to people living in endemic areas. In spite of many years of praziquantel use, the prevalence of infection remains high. The specter of evolving resistance to praziquantel, the only drug available for disease treatment, calls for the identification of new protein targets, the discovery of lead compounds and the development of new drugs for the treatment of the disease. The S. mansoni CYP450 exists as a single gene in the parasite genome. Our work shows that it is essential for parasite survival and could be an ideal drug target. In addition, select anti-fungal azoles could be promising starting points for future studies towards identifying new therapies for schistosomiasis.
Acknowledgments
We thank Dr. David R. Nelson, University of Tennessee Health Science Center, for helpful discussions and Dr. Ruben Abagyan, UCSD, for building the CYP3050A1 structure model. Schistosome-infected mice and snails were provided by the NIAID Schistosomiasis Resource Center at the Biomedical Research Institute (Rockville, MD) through NIH-NIAID Contract HHSN272201000005I for distribution through BEI Resources.
Data Availability
The full-length sequence of Schistosoma mansoni cytochrome P450 was submitted to GenBank and assigned the accession number KT072747.
Funding Statement
Peter Ziniel received a Burroughs Wellcome Fund travel grant. Peter Ziniel received a stipend from the Graduate College at Rush University Medical Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, Habbema JD, et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop 2003. May;86(2–3):125–139. [DOI] [PubMed] [Google Scholar]
- 2. King CH. Health metrics for helminth infections. Acta Trop 2015. January;141(Pt B):150–160. doi: 10.1016/j.actatropica.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cioli D, Pica-Mattoccia L, Basso A, Guidi A. Schistosomiasis control: praziquantel forever? Mol Biochem Parasitol 2014. June;195(1):23–29. doi: 10.1016/j.molbiopara.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 4. Wang W, Wang L, Liang YS. Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol Res 2012. November;111(5):1871–1877. doi: 10.1007/s00436-012-3151-z [DOI] [PubMed] [Google Scholar]
- 5. Aragon AD, Imani RA, Blackburn VR, Cupit PM, Melman SD, Goronga T, et al. Towards an understanding of the mechanism of action of praziquantel. Mol Biochem Parasitol 2009. March;164(1):57–65. doi: 10.1016/j.molbiopara.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabah AA, Fletcher C, Webbe G, Doenhoff MJ. Schistosoma mansoni: chemotherapy of infections of different ages. Exp Parasitol 1986. June;61(3):294–303. [DOI] [PubMed] [Google Scholar]
- 7. Xiao SH, Catto BA, Webster LT Jr. Effects of praziquantel on different developmental stages of Schistosoma mansoni in vitro and in vivo. J Infect Dis 1985. June;151(6):1130–1137. [DOI] [PubMed] [Google Scholar]
- 8. Pica-Mattoccia L, Cioli D. Sex- and stage-related sensitivity of Schistosoma mansoni to in vivo and in vitro praziquantel treatment. Int J Parasitol 2004. March 29;34(4):527–533. [DOI] [PubMed] [Google Scholar]
- 9. Cvilink V, Lamka J, Skalova L. Xenobiotic metabolizing enzymes and metabolism of anthelminthics in helminths. Drug Metab Rev 2009;41(1):8–26. doi: 10.1080/03602530802602880 [DOI] [PubMed] [Google Scholar]
- 10. Brophy PM, Barrett J. Glutathione transferase in helminths. Parasitology 1990. April;100 Pt 2:345–349. [DOI] [PubMed] [Google Scholar]
- 11. Mo AX, Agosti JM, Walson JL, Hall BF, Gordon L. Schistosomiasis elimination strategies and potential role of a vaccine in achieving global health goals. Am J Trop Med Hyg 2014. January;90(1):54–60. doi: 10.4269/ajtmh.13-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valentim CL, Cioli D, Chevalier FD, Cao X, Taylor AB, Holloway SP, et al. Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites. Science 2013. December 13;342(6164):1385–1389. doi: 10.1126/science.1243106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenberg RM. Schistosome ABC multidrug transporters: From pharmacology to physiology. Int J Parasitol Drugs Drug Resist 2014. September 26;4(3):301–309. doi: 10.1016/j.ijpddr.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortiz de Montellano PR editor. Cytochrome P450. 3rd ed. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 15. Nelson DR. The cytochrome p450 homepage. Hum Genomics 2009. October;4(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature 2009. July 16;460(7253):352–358. doi: 10.1038/nature08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saeed HM, Mostafa MH, O'Connor PJ, Rafferty JA, Doenhoff MJ. Evidence for the presence of active cytochrome P450 systems in Schistosoma mansoni and Schistosoma haematobium adult worms. FEBS Lett 2002. May 22;519(1–3):205–209. [DOI] [PubMed] [Google Scholar]
- 18. Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu Rev Nutr 2011. August 21;31:65–87. doi: 10.1146/annurev-nutr-072610-145127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tomaszewski P, Kubiak-Tomaszewska G, Pachecka J. Cytochrome P450 polymorphism—molecular, metabolic, and pharmacogenetic aspects. II. Participation of CYP isoenzymes in the metabolism of endogenous substances and drugs. Acta Pol Pharm 2008. May-Jun;65(3):307–318. [PubMed] [Google Scholar]
- 20. Benenati G, Penkov S, Muller-Reichert T, Entchev EV, Kurzchalia TV. Two cytochrome P450s in Caenorhabditis elegans are essential for the organization of eggshell, correct execution of meiosis and the polarization of embryo. Mech Dev 2009. May-Jun;126(5–6):382–393. doi: 10.1016/j.mod.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 21. Ha-Duong NT, Dijols S, Marques-Soares C, Minoletti C, Dansette PM, Mansuy D. Synthesis of sulfaphenazole derivatives and their use as inhibitors and tools for comparing the active sites of human liver cytochromes P450 of the 2C subfamily. J Med Chem 2001. October 25;44(22):3622–3631. [DOI] [PubMed] [Google Scholar]
- 22. Ha-Duong NT, Marques-Soares C, Dijols S, Sari MA, Dansette PM, Mansuy D. Interaction of new sulfaphenazole derivatives with human liver cytochrome p450 2Cs: structural determinants required for selective recognition by CYP 2C9 and for inhibition of human CYP 2Cs. Arch Biochem Biophys 2001. October 15;394(2):189–200. [DOI] [PubMed] [Google Scholar]
- 23. Basch PF. Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. The Journal of Parasitology 1981;67(2):179–85. [PubMed] [Google Scholar]
- 24. Lewis F. Schistosomiasis. Curr Protoc Immunol 2001. May;Chapter 19:Unit 19.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abagyan R, Totrov M, Kuznetsov D. ICM: A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry 1994;15(5):488–506. [Google Scholar]
- 26. Abagyan R, Batalov S, Cardozo T, Totrov M, Webber J, Zhou Y. Homology modeling with internal coordinate mechanics: deformation zone mapping and improvements of models via conformational search. Proteins 1997;Suppl 1:29–37. [DOI] [PubMed] [Google Scholar]
- 27. Marsden B, Abagyan R. SAD—a normalized structural alignment database: improving sequence-structure alignments. Bioinformatics 2004. October 12;20(15):2333–2344. [DOI] [PubMed] [Google Scholar]
- 28. Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol 1994. January 21;235(3):983–1002. [DOI] [PubMed] [Google Scholar]
- 29. Maiorov V, Abagyan R. Energy strain in three-dimensional protein structures. Fold Des 1998;3(4):259–269. [DOI] [PubMed] [Google Scholar]
- 30. Collins JJ 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol 2010. October 12;8(10):e1000509 doi: 10.1371/journal.pbio.1000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stefanic S, Dvorak J, Horn M, Braschi S, Sojka D, Ruelas DS, et al. RNA interference in Schistosoma mansoni schistosomula: selectivity, sensitivity and operation for larger-scale screening. PLoS Negl Trop Dis 2010. October 19;4(10):e850 doi: 10.1371/journal.pntd.0000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toh S.. Haem Biosynthesis and Uptake in Schistosoma mansoni School of Veterinary Science, The University of Queensland; 2014. [Google Scholar]
- 33. Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel "gene expression's CT difference" formula. J Mol Med (Berl). 2006;84(11):901–10. [DOI] [PubMed] [Google Scholar]
- 34. Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol 1993;12(1):1–51. [DOI] [PubMed] [Google Scholar]
- 35. Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, et al. P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 1996;6(1):1–42. [DOI] [PubMed] [Google Scholar]
- 36. Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol 2000;1(6):REVIEWS3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kemper B. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol Appl Pharmacol 2004. September 15;199(3):305–315. [DOI] [PubMed] [Google Scholar]
- 38. Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, et al. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry 2003. August 12;42(31):9335–9345. [DOI] [PubMed] [Google Scholar]
- 39. You H, Gobert GN, Duke MG, Zhang W, Li Y, Jones MK, et al. The insulin receptor is a transmission blocking veterinary vaccine target for zoonotic Schistosoma japonicum. Int J Parasitol 2012. August;42(9):801–807. doi: 10.1016/j.ijpara.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 40. Turman CM, Hatley JM, Ryder DJ, Ravindranath V, Strobel HW. Alternative splicing within the human cytochrome P450 superfamily with an emphasis on the brain: The convolution continues. Expert Opin Drug Metab Toxicol 2006. June;2(3):399–418. [DOI] [PubMed] [Google Scholar]
- 41. Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 2004. January;14(1):1–18. [DOI] [PubMed] [Google Scholar]
- 42. Pakharukova MY, Ershov NI, Vorontsova EV, Katokhin AV, Merkulova TI, Mordvinov VA. Cytochrome P450 in fluke Opisthorchis felineus: identification and characterization. Mol Biochem Parasitol 2012. February;181(2):190–194. doi: 10.1016/j.molbiopara.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 43. Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 1995. January 15;3(1):41–62. [DOI] [PubMed] [Google Scholar]
- 44. Hatae T, Hara S, Yokoyama C, Yabuki T, Inoue H, Ullrich V, et al. Site-directed mutagenesis of human prostacyclin synthase: Alteration of Cys441 of the Cys-pocket, and Glu347 and Arg350 of the EXXR motif. FEBS Lett 1996. July 8;389(3):268–272. [DOI] [PubMed] [Google Scholar]
- 45. Rupasinghe S, Schuler MA, Kagawa N, Yuan H, Lei L, Zhao B, et al. The cytochrome P450 gene family CYP157 does not contain EXXR in the K-helix reducing the absolute conserved P450 residues to a single cysteine. FEBS Lett 2006. November 27;580(27):6338–6342. [DOI] [PubMed] [Google Scholar]
- 46. Girardini JE, Khayath N, Amirante A, Dissous C, Serra E. Schistosoma mansoni: ferredoxin-NADP(H) oxidoreductase and the metabolism of reactive oxygen species. Exp Parasitol 2005. June;110(2):157–161. [DOI] [PubMed] [Google Scholar]
- 47. Girardini JE, Dissous C, Serra E. Schistosoma mansoni ferredoxin NADP(H) oxidoreductase and its role in detoxification. Mol Biochem Parasitol 2002. Sep-Oct;124(1–2):37–45. [DOI] [PubMed] [Google Scholar]
- 48. Abdel Baset H, O'Neill GP, Ford-Hutchinson AW. Characterization of arachidonic-acid-metabolizing enzymes in adult Schistisoma mansoni. Mol Biochem Parasitol 1995. July;73(1–2):31–41. [DOI] [PubMed] [Google Scholar]
- 49. Angeli V, Faveeuw C, Roye O, Fontaine J, Teissier E, Capron A, et al. Role of the parasite-derived prostaglandin D2 in the inhibition of epidermal Langerhans cell migration during schistosomiasis infection. J Exp Med 2001. May 21;193(10):1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fusco AC, Salafsky B, Kevin MB. Schistosoma mansoni: eicosanoid production by cercariae. Exp Parasitol 1985. February;59(1):44–50. [DOI] [PubMed] [Google Scholar]
- 51. Nevhutalu PA, Salafsky B, Haas W, Conway T. Schistosoma mansoni and Trichobilharzia ocellata: comparison of secreted cercarial eicosanoids. J Parasitol 1993. February;79(1):130–133. [PubMed] [Google Scholar]
- 52. Nirde P, De Reggi ML, Capron A. Fundamental aspects and potential roles of ecdysteroids in schistosomes an update overview. J Chem Ecol 1986. August;12(8):1863–1884. doi: 10.1007/BF01022389 [DOI] [PubMed] [Google Scholar]
- 53. Salafsky B, Fusco AC. Schistosoma mansoni: a comparison of secreted vs nonsecreted eicosanoids in developing schistosomulae and adults. Exp Parasitol 1987. December;64(3):361–367. [DOI] [PubMed] [Google Scholar]
- 54. Mebius MM, van Genderen PJ, Urbanus RT, Tielens AG, de Groot PG, van Hellemond JJ. Interference with the host haemostatic system by schistosomes. PLoS Pathog 2013;9(12):e1003781 doi: 10.1371/journal.ppat.1003781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Da'dara A, Skelly PJ. Manipulation of vascular function by blood flukes? Blood Rev 2011. July;25(4):175–179. doi: :10.1016/j.blre.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Silveira AM, Friche AA, Rumjanek FD. Transfer of [14C] cholesterol and its metabolites between adult male and female worms of Schistosoma mansoni. Comp Biochem Physiol B 1986;85(4):851–857. [DOI] [PubMed] [Google Scholar]
- 57. Briggs MH. Metabolism of steroid hormones by schistosomes. Biochim Biophys Acta 1972. November 30;280(3):481–485. [DOI] [PubMed] [Google Scholar]
- 58. Correia da Costa JM, Vale N, Gouveia MJ, Botelho MC, Sripa B, Santos LL, et al. Schistosome and liver fluke derived catechol-estrogens and helminth associated cancers. Front Genet 2014. December 23;5:444 doi: 10.3389/fgene.2014.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Romero R, Bueno D. Disto-proximal regional determination and intercalary regeneration in planarians, revealed by retinoic acid induced disruption of regeneration. Int J Dev Biol 2001. June;45(4):669–673. [PubMed] [Google Scholar]
- 60. Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB J 1996. July;10(9):993–1001. [DOI] [PubMed] [Google Scholar]
- 61. Freebern WJ, Osman A, Niles EG, Christen L, LoVerde PT. Identification of a cDNA encoding a retinoid X receptor homologue from Schistosoma mansoni. Evidence for a role in female-specific gene expression. J Biol Chem 1999. February 19;274(8):4577–4585. [DOI] [PubMed] [Google Scholar]
- 62. de Mendonca RL, Escriva H, Bouton D, Zelus D, Vanacker JM, Bonnelye E, et al. Structural and functional divergence of a nuclear receptor of the RXR family from the trematode parasite Schistosoma mansoni. Eur J Biochem 2000. June;267(11):3208–3219. [DOI] [PubMed] [Google Scholar]
- 63. Fantappie MR, Freebern WJ, Osman A, LaDuca J, Niles EG, LoVerde PT. Evaluation of Schistosoma mansoni retinoid X receptor (SmRXR1 and SmRXR2) activity and tissue distribution. Mol Biochem Parasitol 2001. June;115(1):87–99. [DOI] [PubMed] [Google Scholar]
- 64. Qiu C, Fu Z, Shi Y, Hong Y, Liu S, Lin J. A retinoid X receptor (RXR1) homolog from Schistosoma japonicum: its ligand-binding domain may bind to 9-cis-retinoic acid. Mol Biochem Parasitol 2013. March;188(1):40–50. doi: 10.1016/j.molbiopara.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 65. Torpier G, Hirn M, Nirde P, De Reggi M, Capron A. Detection of ecdysteroids in the human trematode, Schistosoma mansoni. Parasitology 1982. February;84(1):123–130. [DOI] [PubMed] [Google Scholar]
- 66. Foster JM, Mercer JG, Rees HH. Analysis of ecdysteroids in the trematodes, Schistosoma mansoni and Fasciola hepatica. Trop Med Parasitol 1992. December;43(4):239–244. [PubMed] [Google Scholar]
- 67. Nirde P, Torpier G, De Reggi ML, Capron A. Ecdysone and 20 hydroxyecdysone: new hormones for the human parasite schistosoma mansoni. FEBS Lett 1983. January 24;151(2):223–227. [DOI] [PubMed] [Google Scholar]
- 68. Shiff CJ, Dossaji SF. Ecdysteroids as regulators of host and parasite interactions: a study of interrelationships between Schistosoma mansoni and the host snail, Biomphalaria glabrata. Trop Med Parasitol 1991. March;42(1):11–16. [PubMed] [Google Scholar]
- 69. De Mendonca RL, Bouton D, Bertin B, Escriva H, Noel C, Vanacker JM, et al. A functionally conserved member of the FTZ-F1 nuclear receptor family from Schistosoma mansoni. Eur J Biochem 2002. November;269(22):5700–5711. [DOI] [PubMed] [Google Scholar]
- 70. Wu W, Tak EY, LoVerde PT. Schistosoma mansoni: SmE78, a nuclear receptor orthologue of Drosophila ecdysone-induced protein 78. Exp Parasitol 2008. June;119(2):313–318. doi: 10.1016/j.exppara.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 71. Abdulla MH, Ruelas DS, Wolff B, Snedecor J, Lim KC, Xu F, et al. Drug discovery for schistosomiasis: hit and lead compounds identified in a library of known drugs by medium-throughput phenotypic screening. PLoS Negl Trop Dis 2009. July 14;3(7):e478 doi: 10.1371/journal.pntd.0000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moore DV, Sandground JH. The relative egg producing capacity of Schistosoma mansoni and Schistosoma japonicum. Am J Trop Med Hyg 1956. September;5(5):831–840. [DOI] [PubMed] [Google Scholar]
- 73. Pellegrino J, Coelho PM. Schistosoma mansoni: wandering capacity of a worm couple. J Parasitol 1978. February;64(1):181–182. [PubMed] [Google Scholar]
- 74. Jurberg AD, Goncalves T, Costa TA, de Mattos AC, Pascarelli BM, de Manso PP, et al. The embryonic development of Schistosoma mansoni eggs: proposal for a new staging system. Dev Genes Evol 2009. May;219(5):219–234. doi: 10.1007/s00427-009-0285-9 [DOI] [PubMed] [Google Scholar]
- 75. Mathieson W, Wilson RA. A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: the miracidium, hatch fluid and secretions. Int J Parasitol 2010. April;40(5):617–628. doi: 10.1016/j.ijpara.2009.10.014 [DOI] [PubMed] [Google Scholar]
- 76. Ashton PD, Harrop R, Shah B, Wilson RA. The schistosome egg: development and secretions. Parasitology 2001. March;122(Pt 3):329–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The full-length sequence of Schistosoma mansoni cytochrome P450 was submitted to GenBank and assigned the accession number KT072747.