Abstract
Polarized assembly of actin filaments forms the basis of actin-based motility and is regulated both spatially and temporally. Cells use a variety of mechanisms by which intrinsically slower processes are accelerated, and faster ones decelerated, to match rates observed in vivo. Here we discuss how kinetic studies of individual reactions and cycles that drive actin remodeling have provided a mechanistic and quantitative understanding of such processes. We specifically consider key barbed-end regulators such as capping protein and formins as illustrative examples. We compare and contrast different kinetic approaches, such as the traditional pyrene-polymerization bulk assays, as well as more recently developed single-filament and single-molecule imaging approaches. Recent development of novel biophysical methods for sensing and applying forces will in future allow us to address the very important relationship between mechanical stimulus and kinetics of actin-based motility.
INTRODUCTION
Motile processes develop on time scales of 1 s to several tens of seconds, which reflects the range of relevant kinetic parameters governing intracellular actin assembly dynamics. The chemotactic ability of a cell to respond rapidly to environmental changes depends entirely on the rapid remodeling of its actin cytoskeleton (Carlier et al., 2015). By means of kinetic regulation, intrinsically slower processes are accelerated, and faster ones decelerated, to match rates observed in vivo. Identifying these kinetic mechanisms is therefore a principal step in reconstituting motile processes from individual cell components and mathematical modeling of cellular behavior for gaining a quantitative understanding of biological processes (Shekhar et al., 2014).
A simple example of kinetic regulation of actin dynamics is the 100-fold difference in rate of turnover/treadmilling of actin filaments in vitro compared with the in vivo rate in motile processes. Similar to the acceleration of the rate-limiting step in the ATPase cycle of myosin by actin (Lymn and Taylor, 1971), actin-depolymerizing factor (ADF)/cofilin enhances pointed-end depolymerization, which is the rate-limiting step of the filament turnover cycle. ADF cooperatively binds to and destabilizes actin–actin bonds in ADP–F-actin, resulting in enhanced disassembly of ADP–F‑actin from pointed ends (Figure 1). ADF/cofilin thus establishes a larger stationary pool of polymerizable ATP–actin monomers (CSS), leading to higher monomer flux associating to barbed ends (k+B.CSS, where k+B is the association rate constant of actin monomers at the barbed end and CSS is the steady-state actin monomer concentration) and hence faster protrusion rates. The destabilization of actin–actin bonds also facilitates filament severing (Maciver et al., 1991). Note, however, that severing by itself (e.g., mediated by sonic vibration or by a potent severer like Cordon-bleu) does not affect the value of CSS. If a kinetic screen for a protein-enhancing treadmilling had been designed before ADF/cofilin’s discovery, ADF/cofilin would certainly have been found (Le Clainche and Carlier, 2008). It has recently been shown that ADF further enhances filament disassembly by synergizing with other factors like Aip1 (Nadkarni and Brieher, 2014; Gressin et al., 2015), Coronin (Mikati et al., 2015), and Twinfilin and Srv2/CAP (Johnston et al., 2015).
FIGURE 1:
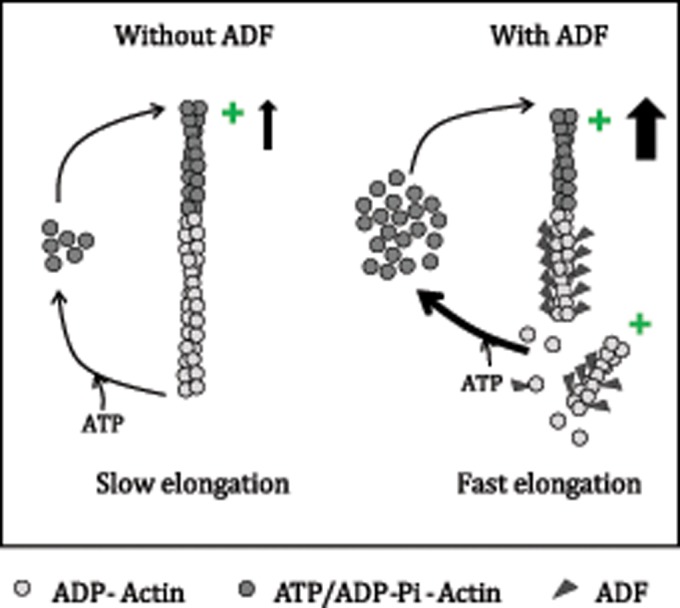
ADF enhances filament turnover rate. ADF enhances the rate-limiting of step of filament depolymerization during the treadmilling cycle. It does so by cooperatively binding the ADP–F-actin subunits and enhancing their disassembly at pointed ends by destabilizing actin–actin bonds in the filament. Similarly, ADF enhances spontaneous filament fragmentation. As a result of enhanced depolymerization, ADF enhances the stationary pool of monomeric ATP–actin, leading to a faster elongation rate.
Other examples of differences between in vivo and in vitro rates include dissociation of capping protein (CP) from barbed ends, which is intrinsically very slow but three orders of magnitude faster in lamellipodia (Miyoshi et al., 2006), consistent with a lowered affinity. This was demonstrated by the discrepancy between apparent Kd (∼100 nM) of Dictyostelium CP in whole-cell extracts and 100-fold lower Kd for purified Dictyostelium CP (Schafer et al., 1996). Similarly, formins exhibit long dwell times at the barbed ends in vitro, resulting in much longer filaments than observed in formin-mediated cellular processes. We will show examples in which kinetic control of the duration of formin and CP residence at the barbed end is elicited either by an allosteric mechanism or competition between various actin-binding motifs present in a variety of proteins.
The regulation of rates at the single-filament level has profound implications for defining the functional diversity of filament networks. Coordinated turnover of various actin arrays in the same cell suggests the existence of timers, phasing kinetic steps, and retroactive loops. It is therefore very important to study the kinetics of individual reactions before exploring how they are integrated into more complex cycles. Here we present appropriate kinetic approaches for analysis of the mechanisms that govern reactivity of actin filament barbed ends, comparing their respective strengths and drawbacks and providing a few illustrative cases. We also discuss the role of kinetics in quantitative understanding of motile processes.
EXAMPLES OF PROCESSES THAT NEED ACCELERATION OR DECELERATION
Some intrinsic reactions occurring at the barbed end of an actin filament are so slow that their kinetic up-regulation must occur to explain faster dynamics seen in vivo. For example capping protein binds barbed ends with very high affinity, dissociating from barbed ends with half-life of ∼25 min in vitro (Schafer et al., 1996). Long dwell times can be both an advantage and a disadvantage. In the bulk cytoplasm and in quiescent nonmotile cells, stable capping prevents unproductive energy consumption due to actin monomer–polymer exchange. In motile regions of the cell, however, a more dynamic interaction of CP with barbed ends is required to allow efficient growth of dendritic filament arrays.
A search for cellular factors to enhance dissociation of CP from barbed ends first led to polyphosphoinositides like phosphatidylinositol 4,5-bisphosphate (Schafer et al., 1996). Recently a class of proteins referred to as “uncappers” have been shown to rapidly release CP from capped filaments. By allosterically binding CP bound to the barbed end, uncappers reduce CP’s affinity for the barbed end, thus enhancing its dissociation (Figure 2a). Proteins containing the uncapper CapZIP motifs include CARMIL, CIN85, Duboraya, and FAM21. These proteins act in a site‑directed manner (Fujiwara et al., 2014) in close association with machineries assembling branched filaments with Arp2/3 complex.
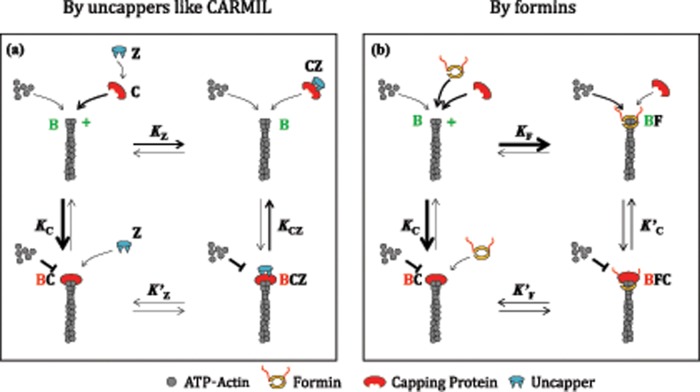
FIGURE 2:
Two mechanisms for rapid uncapping of CP-capped barbed ends. (a) Scheme 1: uncapping by uncappers. Filament barbed ends (B) bind CP (C) with high affinity (KC = 0.1 nM). CP makes a complex, CZ, with CapZIP (Z), which also caps barbed ends, albeit with affinity lower than CP (KCZ = 38 nM). CP dissociates very slowly from barbed ends, k-C = 0.0003 s−1, whereas CZ dissociates much more rapidly, kB-CZ = 0.095 s−1 (Fujiwara et al., 2010). Although KCZ was not experimentally determined, since the other equilibrium constants were measured, KCZ was calculated from detailed balance (KCK′Z = KZKCZ). At low concentration of CP, addition of Z to capped filaments (BC) leads to formation of a transient BCZ complex, followed by dissociation of CZ, leaving free uncapped barbed ends. At higher CP concentrations, adding Z leads to formation of CZ in amounts sufficient to bind barbed ends, and barbed ends stay capped by CZ (BCZ) in a more dynamic equilibrium than by C alone. Red indicates paused state, and green indicates elongating state. Arrow thickness signifies the magnitude of the reaction rate. (b) Scheme 2: uncapping by a barbed-end tracker like formin. Barbed ends (B) bind CP (C) and formin (F) with high affinity and slow dissociation rates. Both proteins can be bound simultaneously in the ternary complex BFC, with enhanced dissociation rates of both F (k′-F) and C (k′-C) within this complex. On addition of either C to BF or F to BC, the distribution of conformational states after transient formation of BFC depends on the relative values of k′-F and k′-C (Shekhar et al., 2015). Note that in the standard mutual exclusion scheme (direct competition) only B, BC, and BF states exist.
Another recently discovered uncapping mechanism is via the formation of a transient ternary complex, in which CP and a barbed end–tracking protein simultaneously bind a barbed end. In the process, each of them lowers the other’s affinity, thus enhancing CP’s dissociation from the barbed end (Figure 2b). This form of uncompetitive inhibition, opposed to the mutually exclusive binding scheme, is kinetically effective in displacing CP by other barbed end trackers. Examples include uncapping of CP by WH2 domain–containing VopF and Enabled/vasodilator-stimulated phosphoprotein or of FH2 domain–containing formins (Pernier et al., 2013; Bombardier et al., 2015; Shekhar et al., 2015). In a reciprocal manner, CP can also displace formin from the barbed end. This recent discovery disproved the previously held view that CP and formin bind to barbed ends in a mutually exclusive manner (Zigmond et al., 2003; Moseley et al., 2004; Kovar et al., 2005; Bartolini et al., 2012). Formin-anchored filament elongation is essential in organelles like filopodia. However, formin detachment from filaments has to be accelerated to prevent uncontrolled elongation of filaments due to the long dwell times of formins on barbed ends. In yeast, Bud14 rapidly displaces formin Bnr1 from growing barbed ends (Chesarone et al., 2009), and Smy1 dampens elongation by interacting with the FH2 domain of formin Bnr1 (Chesarone-Cataldo et al., 2011). Similarly, CP association to a formin‑bound barbed end has recently been shown to accelerate formin dissociation from the barbed end (Bombardier et al., 2015; Shekhar et al., 2015). This reaction may underlie the reported filopodial regulation by CP (Sinnar et al., 2014).
Another unexpected and interesting case is the synergy between Formin2 and Spire in actin assembly. These two proteins should antagonize and compete at barbed ends. Spire uses its WH2 domains to cap barbed ends. Formin2 by itself is a poor nucleator of actin filament and associates to barbed ends unusually slowly. However, Formin2 nucleates efficiently in the presence of Spire. When Spire is bound to barbed ends, its exposed N-terminal KIND domain associates with the C-terminal tail of Formin2, which allows fast recruitment of Formin2 at barbed ends and immediate onset of processive assembly coupled to displacement of Spire from barbed ends (Montaville et al., 2014). The group of Bruce Goode has reported other examples of such a synergy between a nucleation-promoting factor and an elongator, including pairs of mDia1 and APC (Breitsprecher et al., 2012), Bud6 and formin Bni1 (Graziano et al., 2011).
Whereas some reactions need to be speeded up to match in vivo rates, others need to be slowed down. Proteins like CP or formins often associate extremely rapidly to filament barbed ends in a diffusion-limited manner. Their rate of association can be reduced by an inhibitor that binds in rapid equilibrium to barbed ends. Our recent experiments identified this function in the ubiquitous protein profilin. Profilin binds both G-actin (with high affinity) and F-actin (with lower affinity). Profilin binds barbed ends of ADP-F-actin filaments with Kd = 1–25 μM (Kinosian et al., 2002; Jegou et al., 2011) and enhances depolymerization (Bubb et al., 2003). Similar Kd values have been found for profilin binding to ATP and ADP-Pi barbed ends (Jegou et al., 2011), and a higher value of ∼225 μM was found for AMPPNP barbed ends (Courtemanche and Pollard, 2013). Thus profilin competes with CP and barbed end–tracking proteins (Pernier et al., 2016), thus slowing down their association to barbed ends.
BULK-SOLUTION KINETICS AND SINGLE- FILAMENT KINETICS: COMPLEMENTARY APPROACHES TO ADDRESS THE SAME QUESTIONS
Traditionally, bulk kinetic assays have been used for quantifying rate constants of interaction between two or more proteins. Commonly used bulk approaches include changes in light scattering, fluorescence intensity, and anisotropy. The behavior of all molecules is averaged out in the monitored output. The advantage of these methods is that rate constants are easily and rapidly derived, assuming that all molecules are identical. A commonly used assay for F-actin assembly exploits the 20-fold increase in fluorescence intensity of pyrenyl-labeled actin (Kouyama and Mihashi, 1981) or 7-chloro-4-nitrobenzeno-2-oxa-1,3-diazole–labeled actin (Detmers et al., 1981) associated with the transition from the G-actin to the F-actin state. The increase in fluorescence intensity is proportional to total polymer mass and therefore provides quantitative evaluation of time-dependent rate of assembly of G-actin into F-actin. Rate parameters can then be extracted using appropriate mathematical modeling of the assembly kinetics.
Notwithstanding its immense contributions to kinetics, this technique has certain limitations. First, some proteins, such as ADF or profilin, bind labeled actin with reduced affinity and may affect its fluorescence (Malm, 1984; Carlier et al., 1997). Second, because this method only measures the amount of polymerized actin, it is difficult to identify events like nucleation, annealing, and severing. The method applies to reactions in which F-actin is a soluble polymer. However, reaction rates may differ when the polymer is side bound or end anchored to a membrane. Third and most important, bulk-solution “self-averaging” approaches fall short of identifying molecular mechanisms that occur at the scale of individual filaments (like length fluctuations) or in analysis of vectorial/processive reactions. Their application is also limited when nonhomogeneous populations (or rare subpopulations) exist. In these cases, what was an advantage turns out to blur the real thing. Single-filament studies, which emerged in the cytoskeleton field 15 years ago, overcame these limitations using total internal reflection fluorescence microscopy for observing real-time branching of actin filaments (Amann and Pollard, 2001; Mahaffy and Pollard, 2006) and assembly dynamics at barbed and pointed ends (Kuhn and Pollard, 2005).
Over the years, single-filament kinetics has evolved into a choice tool to analyze how individual regulators regulate barbed ends. Either the presence of a regulator at the barbed end can be identified by its effect on the elongation rate or the protein can be directly labeled fluorescently and observed by single-molecule fluorescence approaches. Each approach has its strengths and limitations.
In the first approach, changes in filament elongation rate provide a kinetic probe to detect the binding of a ligand. These changes are used to characterize the underlying kinetic mechanisms. A detailed kinetic analysis at several ligand concentrations is required to establish whether the monitored change in growth rate is strictly coupled to association-dissociation of the ligand at filament ends or occurs in a subsequent isomerization step. Thus a wealth of information is derived regarding the molecular mechanism of interaction of the ligand with barbed ends. However, not all barbed end–binding proteins cause a drastic change in elongation rate, making their detection difficult. Examples include barbed end–tracking proteins such as VopF, which does not exhibit a detectable change in elongation rate but is detected by its uncapping activity (Pernier et al., 2013). The classical use of competitive inhibitors thus can reveal the interaction of a “mute” ligand with barbed ends. In addition, two proteins bound simultaneously to the barbed end might show the same phenotype as when only one of them is bound; for example, simultaneous binding of CP and formin to barbed ends arrests filament growth in the same manner as CP alone does (Shekhar et al., 2015).
In the second approach, single-molecule fluorescence imaging is used to visualize interaction of fluorescently labeled proteins with the filaments (Smith et al., 2013a). Direct observation of uncapping of CP-capped filaments by a labeled CARMIL fragment (Fujiwara et al., 2010) and measurement of filament branching kinetics by Arp2/3 (Smith et al., 2013a, b) have exploited this approach. However, it should be kept in mind that due to lack of single actin subunit–level resolution (resolution limited to 160 nm or 50 subunits), just visualizing a fluorescent spot at the end of a filament by itself does not necessarily mean that the protein is actually interacting at the barbed face of the last terminal subunits at the barbed end. The protein could actually be bound in a 50 subunits–range away from the end, on the side of the filament. In addition, fluorescence labeling by itself might affect the protein’s binding activity to actin. Finally, in single-molecule imaging, only low amounts of labeled protein can be used to avoid nonspecific adsorption of molecules to the coverslip, as well as to ensure that there is only one fluorescent molecule per unit diffraction‑limited detection volume (approximately nanomolar concentration; Loveland et al., 2012). These drawbacks limit its application. In standard open-flowcell setups, analysis of rapid reactions is prevented by the large dead time (tens of seconds) between perturbing the sample conditions and recording observations.
Microfluidics-assisted fluorescence microscopy has helped overcome majority of these limitations and facilitated a high-throughput study of actin kinetics, in particular elucidating the mechanism of inorganic phosphate release in ATP hydrolysis on F-actin (Jegou et al., 2011). First, the biochemical conditions to which the filaments are being exposed can be changed in <1 s dead time, at least 10-fold faster than in a traditional open-flowcell; second, growth rates can be monitored in a large range of ligand concentrations. The rapid simultaneous observation of a large filament population (n > 100) leads to straightforward and accurate evaluation of rate constants. Compared to single-molecule fluorescence imaging, this approach also allows working at higher concentrations, except with proteins that strongly adsorb to the surface and may artifactually bind filaments.
FUTURE OF SINGLE-FILAMENT KINETIC ASSAYS
Combining the microfluidics approach with multicolor single-molecule imaging should prove invaluable in the future. Transient exposure of filaments to high concentrations of labeled molecules will be possible, and the ability to wash out the unbound labeled molecules will eliminate the background of free labeled molecules. Several fluorescently labeled proteins may be monitored simultaneously (Smith et al., 2013b), taking in vitro systems ever closer to in vivo–like situation in which multiple proteins work together. To achieve this, improved passivation and labeling methodologies will have to be developed. More sophisticated designs of microfluidic devices will also be required. Data collected in complex schemes will foster novel modeling approaches that need quantitative assessments of rate constants. Along this line, a successful prediction of the spatiotemporal dynamics of filopodia was made using reaction rate constants of barbed-end regulators (Mogilner and Rubinstein, 2005).
The single-filament assays and bulk-solution assays allow the study of actin assembly dynamics at two extreme size scales. A novel approach might be found at the crossover of the two scales. Observing rare labeled filaments in solutions containing unlabeled filaments has been used to understand actin rheology (Kas et al., 1994; Murrell and Gardel, 2012). Inspired by this assay, the kinetic behavior of individual labeled filaments placed in a flow containing unlabeled actin and a cocktail of defined regulatory proteins is now at hand and would reveal how individual filaments behave when placed in in vivo mimicking conditions.
Single-filament assays have also enabled the study of mechanical properties at the scale of individual filaments (Jegou et al., 2013). Biophysical methods designed both to measure and apply forces in the pico- to nano-Newton range have renewed the interest in the mechanochemical basis of cell motility, allowing studies of force-dependent binding strengths. A number of actin-binding proteins interact with the sides of the filaments, either stabilizing (e.g., tropomyosin) or destabilizing (e.g., ADF/cofilin) the polymer. Most single-filament kinetic measurements have been done in the absence of load, on unstretched/uncompressed filaments. However, filaments in cells often grow under tension. Tensile forces might affect actin assembly, as well as the association/dissociation reactions of regulators with filaments. Specifically, the elongation rate of a formin-bound barbed end increases under a pulling force (Jegou et al., 2013). How the complexes dissociate upon application of force will provide insights into the molecular mechanism of complex formation: either a simple bimolecular reaction or a two-step process in which isomerization of a first low-affinity complex in rapid equilibrium strengthens the interaction. Corresponding slip bonds and catch bonds have been defined (Marshall et al., 2003), as well as catch–slip bonds (Sundd et al., 2011), which govern actin disassembly (Lee et al., 2013). Applying a pulling force on the filament increases the dissociation of formin from the barbed end (unpublished results). In contrast, applying a pulling force on cadherin–catenin complexes on filaments stabilizes the binding (Buckley et al., 2014). Tension generated in the actin cytoskeleton can have secondary indirect effects on the kinetics of interaction between filaments and other actin-binding proteins. For example, applying tension to an actin filament has been shown to cause reduction in severing by ADF (Hayakawa et al., 2011). As another example, tension generated by actomyosin stretches talin in focal adhesions, enhancing its binding to vinculin (Ciobanasu et al., 2014). In exploring how the binding kinetics of other important side-binding proteins such as tropomyosins or the formin processive walk are affected by mechanical strain on the filament, either tension or torque should reveal as-yet-unknown aspects of their binding mode to actin. Gelsolin and Spire, the actin filament cappers, bind the side of a filament, followed by filament severing and capping of the newly formed barbed end. It will be interesting to know whether applying tension on the filament affects association of gelsolin to the filament. Alternatively, filaments can be bent mechanically. Local filament curvature was found to affect Arp2/3 based filament branching (Risca et al., 2012).
In conclusion, a wealth of new information is expected to come from the application of novel and more extensive kinetic approaches to actin dynamics. Obvious consequences in the structural biology of actin and quantitative modeling of normal and pathological cell processes are anticipated.
Acknowledgments
We thank Guillaume Romet-Lemonne, Antoine Jégou, Christophe Le Clainche, Sonja Kühn, and Jeff Gelles for helpful discussions. We acknowledge the support of European Research Council Advanced Grant “forcefulactin” 249982 to M.-F. C.
Abbreviations used:
- ADF
actin depolymerizing factor
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- CapZIP
CapZ-interacting protein
- CARMIL
capping protein Arp2/3 myosin i linker
- CP
capping protein
- CSS
steady-state actin monomer concentration
- F-actin
filamentous actin
- FH2
formin-homology 2 domain
- G-actin
globular (monomeric) actin
- k+B
association rate constant of actin monomers at the barbed end
- Kd
equilibrium dissociation constant
- WH2
WASP-homology 2 domain.
Footnotes
REFERENCES
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci USA. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–4040. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardier JP, Eskin JA, Jaiswal R, Corrêa IR, Xu M-Q, Gelles J, Goode BL. Single-molecule visualization of a formin-capping protein “decision complex” at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Yarmola EG, Gibson BG, Southwick FS. Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J Biol Chem. 2003;278:24629–24635. doi: 10.1074/jbc.M302796200. [DOI] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pernier J, Montaville P, Shekhar S, Kuhn S, Cytoskeleton Dynamics and Motility Group Control of polarized assembly of actin filaments in cell motility. Cell Mol Life Sci. 2015;72:3051–3067. doi: 10.1007/s00018-015-1914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone-Cataldo M, Guerin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell. 2011;21:217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C, Faivre B, Le Clainche C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat Commun. 2014;5:3095. doi: 10.1038/ncomms4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. Interaction of profilin with the barbed end of actin filaments. Biochemistry. 2013;52:6456–6466. doi: 10.1021/bi400682n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers P, Weber A, Elzinga M, Stephens RE. 7-Chloro-4-nitrobenzeno-2-oxa-1,3-diazole actin as a probe for actin polymerization. J Biol Chem. 1981;256:99–105. [PubMed] [Google Scholar]
- Fujiwara I, Remmert K, Hammer JA., 3rd Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem. 2010;285:2707–2720. doi: 10.1074/jbc.M109.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Remmert K, Piszczek G, Hammer JA. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc Natl Acad Sci USA. 2014;111:E1970–E1979. doi: 10.1073/pnas.1313738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressin L, Guillotin A, Guerin C, Blanchoin L, Michelot A. Architecture dependence of actin filament network disassembly. Curr Biol. 2015;25:1437–1447. doi: 10.1016/j.cub.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegou A, Carlier MF, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- Jegou A, Niedermayer T, Orban J, Didry D, Lipowsky R, Carlier MF, Romet-Lemonne G. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AB, Collins A, Goode BL. High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat Cell Biol. 2015;17:1504–1511. doi: 10.1038/ncb3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas J, Strey H, Sackmann E. Direct imaging of reptation for semiflexible actin filaments. Nature. 1994;368:226–229. doi: 10.1038/368226a0. [DOI] [PubMed] [Google Scholar]
- Kinosian HJ, Selden LA, Gershman LC, Estes JE. Actin filament barbed end elongation with nonmuscle MgATP-actin and MgADP-actin in the presence of profilin. Biochemistry. 2002;41:6734–6743. doi: 10.1021/bi016083t. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Lee CY, Lou J, Wen KK, McKane M, Eskin SG, Ono S, Chien S, Rubenstein PA, Zhu C, McIntire LV. Actin depolymerization under force is governed by lysine 113:glutamic acid 195-mediated catch-slip bonds. Proc Natl Acad Sci USA. 2013;110:5022–5027. doi: 10.1073/pnas.1218407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Habuchi S, Walter JC, van Oijen AM. A general approach to break the concentration barrier in single-molecule imaging. Nat Methods. 2012;9:987–992. doi: 10.1038/nmeth.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm B. Chemical modification of Cys-374 of actin interferes with the formation of the profilactin complex. FEBS Lett. 1984;173:399–402. doi: 10.1016/0014-5793(84)80813-3. [DOI] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Mikati MA, Breitsprecher D, Jansen S, Reisler E, Goode BL. Coronin enhances actin filament severing by recruiting cofilin to filament sides and altering F-actin conformation. J Mol Biol. 2015;427:3137–3147. doi: 10.1016/j.jmb.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol. 2014;12:e1001795. doi: 10.1371/journal.pbio.1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell MP, Gardel ML. F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc Natl Acad Sci USA. 2012;109:20820–20825. doi: 10.1073/pnas.1214753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni AV, Brieher WM. Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr Biol. 2014;24:2749–2757. doi: 10.1016/j.cub.2014.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernier J, Orban J, Avvaru BS, Jegou A, Romet-Lemonne G, Guichard B, Carlier MF. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat Struct Mol Biol. 2013;20:1069–1076. doi: 10.1038/nsmb.2639. [DOI] [PubMed] [Google Scholar]
- Pernier J, Shekhar S, Jegou A, Guichard B, Carlier M-F. Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching and motility. Dev Cell. 2016 doi: 10.1016/j.devcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risca VI, Wang EB, Chaudhuri O, Chia JJ, Geissler PL, Fletcher DA. Actin filament curvature biases branching direction. Proc Natl Acad Sci USA. 2012;109:2913–2918. doi: 10.1073/pnas.1114292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, Kerleau M, Kuhn S, Pernier J, Romet-Lemonne G, Jegou A, Carlier MF. Formin and capping protein together embrace the actin filament in a “ménage à trois.”. Nat Commun. 2015;6:8730. doi: 10.1038/ncomms9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, Zhu L, Mazutis L, Sgro AE, Fai TG, Podolski M. Quantitative biology: where modern biology meets physical sciences. Mol Biol Cell. 2014;25:3482–3485. doi: 10.1091/mbc.E14-08-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnar SA, Antoku S, Saffin JM, Cooper JA, Halpain S. Capping protein is essential for cell migration in vivo and for filopodial morphology and dynamics. Mol Biol Cell. 2014;25:2152–2160. doi: 10.1091/mbc.E13-12-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Daugherty-Clarke K, Goode BL, Gelles J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc Natl Acad Sci USA. 2013a;110:1285–1290. doi: 10.1073/pnas.1211164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife. 2013b;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundd P, Pospieszalska MK, Cheung LS, Konstantopoulos K, Ley K. Biomechanics of leukocyte rolling. Biorheology. 2011;48:1–35. doi: 10.3233/BIR-2011-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]