Tousled-like kinase is required for signaling between polar cells and border cells in the Drosophila ovary, thus controlling their collective migration. Tlk knockdown in polar cells inhibits cytokine expression without affecting polar cell fate or viability. This study shows novel, cell type–specific functions for this ubiquitous nuclear protein.
Abstract
Collective cell migration is emerging as a major contributor to normal development and disease. Collective movement of border cells in the Drosophila ovary requires cooperation between two distinct cell types: four to six migratory cells surrounding two immotile cells called polar cells. Polar cells secrete a cytokine, Unpaired (Upd), which activates JAK/STAT signaling in neighboring cells, stimulating their motility. Without Upd, migration fails, causing sterility. Ectopic Upd expression is sufficient to stimulate motility in otherwise immobile cells. Thus regulation of Upd is key. Here we report a limited RNAi screen for nuclear proteins required for border cell migration, which revealed that the gene encoding Tousled-like kinase (Tlk) is required in polar cells for Upd expression without affecting polar cell fate. In the absence of Tlk, fewer border cells are recruited and motility is impaired, similar to inhibition of JAK/STAT signaling. We further show that Tlk in polar cells is required for JAK/STAT activation in border cells. Genetic interactions further confirmed Tlk as a new regulator of Upd/JAK/STAT signaling. These findings shed light on the molecular mechanisms regulating the cooperation of motile and nonmotile cells during collective invasion, a phenomenon that may also drive metastatic cancer.
INTRODUCTION
Collective cell migration contributes to normal development and disease, and communication among distinct cell types within a moving collective serves key functions during this process. For example, during development of the zebrafish lateral line, interactions between leading and trailing cells establish polarity within the collective that is essential for its directional movement (Dalle Nogare et al., 2014). Paracrine signaling between breast cancer cells and tumor-associated macrophages promotes their dissemination in vivo (Goswami et al., 2005).
Border cell migration in the Drosophila ovary is a well-developed and genetically tractable model for studying collective cell migration in vivo (Montell et al., 1992; Prasad et al., 2011; Montell et al., 2012). Fly ovaries are composed of egg chambers, which contain 16 germline cells surrounded by an epithelium of somatic follicle cells. Early in oogenesis, special follicle cells, the polar cells, develop at each pole of each egg chamber. Polar cells secrete the cytokine Unpaired (Upd; Silver et al., 2001; McGregor et al., 2002), which activates Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling in neighboring follicle cells. Four to eight anterior follicle cells with the highest STAT activity differentiate into border cells in stage 8 egg chambers. At stage 9, the border and polar cells detach as a cluster from the basal lamina surrounding the egg chamber and from neighboring follicle cells and move in between the nurse cells until they arrive at the border of the oocyte by late stage 9 or early stage 10 (Figure 1A).
FIGURE 1:
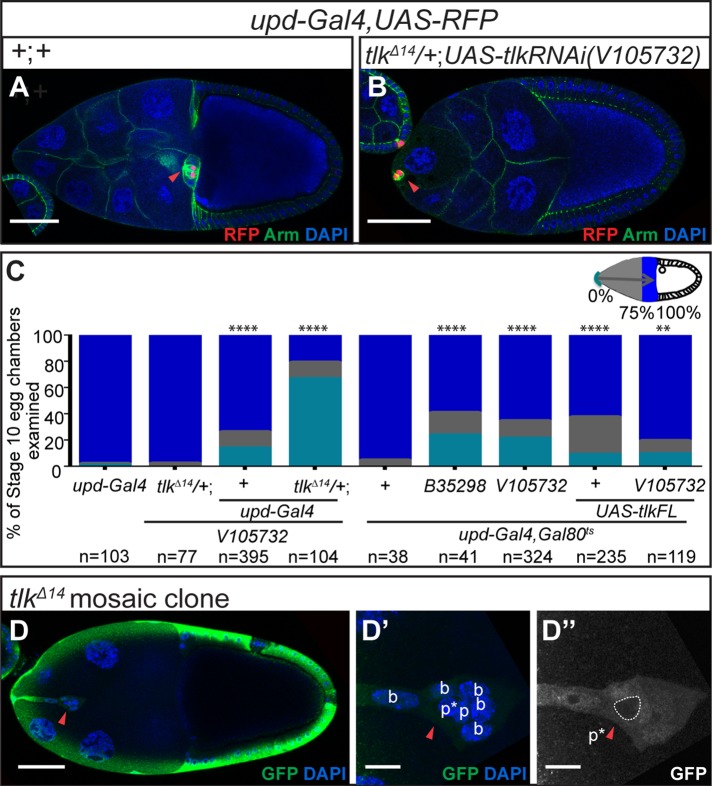
Effect of inhibition of the nuclear protein Tlk on border cell migration. (A, B) Confocal micrographs of stage 10 egg chambers of the indicated genotypes stained for Armadillo (Arm, green) and DAPI (blue). The positions of the border cell clusters are indicated with red arrowheads. (C) Histogram of the spatial distribution of border cell clusters in stage 10 egg chambers of the indicated genotypes. The RNAi line used was V105732. The migration path was divided into three parts. Complete migration to the oocyte is shown in royal blue. Gray indicates clusters that initiated but did not complete migration. Aqua indicates clusters that failed to initiate migration. (D–E′) Anti-Tlk antibody staining (red in D, D′′, and E; white in D′, D′′ insets, and E′). (D–D′′) Wild-type expression of Tlk during oogenesis. (D′′) Higher magnification of the boxed regions in D′. In the migrating border cell cluster, p labels polar cells, and b labels border cells. The insets show anterior polar cells at an early stage (left) and posterior polar cells at stage 9 (right). Red arrowheads indicate polar cells. (E, E′) Mosaic cluster containing wild-type (GFP-negative) cells and cells expressing UAS-tlkRNAi (V105732) driven by AyGal4 in FLP-OUT clones (GFP+, with white dashed line; see Materials and Methods for details). (F) Quantification of Tlk antibody staining in cells of the indicated genotypes. Data are presented as mean ± SD. (G) Quantification of border cell migration defect in stage 10 egg chambers from c306-Gal4, Gal80ts with or without UAS-tlkRNAi. (H, H′) A border cell cluster (red arrowheads) containing a tlk mutant border cell (b*) and polar cell (p*; GFP negative). (H′) Higher- magnification view of the border cell cluster. Scale bars, 50 μm (A, B, D, D′, H), 10 μm (D′′, H′), and 5 μm (E, E′). We adopted the Mann–Whitney U test to analyze the statistical significance of border cell migration defect (C, G), and used the t test for Tlk intensity quantification. ****p < 0.0001; n, number of egg chambers examined.
Border cell migration requires cooperation between immotile polar cells and motile border cells. Polar cells cannot migrate without the border cells (Han et al., 2000; Silver et al., 2001), and border cells fail to move in the absence of Upd secreted from polar cells (Silver et al., 2001). Upd/JAK/STAT signaling is not only necessary but also sufficient for motility. Ectopic expression of Upd or activated JAK is sufficient to specify ectopic border cells (Silver et al., 2004) and cause extra cells to migrate and invade the nurse cell cluster (Silver and Montell, 2001). Therefore regulation of Upd is the critical step in regulating epithelial follicle cell motility. Here we report an RNA interference (RNAi) screen for nuclear proteins required for border cell migration, which revealed a requirement for the gene encoding Tousled-like kinase (Tlk). Further studies show that Tlk is required in polar cells for proper Upd expression and JAK/STAT signaling activity without affecting polar cell viability or fate.
RESULTS AND DISCUSSION
Tlk, a new regulator of collective border cell migration
A transcriptional network containing multiple feedforward and feedback loops coordinates spatial and temporal control of border cell fate specification, differentiation, and migration (Bai et al., 2000; Liu et al., 2001; Schober et al., 2005; Starz-Gaiano et al., 2008; Jang et al., 2009; Yoon et al., 2011; Gunawan et al., 2013; Monahan et al., 2013). Owing to this extensive transcriptional control, we performed an RNAi screen of genes encoding nuclear proteins, using the GAL4/UAS system (Brand et al., 1993; see Materials and Methods). Tlk emerged as a candidate gene required for normal border cell development. In contrast to wild type (Figure 1A), tlk knockdown (KD) resulted in a severe migration defect (Figure 1, B and C). Whereas virtually all wild-type clusters reach the oocyte by stage 10, ∼70% of tlk KD border cell clusters failed to complete the migration at the same stage (Figure 1C).
Tlk is a conserved serine/threonine kinase required in mammalian cells for DNA repair, replication, transcription, and chromosome segregation (Li et al., 2007; De Benedetti, 2010; Ronald et al., 2013; Klimovskaia et al., 2014). In Drosophila, its only well-described function is to promote mitosis during embryogenesis (Carrera, Moshkin, et al., 2003; Li et al., 2009). However, border cells are postmitotic, indicating a different function.
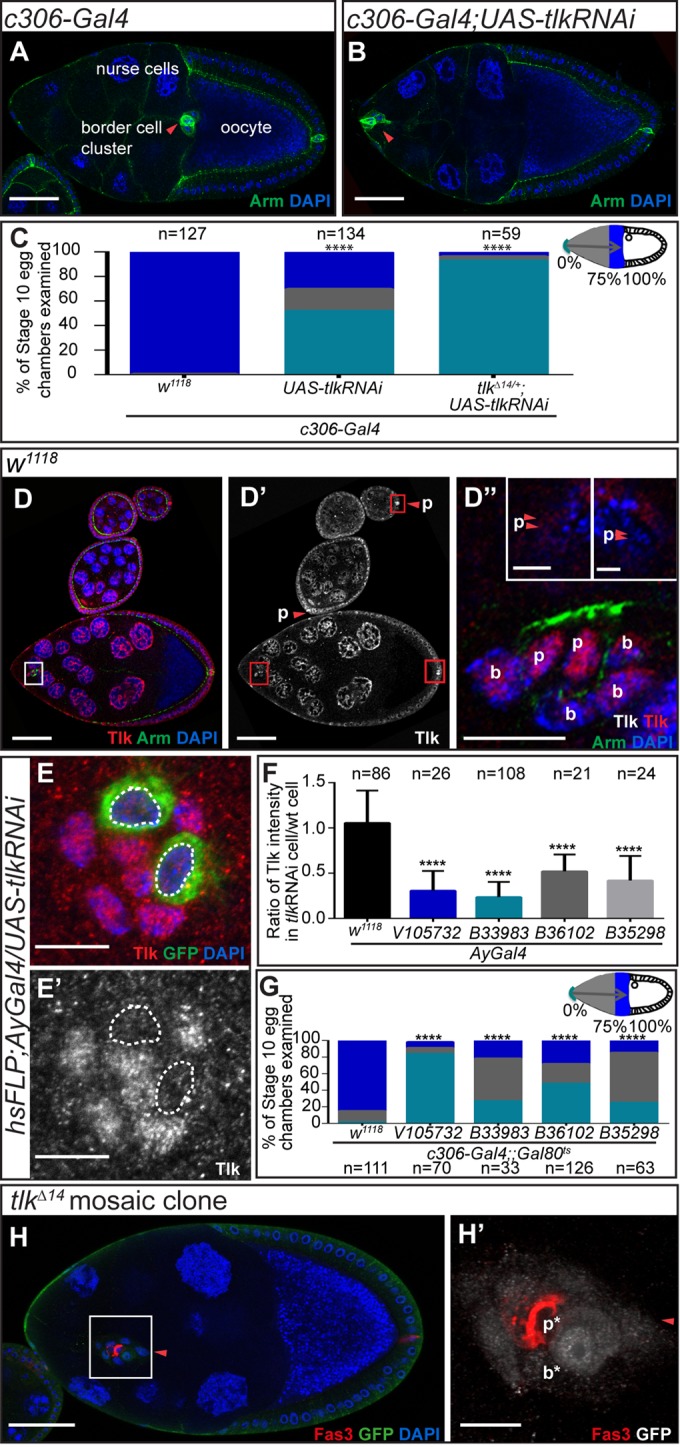
Using an anti-Tlk antibody (Carrera, Moshkin, et al., 2003), we detected Tlk in the nuclei of all egg chamber cells (Figure 1D), with enrichment in polar cells (Figure 1, D–D′′). Clonal analysis via the FLP-OUT technique (Ito et al., 1997) verified the effectiveness of tlk KD by the V105732 RNAi strain, from the Vienna Drosophila RNAi Center (VDRC; Vienna, Austria). Green fluorescent protein (GFP)–positive, tlk KD border cells exhibited a 70% reduction in Tlk staining compared with neighboring wild-type cells (Figure 1, E and F). We ruled out off-target effects associated with some RNAi fly strains from the VDRC (Green et al., 2014; Supplemental Figure S1A), using the published PCR-based diagnosis (see Materials and Methods).
Three additional RNAi fly strains from the Bloomington Drosophila Stock Center (BDSC; Bloomington, IN) strongly reduced Tlk protein (Figure 1F) and inhibited border cell migration (Figure 1G). We included the temperature-sensitive repressor tub-Gal80ts because these strains caused lethality without it. We grew the flies at 18°C and then shifted them to 31°C as adults. Many female adult progeny died, especially for line B33983. In those that survived, stage 10 egg chambers were relatively rare. Therefore, to evaluate border cell migration, we had to use conditions that did not cause the strongest possible Tlk KD.
We confirmed the migration phenotype using the tlkΔ14 mutant allele (Carrera, Moshkin, et al., 2003) in mosaic clones (Figure 1, H and H′). Approximately 40% (n = 78) of mosaic stage 10 egg chambers exhibited incomplete border cell migration, compared with ∼3% (n = 100) of controls of the same genotype without heat shock–induced mitotic recombination. Combining the tlkΔ14 heterozygous mutation with tlkRNAi (V105732) further aggravated the migration defect to ∼96% (Figure 1C). Taken together, these results reveal that Tlk is a new regulator of border cell migration.
Tlk is required in polar cells
To distinguish the cell type(s) in which Tlk functions, we first knocked down tlk (V105732) specifically in polar cells using upd-Gal4 and observed a significant migration defect (Figure 2, A–C). Combining tlk RNAi with a tlkΔ14 heterozygous mutation increased the severity of the migration defect from ∼30 to ∼80% (Figure 2C). Although overexpression of the full-length Tlk protein (tlkFL) in polar cells caused a mild defect on its own, it also significantly rescued the migration defect caused by tlk KD (Figure 2C). We confirmed this result with a second RNAi line. Although some of the tlk RNAi lines caused significant lethality, we were able to obtain sufficient stage 10 egg chambers with the B35298 line to confirm the phenotype (Figure 2C). Mosaic analysis with the tlkΔ14 mutant allele further confirmed that loss of tlk from polar cells inhibited border cell migration (Figure 2, D–D′′). Thus Tlk is essential in polar cells for border cell migration. Tlk KD in outer border cells using slbo-Gal4 also caused a milder migration defect (Supplemental Figure S1, B–D), suggesting multiple functions for Tlk; however, we focused on its function in the polar cells.
FIGURE 2:
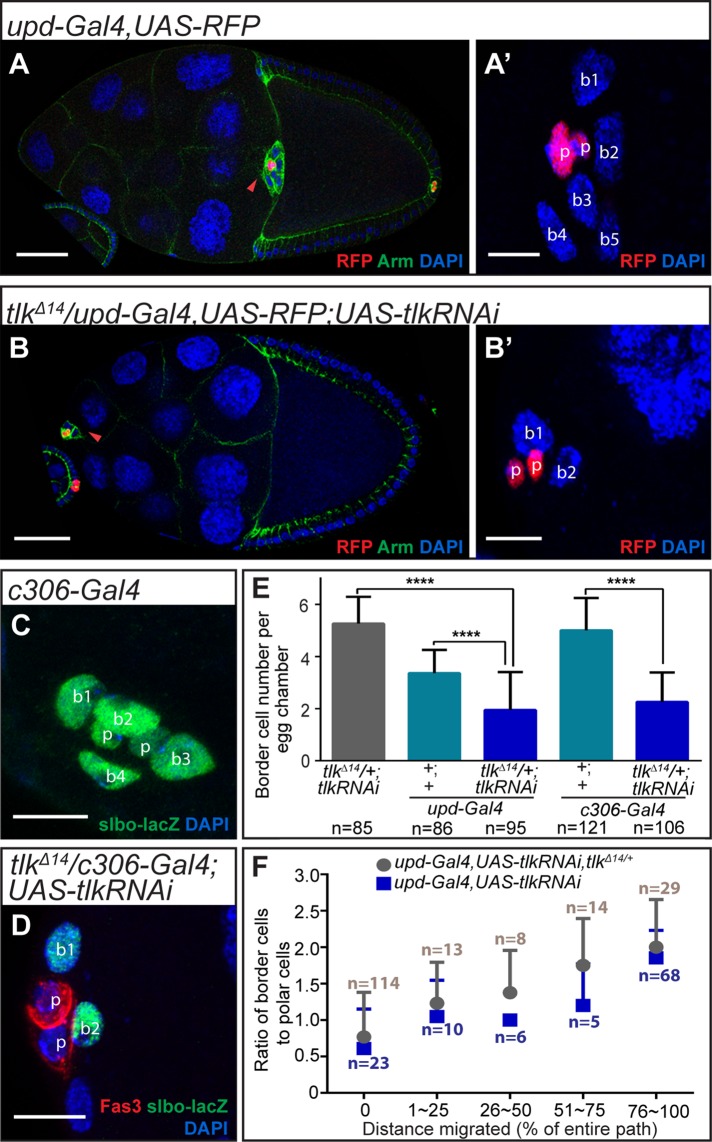
Tlk reduction in polar cells affects border cell migration. (A, B) Confocal micrographs of stage 10 egg chambers of the indicated genotypes. Red fluorescent protein (RFP; red) marks polar cells. (C) Quantification of migration defects in the indicated genotypes. Flies were fattened at 29°C (Gal4 alone) or 31°C (in the presence of Gal80ts ). The Mann–Whitney U test was used to analyze statistical significance of border cell migration defect. ****p < 0.0001, **p < 0.01; n, number of egg chambers examined. (D–D′′) A border cell cluster with one tlk mutant polar cell (GFP negative; p* with a white dashed circle) showing migration defect. (D′, D′′) Higher-magnification views. The polar cells are distinguished from border cells by their smaller nuclear size and central location in the cluster (D′). Red arrowheads indicate border cell clusters in A, B, and D–D′′. Scale bars, 50 μm (A, A′, D) and 10 μm (D′, D′′).
Tlk in polar cells is essential for border cell fate determination
Because polar cells determine the number of border cells and endow them with the ability to migrate, we investigated whether Tlk in polar cells was required to specify the normal number of border cells. Border cell clusters normally contain two polar cells and four to eight outer migratory cells (e.g., Figure 3, A and A′). In the tlkΔ14/+; UAS-tlkRNAi control, the average was 5.3 (Figure 3E). The upd-Gal4 transgene is inserted into the upd locus and causes a partial loss of function, so upd-Gal4, UAS-RFP clusters contain on average 3.4 border cells (Figure 3E). However, knocking down Tlk expression in polar cells using upd-Gal4, UAS-tlkRNAi in combination with tlkΔ14/+ further reduced the average border cell number to 1.9 (Figure 3, B, B′, and E). The border cell number was also reduced from an average of 5 in the c306-Gal4 control to an average of 2.3 when Tlk was knocked down with c306-Gal4 in a tlkΔ14/+ background (Figure 3, C–E). In wild-type clusters, the ratio of outer migratory border cells to polar cells is ∼2.5:1, and previous work suggested that the number of migratory border cells strongly correlates with the ability of the cluster to complete migration by stage 10 (Silver and Montell, unpublished data). Therefore the effect of tlk KD on migration was likely due to the reduction in border cell number. We confirmed that the ratio of outer migratory border cells to polar cells correlated with the extent of migration after polar cell KD of Tlk (Figure 3F). Complete or nearly complete migration was limited to clusters with >1.5 migratory cells/polar cell (Figure 3F).
FIGURE 3:
tlk KD in polar cells reduces border cell number. (A–D) Confocal micrographs showing border cell number (b1–b5). Polar cells (p) are distinguished from border cells by RFP (A–B′), smaller nuclear size, and lower slbo-lacZ intensity (C) or Fas3 expression (D). (A′, B′) Magnified views of border cell clusters in A and B, respectively. Border cells were identified by slbo-lacZ (C, D). (E) Quantification of border cell number per cluster in the indicated genotypes. ****p < 0.0001 determined using a t test. Data are presented as mean ± SD. (F) Comparison of the ratio of border cells to polar cells and cluster migration. The migration path was divided into five segments corresponding to different percentages of the entire posterior migration path. All of the clusters evaluated in this analysis contained two polar cells (see Materials and Methods for details). n, number of egg chambers examined. All flies were incubated at 29°C. Scale bars, 50 μm (A, B) and 10 μm (A′, B′, C, D).
tlk is not required for polar cell viability or differentiation
Because Tlk is required for cell viability during Drosophila embryogenesis (Carrera, Moshkin, et al., 2003), we tested whether it was required for viability of follicle cells. Of 142 anterior polar cells examined in stages 8–10 egg chambers, we observe no cleaved caspase-3 (cCasp-3) staining (Figure 4, A and A′, and Supplemental Table S1). Nor did tlk KD in polar cells affect the viability of border cells (Figure 4, A and A′, and Supplemental Table S1). In contrast, we observed frequent cCasp-3 staining in other follicle cells in homozygous mutant tlkΔ14 clones (Figure 4, B and C, and Supplemental Table S1). Thus, although Tlk is a ubiquitously expressed protein, it serves distinct functions in different cell types, and polar and border cells do not require Tlk for viability.
FIGURE 4:
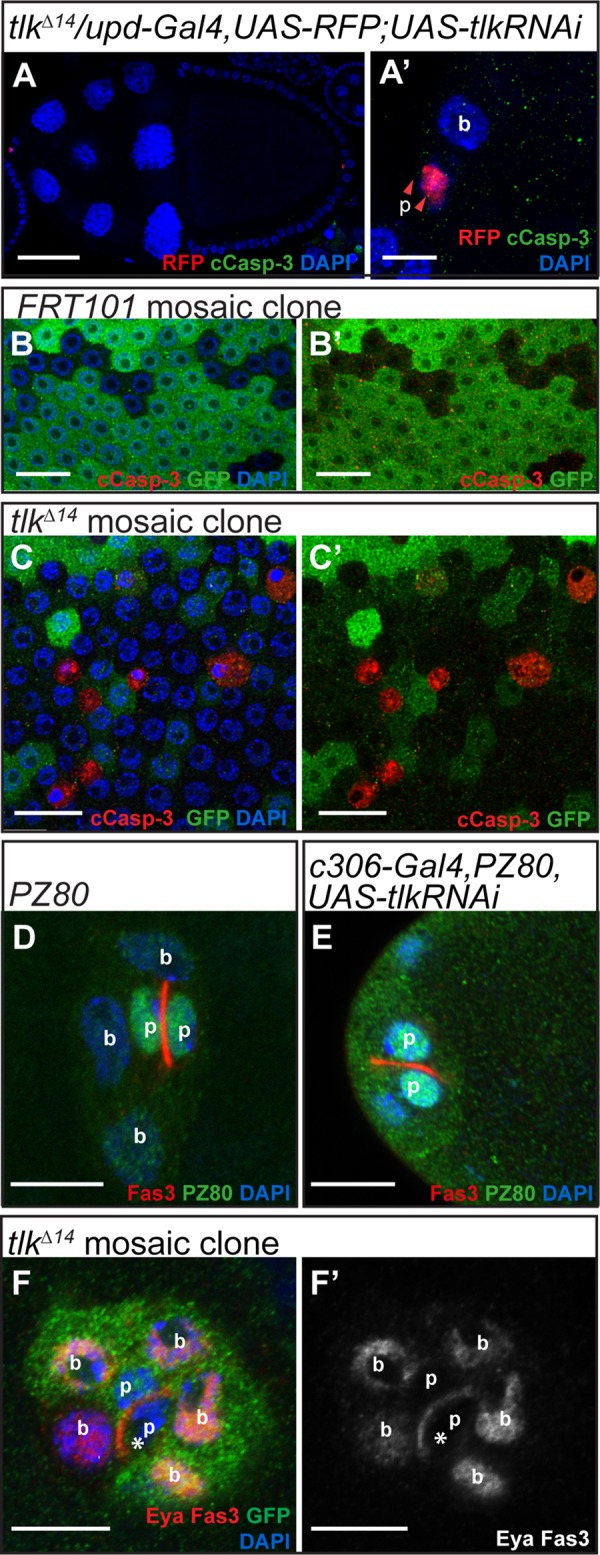
Polar cells lacking Tlk show normal viability and differentiation. Confocal micrographs of egg chambers of the indicated genotypes. (A, A′) Polar cells (p) lacking Tlk do not express cleaved caspase 3 (cCaps-3; green) at stage 10. (A′) Higher-magnification image. (B–C′) Mosaic clones in main body epithelial follicle cells. GFP-negative cells represent control cell clones (B, B′) or tlkΔ14 mutant cell clones (C, C′). Apoptosis signal is shown by anti–cCaps-3 signal (red). (D, E) Anti–b-gal staining (green) for the PZ80 enhancer trap insertion in otherwise wild-type (D) and tlk KD (E) polar cells. Fas3 is enriched in and Eya is absent from both wild- type and tlk KD polar cells (F, F′). p and b label polar and border cells, respectively. Asterisks label GFP-negative, tlk mutant polar cells in F and F′. Scale bars, 50 μm (A), 20 μm (B–C′), and 10 μm (A′, D–F′).
We then tested whether Tlk is required for polar cell differentiation, using polar cell markers. Eya is a key repressor of polar cell fate, and thus normally is not expressed in polar cells (Bai et al., 2002). Fasciclin3 (Fas3) and PZ80 (a lacZ enhancer trap inserted into the Fas3 locus) are normally restricted to polar cells after stage 6 (Ruohola et al., 1991; Karpen et al., 1992). We found that tlk KD polar cells exhibited normal patterns of Eya, Fas3, and PZ80 (Figure 4, D–F′). Thus Tlk is not required for polar cell fate, viability, or general differentiation.
Tlk in polar cells is required for Upd expression and JAK/STAT activation
The phenotype caused by tlk KD in polar cells resembled that of upd/jak/stat loss of function, as these mutations all cause defects in border cell number and migration (Silver et al., 2001; McGregor et al., 2002). Upd/JAK/STAT is also required for polar cell development, in particular for the apoptosis of extra polar cells (Borensztejn et al., 2013).
There are normally precisely two polar cells at each end of each egg chamber older than stage 5 (Supplemental Figure S2A); however, early in oogenesis, more than two polar cells frequently develop (Besse et al., 2003). Extra polar cells are eliminated by apoptosis during stages 4–5 under the control of the JAK/STAT pathway (Borensztejn et al., 2013). When upd is knocked down, more than two polar cells persist even after stage 5 (Supplemental Figure S2B; Borensztejn et al., 2013). We observed a similar phenotype after tlk inhibition in polar cells (Supplemental Figure S2, C and E). About half of the egg chambers examined (108 of 208) retained more than two polar cells. Furthermore, like upd KD (Borensztejn et al., 2013), tlk KD compromised the ability of the extra polar cells to recruit border cells. Whereas extra polar cells typically induce extra border cells (Liu et al., 1999; Bai et al., 2002; Ghiglione et al., 2002; Grammont et al., 2002; Borensztejn et al., 2013), the extra polar cells caused by tlk KD did not. Regardless of the number of polar cells, border cell number ranged from zero to three when tlk was knocked down in polar cells. The tlkΔ14 mutant allele caused the same phenotype in mosaic clones (Supplemental Figure S2, D and D′). Because extra polar cells develop early in oogenesis, by manipulating the timing of incubation at 29°C, we identified conditions in which KD of Tlk in polar cells caused only the border cell specification and migration phenotype without extra polar cells. In the studies of the role of Tlk on border cell specification and migration, we only analyzed clusters with the correct number of polar cells.
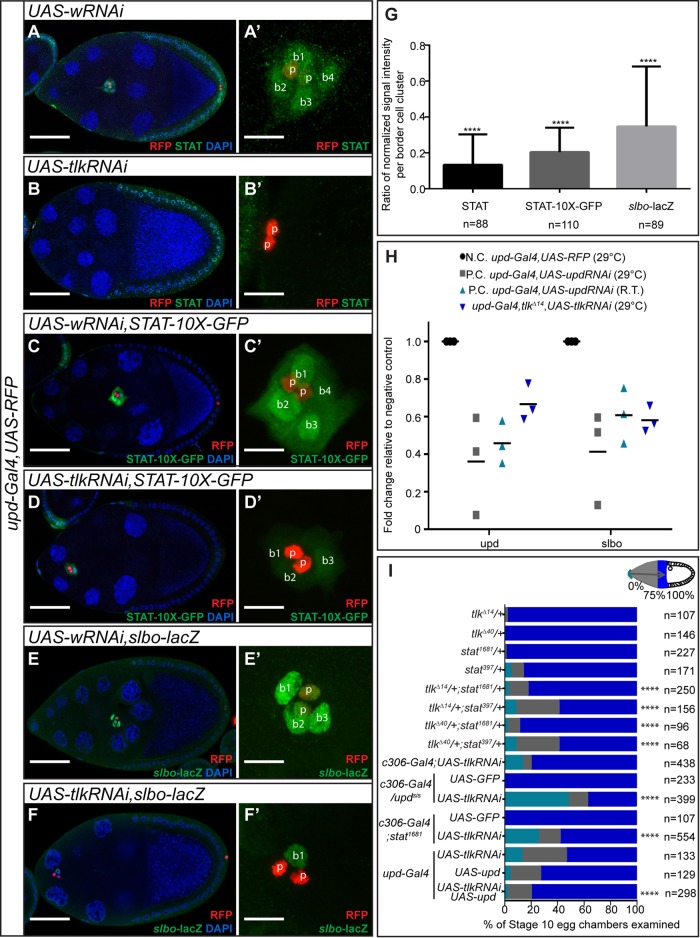
The phenotypic similarities led us to test whether Tlk in polar cells affects JAK/STAT activation in border cells, using three established reporters: the intensity of the nuclear-localized STAT protein (Silver et al., 2005) (Figure 5, A–B′), STAT-10X-GFP (Bach et al., 2007; Figure 5, C–D′), and slbo-lacZ (Silver et al., 2001; Figure 5, E–F′). We found that Tlk KD in polar cells significantly reduced all three markers of STAT activity (Figure 5G).
FIGURE 5:
Effect of Tlk KD on the Upd/JAK/STAT pathway. (A–F′) Confocal micrographs of egg chambers and border cell clusters. RFP labels polar cells. (A–B′) anti-STAT antibody staining (green), (C–D′) 10X-STAT-GFP (green), and (E–F′) anti–β-galactosidase antibody staining from slbo-lacZ (green). Scale bars, 50 μm (A–F) and 10 μm (A′–F′). (G) Quantification of the ratio of normalized nuclear STAT, STAT-10X-GFP, and slbo-lacZ per cluster, comparing tlk KD polar cells (upd-Gal4, UAS-tlkRNAi) to the negative control (upd-Gal4, UAS-wRNAi). All egg chambers analyzed were late stage 9. We used a t test to assess statistical significance of the differences in staining intensities; ****p < 0.0001. (H) Effect of tlk KD on upd and slbo mRNA expression levels determined by qRT-PCR, expressed relative to the negative control (N.C.), and compared with the positive control (P.C.) which was upd KD at two temperatures (29°C and room temperature [R.T.]). Each test included three biological replicates. (I) A histogram showing genetic interactions between tlk, upd, and stat. Border cell migration was evaluated in stage 10 egg chambers of the indicated genotypes. Flies with c306-Gal4 were fattened at room temperature for 16 h before dissection; all others were fattened at 29°C. The percentage of clusters that complete migration is shown in royal blue. Clusters that initiate but do not complete migration are indicated in gray, and those that fail to initiate migration are indicated in aqua. We used the Mann–Whitney U test to determine statistical significance of border cell migration defects; ****p < 0.0001; n, number of egg chambers examined.
These results suggested that Tlk might function in polar cells to regulate production of Upd. To test directly for an effect on Upd mRNA abundance, we performed real-time quantitative reverse transcription PCR (qRT-PCR) and found that upd mRNA abundance was reduced in ovaries containing tlk KD polar cells relative to the control (Figure 5H). This is a likely underestimate of the magnitude of the effect because many Tlk RNAi lines are lethal in combination with upd-Gal4, necessitating use of somewhat less effective lines to obtain viable adults.
Upd expressed in polar cells activates JAK/STAT in neighboring cells, and a key target of JAK/STAT required for border cell development is the gene slow border cells (slbo; Silver and Montell, 2001). We therefore also evaluated the transcription of slbo by qRT-PCR and found significant reduction after tlk KD in polar cells (Figure 5H). Therefore Tlk is required for normal upd mRNA abundance and STAT pathway activation and thus for border cell specification and cluster migration.
We further confirmed the functional relationship between Tlk and Upd/JAK/STAT during border cell migration by testing for genetic interactions. Combining heterozygous mutant alleles of tlk (tlkΔ14 or tlkΔ40) and stat (stat1681 or stat397) yielded migration defects significantly stronger than additive (Figure 5I). A heterozygous mutation of either updsis or stat1681 also enhanced the phenotype caused by tlk KD by RNAi (Figure 5I). Overexpression of upd using upd-Gal4 caused border cell migration defects on its own, and yet it also partially but significantly rescued the border cell migration defect caused by tlk KD (Figure 5I). Taken together, these findings suggested that Tlk in polar cells is necessary for proper activation of JAK/STAT signal in border cells.
We also observed border cell migration defects, albeit somewhat weaker, when we knocked tlk down in the outer migratory cells using slbo-Gal4 (Supplemental Figure S1, B–D). Because border cell viability was not affected and border cells do not express Upd, this suggests yet another function for Tlk. Thus, although Tlk is a ubiquitously expressed nuclear protein, it serves distinct functions in different cell types. Similar results were reported for Hippo and Warts kinases (Lin et al., 2014). Like Tlk, Hippo and Warts serve different functions in polar cells and border cells; however, Hippo and Warts are required in polar cells for proper differentiation, and therefore Tlk is the only nuclear protein known to regulate Upd expression without affecting polar cell differentiation.
MATERIALS AND METHODS
Drosophila strains and genetics
We crossed c306-Gal4 flies to 363 RNAi fly strains from the VDRC and the National Institute of Genetics (NIG; Kyoto, Japan), covering 172 nuclear proteins with unknown function during border cell development. The nine genes that caused a migration defect in >20% of egg chambers examined are shown in Supplemental Table S2. We used c306-Gal4 because it drives expression beginning early in egg chamber development in both polar and border cells (Manseau et al., 1997; Silver et al., 2001). Subsequently, we focused on Tlk and tested additional RNAi lines, as well as mutant alleles.
We used w1118 (BDSC) for wild-type controls and the following four tlk RNAi fly strains: V105732 (VDRC), B33983, B36102, and B35298 (BDSC). Because overexpression of tlkRNAi sequences from the three BDSC RNAi fly strains by either c306-Gal4 or upd-Gal4 caused lethality, V105732 was the default line in this study, unless specified otherwise. François Karch (University of Geneva, Geneva, Switzerland) kindly provided us with tlkΔ14 FRT101/FM7, tlkΔ40/FM7, FRT101, ubi-GFP; hsFLP, and UAS-tlk full-length flies (Carrera, Moshkin, et al., 2003). The stat397/TM3, stat1681/TM3, updsisC5/FM7, slbo-lacZ/CyO, PZ80, and hsFLP; AyGal4, UAS-GFP fly strains were described previously (Silver et al., 2001; Adam et al., 2004). Other BDSC flies include UAS-upd RNAi (B28722), tub-Gal80ts (B7108, B7017) and yw, FRT101 (B1844).
We kept all lines and crosses at 25°C, except for crosses with tub-Gal80ts, which we incubated at 18°C until the progeny hatched. Before dissection, we treated flies in the following conditions: to knock down tlk or upd function during border cell development, we put newly eclosed females on fresh food supplemented with wet yeast paste at 29°C or room temperature for 16 h; to block gene function during early oogenesis, we kept adult progeny at 29°C for 3–5 d; for crosses involving Gal80ts, we transferred newly eclosed adults from 18 to 25°C for 12 h and then shifted them to the nonpermissive temperature 31°C for 24 h to inactivate Gal80 and derepress Gal4 expression. For the FLP-OUT technique, we placed F1 females at 25°C for 12 h, heat shocked them at 37°C for 1 h, and then switched them to 29°C for another 12 h. To make tlk mosaic clones, we heat shocked flies at 37°C three times per day for 1 h for 2 d and fattened them at 25°C for 3–5 d.
Immunohistochemistry
We dissected ovaries in Schneider’s medium (Thermo Fisher Scientific, Hudson, NH) with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO), fixed them in 3.7% formaldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS; Thermo Fisher Scientific) at room temperature for 15 min, washed them three times (15 min each) with PBS with 0.3% Triton (Sigma-Aldrich), incubated samples in the primary antibodies at 4°C overnight, rinsed three times, and stained them with fluorescence-conjugated secondary antibodies (Molecular Probes, Eugene, OR) at 1:400 for 2 h at room temperature, followed by nuclear dye 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) at 1:1000 for 15 min. After washing, ovaries were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were captured with either a Zeiss LSM 780 or a Zeiss LSM 510 Confocal microscope (Carl Zeiss, Irvine, CA) and then processed or analyzed with Photoshop (Adobe, San Jose, CA) and Illustrator (Adobe), Imaris (Bitplane, South Windsor, CT), Zen (Carl Zeiss), or ImageJ (National Institutes of Health, Bethesda, MD).
We used the following primary antibodies from the Developmental Studies Hybridoma Bank (DSHB; lowa City, IA): mouse anti-Arm (N2 7A1, 1:50), mouse anti-Fas3 (7G10, 1:10), mouse anti-Eya (10H6, 1:25), and mouse anti–β-galactosidase (40-1A, 1:50). Rabbit anti-Tlk antibody (1:1000) was a gift from François Karch (Carrera, Moshkin, et al., 2003). Other primary antibodies we used were rabbit anti–β-galactosidase (1:100; Cappel Laboratories, Cochranville, PA), mouse anti-GFP (1:500; Life Technologies, Grand Island, NY), rabbit anti-GFP (1:1000; Invitrogen, Grand Island, NY), rabbit anti-cCaps-3 (1:1000; Cell Signaling Technology, Beverly, MA), and rabbit anti-STAT (1:1000; Starz-Gaiano et al., 2008).
Single-fly genome DNA extraction and PCR test
To PCR diagnose the V105732 fly strain, we extracted genomic DNA from a single fly by using a protocol adapted from Justin P. Kumar’s lab (Indiana University, Bloomington, IN; www.indiana.edu/~kumarlab/7_labresources/protocols/016%20Single%20Fly%20Genomic%20DNA%20Extraction.pdf).
RNAi fly strains from the KK library of the VDRC can induce a Gal4-dependent toxicity if the shRNA carrier vector pKC26 inserts in a specific site in the genome known as the “annotated site” (Green et al., 2014). This possibility was excluded using the PCR test with four sets of primer pairs as described (Green et al., 2014). This strategy demonstrated that the tlk shRNA carrier pKC26 vector integrated only into the host pKC43 vector inserting at the nonannotated site, which does not cause nonspecific toxicity.
Relationship between border cell/polar cell ratio and cluster migration
We counted the number of border cells and polar cells for each border cell cluster in the indicated numbers of stage 10 egg chambers of different genotypes shown in Figure 3F. We also measured the distance the clusters had migrated toward the oocyte as a percentage of the total migration path and divided the samples into five categories. We entered the data into Prismpad software and used the “XY table and graph” function. The plot shows the mean value of the ratio with the error bar (SD).
Real-time quantitative reverse transcription PCR
The qRT-PCR experiment was performed with the Power SYBR Green PCR Master Mix (Life Technologies) and the QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, Grand Island, NY). We tested the mRNA level of upd and slbo in four different genomic backgrounds with three biological replicates. For each group, six or seven fattened females were dissected and kept in TRIzol (Life Technologies) at −80°C immediately until we finished dissecting all 12 groups. We extracted ovary RNA using the TRIzol RNA isolation and purification protocol (http://tools.lifetechnologies.com/content/sfs/manuals/trizol_reagent.pdf) and removed DNA and DNase by the TURBO DNA-free Kit (Life Technologies). For cDNA synthesis, 0.7 μg of purified RNA of each group was reverse transcribed per 20-μl reaction system by using the SuperScript III First-Strand Synthesis System (Invitrogen). Because upd, slbo, and rp49 (endogenous reference gene) have different transcription levels in ovaries, cDNA was diluted to 1:10, 1:100, and 1:1000 for each, respectively. For real-time PCR, we used a 20-μl reaction system, containing 8 μl of cDNA template with a specific dilution for each gene as shown, 2 μl 2.5 mM forward and reverse primer mixture, and 10 μl of 2× SYBR Green PCR Master Mix. we used the following primer sequences:
upd forward primer: TGGATCGACTATCGCAACTTCG
upd reverse primer: TGGATCGACTATCGCAACTTCG
slbo forward primer: CATGCAGCTAATGAACCACGCCAA (Wang et al., 2006)
slbo reverse primer: TCAAGCATTCAAGCACTCACGCAC (Wang et al., 2006)
rp49 forward primer: AAGAAGCGCAAGGAGATTGT (Borghese et al., 2006)
rp49 reverse primer: AATGTGTATTCCGACCACGTT (Borghese et al., 2006)
Supplementary Material
Acknowledgments
We thank Yasmin Sallak (University of California, Santa Barbara, CA) for assistance and Mary Raven (University of California, Santa Barbara), Kenneth Kosik (University of California, Santa Barbara), and Elmer Guzman (University of California, Santa Barbara) for access to and technical support for Imaris and real-time PCR. We thank lab members for helpful discussion. We also thank the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for reagents. This work was supported by National Institutes of Health Grant GM46425 to D.J.M. W.X. was partially supported by a scholarship provided by Shanghai Jiao Tong University (SJTU; Shanghai, China).
Abbreviations used:
- cCasp-3
cleaved caspase-3
- Fas3
fasciclin 3
- JAK
Janus kinase
- STAT
signal transducer and activator of transcription
- Upd
unpaired.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0327) on October 28, 2015.
REFERENCES
Boldface names denote co–first authors.
- Adam JC, Montell DJ. A role for extra macrochaetae downstream of Notch in follicle cell differentiation. Development. 2004;131:5971–5980. doi: 10.1242/dev.01442. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bai J, Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002;129:5377–5388. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Besse F, Pret AM. Apoptosis-mediated cell death within the ovarian polar cell lineage of Drosophila melanogaster. Development. 2003;130:1017–1027. doi: 10.1242/dev.00313. [DOI] [PubMed] [Google Scholar]
- Borensztejn A, Boissoneau E, Fernandez G, Agnes F, Pret AM. JAK/STAT autocontrol of ligand-producing cell number through apoptosis. Development. 2013;140:195–204. doi: 10.1242/dev.079046. [DOI] [PubMed] [Google Scholar]
- Borghese L, Fletcher G, Mathieu J, Atzberger A, Eades WC, Cagan RL, Rorth P. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene-expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Carrera, Gronke S, Sillje HH, Nigg EA, Jackle H, Karch F. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev. 2003;17:2578–2590. doi: 10.1101/gad.276703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare D, Somers K, Rao S, Matsuda M, Reichman -Fried M, Raz E, Chitnis AB. Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium. Development. 2014;141:3188–3196. doi: 10.1242/dev.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A. Tousled kinase TLK1B mediates chromatin assembly in conjunction with Asf1 regardless of its kinase activity. BMC Res Notes. 2010;3:68. doi: 10.1186/1756-0500-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medioni C, Cerezo D, Noselli S. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- Grammont M, Irvine KD. Organizer activity of the polar cells during Drosophila oogenesis. Development. 2002;129:5131–5140. doi: 10.1242/dev.129.22.5131. [DOI] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11:222-+. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- Gunawan F, Arandjelovic M, Godt D. The Maf factor Traffic jam both enables and inhibits collective cell migration in Drosophila oogenesis. Development. 2013;140:2808–2817. doi: 10.1242/dev.089896. [DOI] [PubMed] [Google Scholar]
- Han DD, Stein D, Stevens LM. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development. 2000;127:573–583. doi: 10.1242/dev.127.3.573. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jang AC, Chang YC, Bai J, Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol. 2009;11:569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single-P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimovskaia IM, Young C, Stromme CB, Menard P, Jasencakova Z, Mejlvang J, Ask K, Ploug M, Nielsen ML, Jensen ON, et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun. 2014;5:3394. doi: 10.1038/ncomms4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Chiang CS, Huang HY, Liaw GJ. mars and tousled-like kinase act in parallel to ensure chromosome fidelity in Drosophila. J Biomed Sci. 2009;16:51. doi: 10.1186/1423-0127-16-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gourguechon S, Wang CC. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J Cell Sci. 2007;120:3883–3894. doi: 10.1242/jcs.007955. [DOI] [PubMed] [Google Scholar]
- Lin TH, Yeh TH, Wang TW, Yu JY. The Hippo pathway controls border cell migration through distinct mechanisms in outer border cells and polar cells of the Drosophila ovary. Genetics. 2014;198:1087–1099. doi: 10.1534/genetics.114.167346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Montell DJ. Identification of mutations that cause cell migration defects in mosaic clones. Development. 1999;126:1869–1878. doi: 10.1242/dev.126.9.1869. [DOI] [PubMed] [Google Scholar]
- Liu Y, Montell DJ. Jing: a downstream target of slbo required for developmental control of border cell migration. Development. 2001;128:321–330. doi: 10.1242/dev.128.3.321. [DOI] [PubMed] [Google Scholar]
- Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- McGregor JR, Xi RW, Harrison DA. JAK signaling is somatically required for follicle cell differentiation in Drosophila. Development. 2002;129:705–717. doi: 10.1242/dev.129.3.705. [DOI] [PubMed] [Google Scholar]
- Monahan AJ, Starz-Gaiano M. Socs36E attenuates STAT signaling to optimize motile cell specification in the Drosophila ovary. Dev Biol. 2013;379:152–166. doi: 10.1016/j.ydbio.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. Slow border cells, a locus required for a developmentally regulated cell-migration during oogenesis, encodes Drosophila C/Ebp. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Wang X, He L, Montell DJ. Border cell migration: a model system for live imaging and genetic analysis of collective cell movement. Methods Mol Biol. 2011;769:277–286. doi: 10.1007/978-1-61779-207-6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald S, Awate S, Rath A, Carroll J, Galiano F, Dwyer D, Kleiner-Hancock H, Mathis JM, Vigod S, De Benedetti A. Phenothiazine inhibitors of TLKs affect double-strand break repair and DNA damage response recovery and potentiate tumor killing with radiomimetic therapy. Genes Cancer. 2013;4:39–53. doi: 10.1177/1947601913479020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Schober M, Rebay I, Perrimon N. Function of the ETS transcription factor Yan in border cell migration. Development. 2005;132:3493–3504. doi: 10.1242/dev.01911. [DOI] [PubMed] [Google Scholar]
- Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- Starz-Gaiano M, Melani M, Wang XB, Meinhardt H, Montell DJ. Feedback inhibition of JAK/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Bo JY, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat Cell Biol. 2011;13:1062–1069. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.