GRASP65 plays a role in Golgi ribbon formation. Because the gaps between Golgi stacks are heterogeneous and large, it is possible that other proteins may help GRASP65 in ribbon linking. Mena is a novel GRASP65-binding protein that promotes actin elongation and enhances GRASP65 oligomerization to link Golgi stacks into a ribbon.
Abstract
In mammalian cells, the Golgi reassembly stacking protein 65 (GRASP65) has been implicated in both Golgi stacking and ribbon linking by forming trans-oligomers through the N-terminal GRASP domain. Because the GRASP domain is globular and relatively small, but the gaps between stacks are large and heterogeneous, it remains puzzling how GRASP65 physically links Golgi stacks into a ribbon. To explore the possibility that other proteins may help GRASP65 in ribbon linking, we used biochemical methods and identified the actin elongation factor Mena as a novel GRASP65-binding protein. Mena is recruited onto the Golgi membranes through interaction with GRASP65. Depleting Mena or disrupting actin polymerization resulted in Golgi fragmentation. In cells, Mena and actin were required for Golgi ribbon formation after nocodazole washout; in vitro, Mena and microfilaments enhanced GRASP65 oligomerization and Golgi membrane fusion. Thus Mena interacts with GRASP65 to promote local actin polymerization, which facilitates Golgi ribbon linking.
INTRODUCTION
In mammalian cells, the Golgi apparatus displays a unique structure and localizes to the center of the cell, adjacent to the centrosome and the nucleus. It consists of dozens of Golgi stacks, each containing five to seven flattened Golgi cisternae that are parallel aligned and tightly stuck to each other. These stacks are then laterally connected to form a ribbon (Lowe, 2011). During cell division, the Golgi apparatus undergoes a unique disassembly and reassembly process. At the onset of mitosis, the Golgi complex undergoes ribbon unlinking, cisternae unstacking, and membrane vesiculation, yielding thousands of vesicles that are dispersed throughout the cytoplasm and subsequently distributed into the two daughter cells. During telophase and cytokinesis, the Golgi is reassembled, including cisterna regeneration by vesicle fusion, cisterna restacking, and ribbon reformation (Wei and Seemann, 2010; Wang and Seemann, 2011; Tang and Wang, 2013). The exact mechanisms mediating Golgi structure formation remain largely unknown, but the Golgi reassembly stacking protein 65 (GRASP65) has been indicated to play significant roles in both Golgi cisternal stacking and ribbon linking (Barr et al., 1997; Wang et al., 2003; Puthenveedu et al., 2006; Veenendaal et al., 2014).
GRASP65 and its homologue, GRASP55, are the only two proteins shown to directly participate in Golgi cisternal stacking (Barr et al., 1997; Shorter et al., 1999). GRASP65 localizes to the cis-Golgi and GRASP55 to the medial/trans-Golgi (Shorter et al., 1999). Depletion of either GRASP65 or GRASP55 reduced the number of cisternae in the Golgi stack, and simultaneous depletion of both GRASPs led to complete disassembly of Golgi stacks (Sutterlin et al., 2005; Tang et al., 2010b; Xiang and Wang, 2010). GRASPs from adjacent Golgi cisternae form trans-oligomers through the N-terminal GRASP domain and zipper the cisternae into a stack. Recent structural studies revealed that the GRASP domain is globular and 6.5 nm in length (Truschel et al., 2012; Feng et al., 2013). The size of GRASP65 trans-oligomers fits well with the 11-nm gap between cisternae (Cluett and Brown, 1992). During mitosis, phosphorylation of GRASPs inhibits their oligomerization and leads to cisterna separation. At the end of mitosis, dephosphorylation allows the reformation of GRASP trans-oligomers and restacking of newly formed cisternae (Wang et al., 2003; Tang et al., 2010b; Xiang and Wang, 2010).
GRASP65 and GRASP55 are also involved in Golgi ribbon formation. Depletion of either GRASP caused Golgi ribbon fragmentation (Puthenveedu et al., 2006; Duran et al., 2008; Feinstein and Linstedt, 2008; Veenendaal et al., 2014). It has been suggested that GRASPs link Golgi stacks into a ribbon by forming trans-oligomers at the rims of neighboring Golgi stacks, similar to their mechanism in cisternal stacking (Feinstein and Linstedt, 2008; Bachert and Linstedt, 2010; Truschel et al., 2011). However, because the gaps between Golgi stacks are much larger and relatively more heterogeneous (tens to hundreds of nanometers; Cluett and Brown, 1992), it is possible that GRASPs tether Golgi stacks via other bridging proteins, of which golgins are ideal candidates because of their long coiled-coil domains, which are known in membrane tethering. Depletion of any one of many golgins causes Golgi fragmentation (Munro, 2011). An ideal example is GM130, a golgin that interacts with GRASP65 (Barr et al., 1998; Nakamura, 2010). However, the level and localization of GM130 are not significantly affected by GRASP65 depletion (Sutterlin et al., 2005; Wang et al., 2008), indicating that GM130 is required for Golgi integrity by itself and that other factors may cooperate with GRASP65 in Golgi ribbon formation.
The architecture and central positioning of the Golgi ribbon depend on the cellular cytoskeleton (Egea et al., 2006; Mironov and Beznoussenko, 2011). In mammalian cells, the formation of the Golgi ribbon requires an intact microtubule network and the minus end–directed motor, the dynein/dynactin complex (Brownhill et al., 2009; Yadav and Linstedt, 2011). In addition to microtubules, the actin microfilament is also involved in Golgi trafficking and structure formation (Egea et al., 2006). Actin polymerization facilitates membrane deformation to drive vesicle formation, fission and fusion, and short-range movements (Lanzetti, 2007). Depolymerization and stabilization of actin filaments both induced a compaction phenotype of the Golgi by fluorescence microscopy, which, however, was actually a fragmented and dilated Golgi by electron microscopy (EM; Lazaro-Dieguez et al., 2006), suggesting that a dynamic actin network is required to maintain Golgi ribbon connection. Several Golgi-localized actin regulators, including ADF/cofilin, mDia1, FMNL1γ, INF2, and WHAMM, have been identified, and most of them regulate Golgi transport in different ways (Campellone et al., 2008; von Blume et al., 2009; Colon-Franco et al., 2011; Ramabhadran et al., 2011; Zilberman et al., 2011). How actin maintains the integrity of the Golgi ribbon structure, however, remains largely unknown.
The role of actin filaments in Golgi formation has been indicated in Drosophila cells. Unlike in mammalian cells, the Golgi in Drosophila cells exists as dispersed Golgi stacks. Although not connected into a ribbon, the Golgi elements exist as pairs in Drosophila S2 cells. Actin depolymerization induces the scission of Golgi pairs at late G2 phase, which is mediated by the inactivation of actin nucleation–promoting factors WAVE/Scar/Abi (Kondylis et al., 2007). Similar Golgi pairs were observed in HeLa cells in the same study upon nocodazole treatment, and treatment with an actin-depolymerizing drug caused the scission of one Golgi element into two, indicating that actin filaments also help to link Golgi stacks in mammalian cells (Kondylis et al., 2007).
In this study, we identified the mammalian Enabled homologue (Mena) as a novel GRASP65-interacting protein. Mena is the mammalian homologue of Drosophila Enabled (Ena; Higashi et al., 2009), a member of the Ena/vasodilator-stimulated phosphoprotein (VASP) family that directly enhances actin filament elongation by interacting with the barbed end of the actin filament, preventing capping and recruiting monomeric actin for polymerization (Breitsprecher et al., 2011). Mena plays a role in filopodia and lamellipodia formation in mammalian cells (Rottner et al., 1999; Bear et al., 2002; Applewhite et al., 2007). Of interest, its Drosophila homologue, Ena, has been shown to localize to the cis-Golgi to regulate Golgi membrane fusion and fission in developing photoreceptor cells (Kannan et al., 2014). Consistent with that study, here we find that mammalian Mena, but not its homologue VASP, is targeted to the Golgi through interaction with GRASP65, and Mena facilitates GRASP65 oligomerization and promotes local actin polymerization to link Golgi stacks into a ribbon.
RESULTS
Identification of GRASP65-interacting proteins in the interphase cytosol
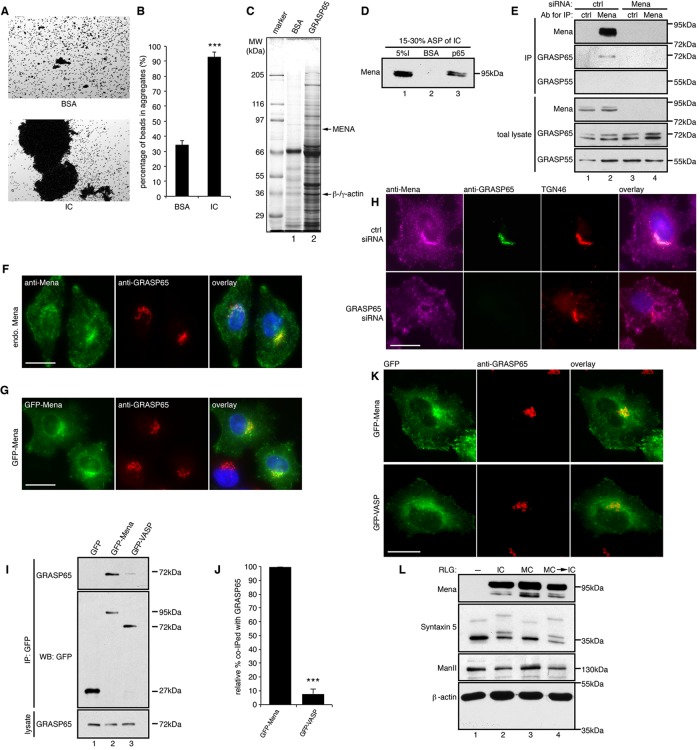
In our previous studies, we used a semiquantitative beads aggregation assay to visualize GRASP65 oligomerization. We coated Dynal M-500 magnetic beads with purified GRASP65 and examined the degree of bead aggregation in different conditions (Wang et al., 2003, 2005; Tang et al., 2010b). When GRASP65-coated beads were incubated in a physiological buffer at 37ºC, the beads formed aggregates to some extent, suggesting that GRASP65 forms trans-oligomers that are sufficient to link neighboring surfaces together (Wang et al., 2003). Of interest, when the beads were incubated with interphase HeLa cell cytosol, aggregation was strongly enhanced in comparison to that with a buffer containing the same concentration of bovine serum albumin (BSA; 92.7 ± 3.5% vs. 34.2 ± 2.8% beads in aggregates; Figure 1, A and B), suggesting that interphase cytosol contains active components that enhance GRASP65 oligomerization.
FIGURE 1:
Mena localizes to the cis-Golgi through interaction with GRASP65. (A) GRASP65- coupled Dynal M-500 beads were incubated with either buffer containing BSA or HeLa interphase cytosol (IC). After incubation, the beads were placed on glass slides, and random fields were photographed. Scale bar, 100 μm. (B) Quantification of A for the percentage of beads in aggregates. Results are expressed as mean ± SEM. Statistical significance was assessed by comparison to BSA-treated beads. The p value was determined by Student’s t test; ***p < 0.001. (C) Affinity purification of GRAS65-interacting proteins. Interphase cytosol fractionated by 15–30% ammonium sulfate precipitation was incubated with either BSA or His-GRASP65–coupled CNBr beads, and bound proteins were analyzed by SDS–PAGE and Coomassie blue staining. Arrows indicate the Mena and β/γ-actin bands that were excised and identified by mass spectrometry. (D) Beads coated with BSA or GRASP65 were incubated with 15–30% ammonium sulfate–precipitated proteins from interphase cytosol (I, input), washed, and analyzed by Western blot for Mena. (E) Immunoprecipitation (IP) by either nonspecific IgG (ctrl) or anti-Mena antibodies from lysate of cells transfected with control (ctrl) or Mena-targeting siRNA. (F) WT HeLa cells were immunostained with indicated antibodies to show the Golgi localization of endogenous Mena. (G) HeLa cells expressing GFP-Mena were immunostained for GRASP65. (H) Cells transfected with control or GRASP65 siRNA were immunostained for the indicated proteins. GRASP65 depletion abolished the Golgi localization of Mena. (I) Cells transfected with GFP, GFP-Mena, or GFP-VASP were lysed and immunoprecipitated by GFP antibodies. (J) Quantification of the amount of GFP-Mena or GFP-VASP that was coimmunoprecipitated with GRASP65, with the level of GFP-Mena normalized to 100% . ***p < 0.001. (K) Cells expressing GFP-Mena or GFP-VASP were immunostained for GRASP65. Mena but not VASP is concentrated on the Golgi. Bar, 20 μm (F–H, J). (L) Purified rat liver Golgi (RLG) membranes were incubated with interphase (IC) or mitotic (MC) cytosol or sequentially incubated with MC and then IC (MC → IC), reisolated, and blotted for indicated proteins.
To further confirm the existence of active components in interphase cytosol, we first aggregated GRASP65-coated beads by interphase cytosol and then disaggregated them by treatment with purified mitotic kinases cyclin-dependent kinase 1 (Cdk1) and Polo-like kinase 1 (Plk1), which are known to phosphorylate GRASP65 and disrupt its oligomerization (Wang et al., 2003; 2005), and further treated the resulting single beads with either interphase cytosol or purified protein phosphatase 2A (PP2A), which dephosphorylates GRASP65 (Tang et al., 2008). As shown in Supplemental Figure S1A, both treatments caused bead reaggregation, but treatment with interphase cytosol resulted in much greater aggregation than that with PP2A, strongly suggesting that interphase cytosol contains active components for GRASP65 oligomerization. To characterize the biochemical properties of these factors, we treated interphase cytosol with high salt, heat, or proteases. All of these treatments abolished the activity, suggesting that the cytosolic factors are proteins in nature (Supplemental Figure S1B). Further experiments using a variety of nucleotides or their analogues showed that this activity requires ATP but not ATP hydrolysis, because addition of nonhydrolyzable ATP analogues such as ATPγS significantly enhanced aggregation similar to ATP (Supplemental Figure S1C).
We then tried to enrich this activity by classical biochemical methods. Using sequential ammonium sulfate precipitation, we found that the activity was enriched in 10–30% ammonium sulfate precipitate (Supplemental Figure S2A). We then fractionated 15–30% ammonium sulfate precipitant of cytosol by a 10–35% glycerol gradient and found that fraction 9, among a total of 12 fractions, contained most of the activity (Supplemental Figure S2B). Comparison with molecular weight standards showed that proteins in fraction 9 were ∼1000 kDa, suggesting that it is a large protein complex.
To affinity purify the active components, we dissolved the 15–30% ammonium sulfate precipitate, dialyzed it into a physiological buffer, and passed it through a column that contained either GRASP65 or BSA cross-linked to CNBr-activated beads, which have higher capacity than Dynal beads. After extensive washing, bound proteins were analyzed by SDS–PAGE and Coomassie blue staining. Proteins that specifically bind to GRASP65 but not BSA beads were excised and identified by mass spectrometry. Mena, the mammalian homologue of Drosophila Ena known to enhance actin filament elongation (Gertler et al., 1996), and β- and γ-actin were identified in this process (Figure 1C and Supplemental Table S1).
Mena is recruited to the Golgi membranes through the interaction with GRASP65
To confirm the interaction between GRASP65 and Mena, we incubated the 15–30% ammonium sulfate cut of interphase cytosol with BSA- or GRASP65-coated CNBr-activated beads and analyzed bound proteins by Western blot. As shown in Figure 1D, GRASP65 pulled down Mena from the cytosol, but BSA did not. To confirm this interaction in cells, we immunoprecipitated Mena from HeLa cell lysate and found that GRASP65, but not its counterpart GRASP55, coprecipitated with Mena (Figure 1E). To ensure the specificity of this interaction, we knocked down Mena with small interfering RNA (siRNA). As a result, GRASP65 was undetectable in the precipitate with Mena antibodies when Mena was depleted (Figure 1E).
We then determined whether Mena is recruited to the Golgi by GRASP65. Immunofluorescence images in previous reports suggest a prominent enrichment of Mena in focal adhesions, lamellipodia, and filopodia. However, most images also show a perinuclear localization of Mena in cells (Bear et al., 2002; Loureiro et al., 2002; Joshi and Wang, 2015). Consistently, Ena and actin filaments recently have been shown to localize to the cis-Golgi to regulate Golgi membrane fusion and fission in developing photoreceptor cells in Drosophila (Kannan et al., 2014). By immunofluorescence microscopy, we found that endogenous Mena is enriched on the Golgi and colocalized with GRASP65 (Figure 1F). Similarly, exogenously expressed green fluorescent protein (GFP)–Mena also concentrated on the Golgi (Figure 1G). The Golgi localization of Mena is GRASP65 dependent, because depletion of GRASP65 by RNA interference (RNAi) dispersed Mena from the Golgi to the cytosol (Figure 1H). To determine whether the interaction between Mena and GRASP65 is specific, we also examined VASP, another member of the Ena/VASP family of proteins. GFP-Mena coimmunoprecipitated with endogenous GRASP65, but GFP-VASP had much lower affinity to GRASP65 (8.2 ± 3.3% relative to GFP-Mena; Figure 1, I and J). In addition, unlike Mena, GFP-VASP was not enriched on the Golgi (Figure 1K), indicating that the interaction with GRASP65 and Golgi localization are specific to Mena. When purified Golgi membranes were incubated with interphase or mitotic HeLa cell cytosol and reisolated, cytosolic Mena was copurified with the Golgi membranes under both interphase and mitotic conditions (Figure 1L), demonstrating that the interaction between GRASP65 and Mena is cell cycle independent.
Mena is required for Golgi ribbon formation in an actin-dependent manner
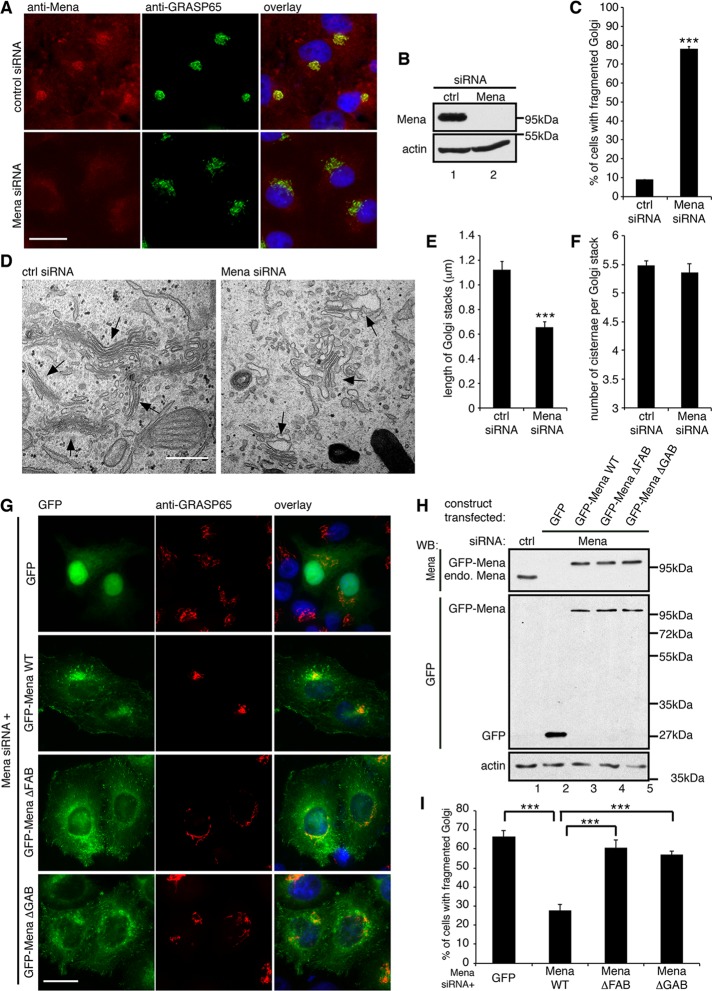
To investigate the function of Mena in Golgi organization, we knocked down Mena in HeLa cells (Figure 2). Transfection with control nonspecific siRNA had no significant effect on the Golgi morphology in the cell, as indicated by GRASP65 staining, whereas depletion of Mena caused Golgi fragmentation, with Golgi elements detached from each other and dispersed into the cytoplasm (Figure 2, A–C). Quantification of these cells showed that the Golgi was fragmented in 78.2 ± 1.5% of Mena-depleted cells, significantly higher than in control siRNA–treated cells (9.0 ± 0.2%; Figure 2C). We then examined the Golgi structure more closely by EM. In control siRNA–transfected cells, the Golgi stacks were well organized and laterally connected in a ribbon (Figure 2D). In contrast, in Mena-depleted cells, the Golgi stacks were disconnected, dispersed throughout the cytoplasm, and significantly shorter than in control cells (Figure 2E). The average length of Golgi stacks was reduced from 1.23 ± 0.08 μm in control cells to 0.72 ± 0.05 μm in Mena-depleted cells (Figure 2E), whereas Golgi stacking was not affected (Figure 2F). In addition, the Golgi cisternae in Mena-depleted cells were mildly dilated, similar to the effect of actin disruption, as previously reported (Lazaro-Dieguez et al., 2006). These results suggest that Mena depletion reduces the connectivity between Golgi stacks in the ribbon.
FIGURE 2:
Mena regulates Golgi integrity through actin polymerization. (A) HeLa cells treated with indicated siRNA were stained for Mena and GRASP65. (B) Immunoblot of HeLa cells from A. (C) Quantification of A for the percentage of cells with fragmented Golgi. (D) Representative EM images of the cells described in A. Arrows, Golgi stacks. Bar, 500 nm. (E) Quantification of D for the average cisternal length of Golgi stacks. (F) Quantification of D for the average number of cisternae per Golgi stack. (G) Mena-depleted HeLa cells were transfected with a cDNA for GFP or GFP-tagged, RNAi-resistant Mena WT or mutants and stained for GRASP65. Bar, 20 μm (A, G). (H) Cells were transfected first with nonspecific (lane 1) or Mena-specific siRNA (lanes 2–5) and then with plasmid of GFP (lane 2), GFP-tagged, RNAi-resistant Mena WT (lane 3), ΔFAB (lane 4), and ΔGAB (lane 5) mutants and analyzed by Western blot. endo. Mena, endogenous Mena. (I) Quantification of the percentage of GFP-positive cells with fragmented Golgi in G. ***p < 0.001.
To ensure that the Golgi fragmentation phenotype was specific for Mena depletion, we expressed an RNAi-resistant form of GFP-Mena, or GFP as control, in cells in which endogenous Mena was knocked down. As shown in Figure 2, G–I, GFP expression had no effect on the fragmented Golgi, whereas in GFP-Mena expressing cells, the Golgi became intact and compact. We then asked whether the function of Mena in Golgi integrity is through actin filament formation. We expressed the Mena ΔFAB or ΔGAB mutants, which cannot bind F-actin or G-actin, respectively (Loureiro et al., 2002; Breitsprecher et al., 2011), in Mena-depleted cells. Both mutants, although localized to the Golgi, failed to rescue the Golgi ribbon from fragmentation (Figure 2, G–I). These results indicate that Mena is required for maintaining Golgi integrity, and its activity depends on actin.
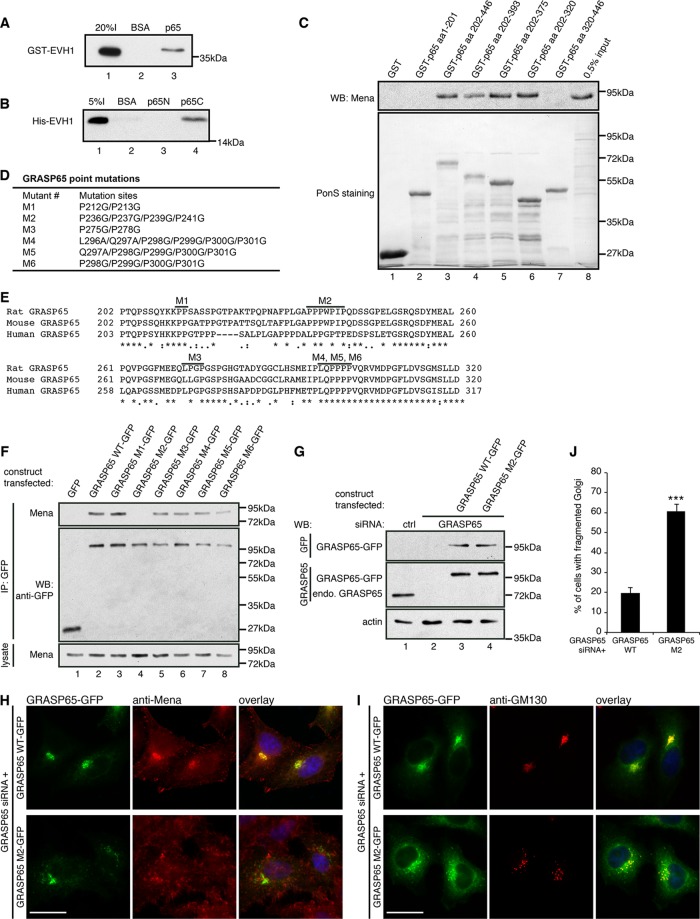
We further mapped the interaction domains on both proteins with pull-down assays. Like other members in the Ena/VASP protein family, Mena contains an N-terminal Ena/VASP homology 1 (EVH1) domain, which binds the proline-rich motif of a number of proteins for subcellular targeting, a middle proline-rich domain, and a C-terminal EVH2 domain that interacts with both G-actin and F-actin and thus enhances F-actin elongation. The EVH2 domain also has a C-terminal coiled-coil domain that mediates tetramerization of Mena (Bear and Gertler, 2009; Breitsprecher et al., 2011). GRASP65, on the other hand, contains an N-terminal GRASP domain and a C-terminal serine/proline-rich (SPR) domain (Wang et al., 2005). To determine whether Mena interacts with the proline-rich domain of GRASP65 through its EVH1 domain, we performed a pulldown assay using purified recombinant human Mena EVH1 and rat GRASP65 fragments. The EVH1 domain of Mena copurified with full-length GRASP65 (Figure 3A), as well as with its C-terminal SPR domain, but not the N-terminal GRASP domain (Figure 3B), demonstrating that the interaction is directly mediated by the EVH1 domain of Mena and the SPR domain of GRASP65 and is actin independent.
FIGURE 3:
Mena–GRASP65 interaction is required for Golgi integrity. (A) BSA- or GRASP65-coated beads were incubated with purified GST-Mena EVH1, washed, and analyzed by Western blot for GST. (B) Beads coupled with BSA or purified GRASP65 aa 1–201 (p65N) or 202–446 (p65C) fragment were incubated with purified His-Mena EVH1, washed, and analyzed by Western blot for His tag. (C) Beads coupled with GST or purified GST-tagged GRASP65 fragments were incubated with HeLa cell lysate, washed, and analyzed by Western blot for Mena. (D) GRASP65 point mutations, with indicated amino acids mutated. (E) Alignment of rat, mouse, and human GRASP65 amino acid sequences in the indicated region. Bold lines indicate the sites where conserved prolines were mutated in the mutants (M1–M6) as in D. (F) Cells transfected with GFP, GFP-tagged GRASP65 WT, or indicated mutants were immunoprecipitated with anti-GFP antibodies and analyzed by Western blot. (G–J) GRASP65-depleted cells were transfected with WT GRASP65 or its M2 mutant and analyzed by Western blot (G) and microscopy (H, I). Bar, 20 μm. (J) Quantitation of cells in I with fragmented Golgi. ***p < 0.001.
Using glutathione S-transferase (GST)–tagged rat GRASP65 fragments to pull down Mena from HeLa cell lysate, we found amino acids (aa) 202–320 as the minimal fragment of GRASP65 for Mena binding (Figure 3C). There are four proline-rich stretches in this region conserved among species. Therefore we mutated the conserved prolines into glycines (Breitsprecher et al., 2011; Figure 3, D and E), expressed them in cells, and tested their interactions with Mena by coimmunoprecipitation. As shown in Figure 3F, mutation of P236/P237/P239/P241 to glycines (mutant M2) in rat GRASP65 abolished the interaction with Mena, whereas mutation of other prolines had no effect. This Mena EVH1-binding sequence on GRASP65, L (human)/P (rat and mouse) PPxPxP, although close to the common F/LPPXP sequence shared by several different Drosophila Ena and mammalian Mena-interacting proteins (Ball et al., 2002), is nontraditional, similar to another Mena EVH1-binding motif in human Abi (Breitsprecher et al., 2011).
To determine whether GRASP65–Mena interaction is required for Mena Golgi localization and Golgi ribbon integrity, we transfected GFP-tagged wild-type (WT) rat GRASP65 or the Mena interaction-deficient mutant M2 in GRASP65-depleted HeLa cells (Figure 3, G and J). Unlike WT GRASP65, the M2 mutant failed to recruit Mena to the Golgi (Figure 3H) or rescue the fragmented Golgi (Figure 3, I and J).
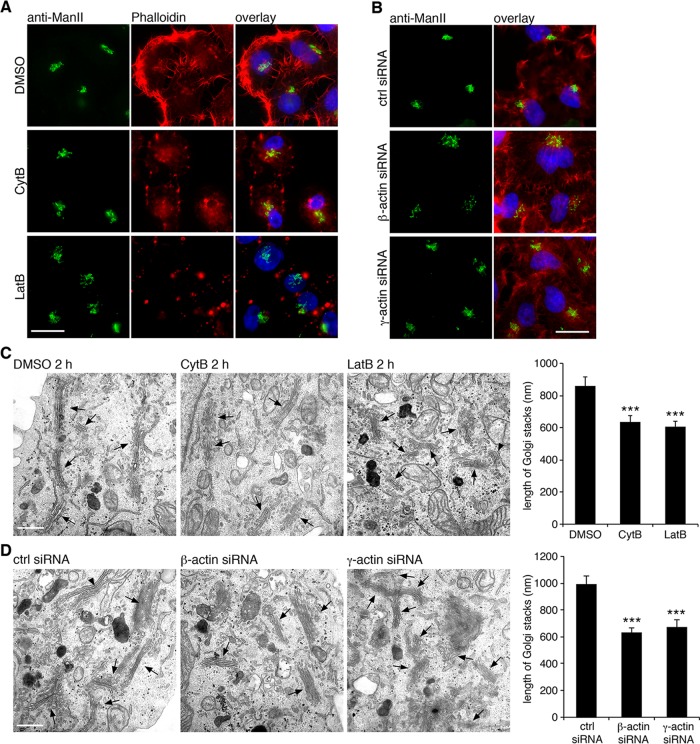
Because Mena regulates Golgi structure by interacting with actin filaments, we then asked whether disturbing actin filaments by pharmacological treatment would affect Golgi structure. Cytochalasin B is known to cap actin filament and therefore inhibits actin filament elongation, antagonizing Mena in preventing actin filament capping (Braet et al., 1996). Latrunculin B binds actin monomer and thus depolymerizes actin filaments (Wakatsuki et al., 2001). In these experiments, we did not observe an obvious condensed Golgi as previously reported (Lazaro-Dieguez et al., 2006); instead, we found that cells treated with cytochalasin B or latrunculin B exhibited a phenotype of loose and unlinked Golgi in comparison with an intact Golgi in dimethyl sulfoxide (DMSO)–treated cells (Figure 4A). In addition, depletion of cellular β- or γ-actin by siRNA resulted in Golgi fragmentation (Figure 4B), similar to Mena depletion. By EM, we found that disruption of actin filaments by drug treatments (Figure 4C) and depletion of β- or γ-actin (Figure 4D) both resulted in dispersed ministacks, apparently caused by Golgi ribbon unlinking. Taken together, these results revealed that Mena and actin filaments regulate Golgi ribbon integrity.
FIGURE 4:
Actin filaments are required for Golgi integrity. (A) HeLa cells treated with DMSO, cytochalasin B (CytB, 40 μM), or latrunculin B (LatB, 0.5 μM) for 2 h were stained for α-mannosidase II (ManII) and rhodamine-labeled phalloidin. (B) Cells transfected with indicated siRNA were stained for ManII and rhodamine–phalloidin. Bar, 20 μm (A, B). (C) Cells treated with DMSO, cytochalasin B, or latrunculin B for 2 h were processed for EM and imaged. Arrows indicate Golgi stacks. The average cisternal length of Golgi stacks was quantified. Statistical significance was assessed by comparison to DMSO-treated cells. (D) Cells transfected with control, β-actin, or γ-actin siRNA for 48 h were processed for EM and analyzed as in C. Statistical significance was assessed by comparison to the control siRNA-treated cells. Bar, 500 nm (C, D). ***p < 0.001.
Actin filaments have been implicated in intracellular trafficking (Campellone et al., 2008; Miserey-Lenkei et al., 2010; Egea et al., 2013; Gurel et al., 2014), and therefore we determined whether Mena as an actin filament regulator also affects protein trafficking, using the vesicular stomatitis virus glycoprotein (VSV-G) as a cargo. Cells first transfected with control or Mena siRNA were infected by adenoviruses that were prepacked with VSV-G-ts045-GFP plasmids (Xiang et al., 2013). Cells were kept at nonpermissive temperature (40.5ºC) overnight to trap VSV-G-ts045 in the endoplasmic reticulum (ER) and then moved to permissive temperature (32ºC) to allow VSV-G export from the ER through the Golgi to the plasma membrane (Presley et al., 1997; Hirschberg et al., 1998). As shown in Supplemental Figure S3, Mena depletion did not significantly affect VSV-G trafficking. Thus Mena plays a critical role in Golgi ribbon formation but not protein trafficking.
Mena and actin filaments facilitate Golgi ribbon formation after nocodazole washout
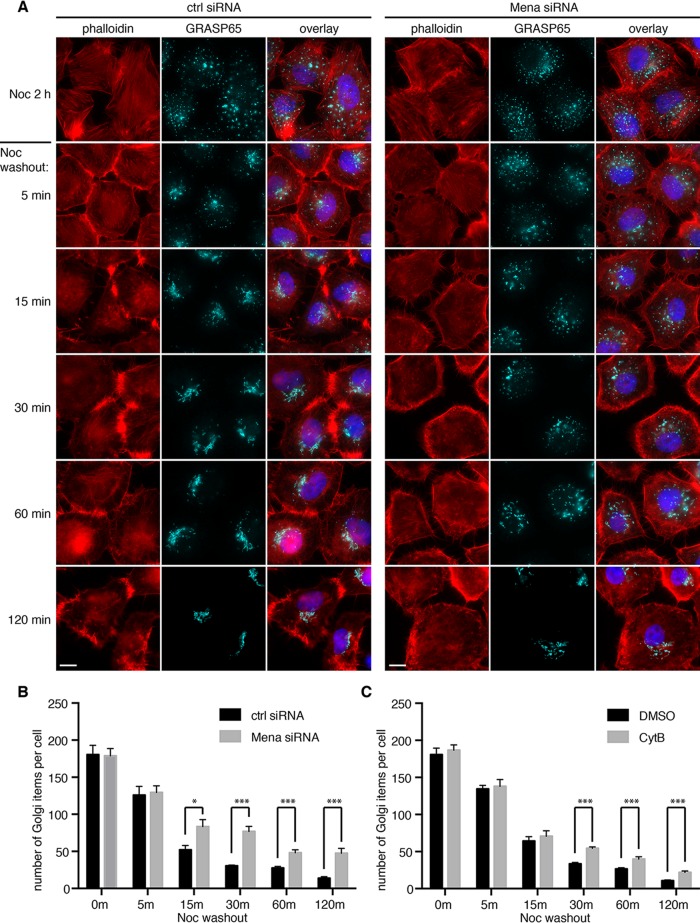
In mammalian cells, Golgi ribbon formation requires an intact microtubule network. Depolymerizing microtubules with nocodazole results in Golgi ministacks dispersed in the cytoplasm (Minin, 1997; Thyberg and Moskalewski, 1999). We then washed away nocodazole to allow the reformation of Golgi ribbon to mimic postmitotic Golgi assembly in the presence or absence of Mena. As shown in Figure 5, the reformation of the Golgi ribbon was significantly delayed in Mena-depleted cells compared with control cells (Figure 5A). At each time point, there were more free Golgi elements not fused with the Golgi core in Mena-depleted cells. At 120 min after nocodazole removal, all of the Golgi elements were concentrated at the cell center, and the Golgi ribbon became intact in all control cells; however, in Mena-depleted cells, although the Golgi elements were transported to the cell center, most of them remained unconnected. Confocal imaging and quantification results showed that Mena depletion increased the number of Golgi elements per cell, indicating a defect in fusion of the Golgi elements (Figure 5B). Similar to Mena depletion, inhibition of actin elongation by cytochalasin B treatment also delayed Golgi ribbon reformation after nocodazole removal (Figure 5C and Supplemental Figure S4). Thus Mena and actin filament elongation are essential in linking Golgi stacks into the ribbon.
FIGURE 5:
Mena depletion impairs Golgi reassembly after nocodazole washout. (A) HeLa cells transfected with control or Mena siRNA were incubated with nocodazole for 2 h. After nocodazole removal for indicated times (minutes), cells were fixed and stained by phalloidin and GRASP65 antibodies. Bar, 10 μm. (B) Experiments in A were analyzed by confocal microscopy, and the number of Golgi particles in the indicated cells was quantified (mean ± SEM). *p < 0.05; ***p < 0.001. (C) Cells were incubated first with nocodazole for 2 h and then with the addition of DMSO or cytochalasin B for another 30 min. Cells were washed and further incubated in growth medium containing DMSO or cytochalasin B, but no nocodazole, for the indicated times. Cells were stained for GRASP65 and by phalloidin (images shown in Supplemental Figures S4 and S5) and analyzed by confocal microscopy and quantified as in B. Quantitation results.
As suggested by Kondylis et al. (2007), when dispersed by nocodazole treatment, Golgi stacks exist as pairs in mammalian cells, similar to those observed in Drosophila S2 cells, and actin filaments are required for the formation of the Golgi pairs. Therefore depolymerizing actin filaments by latrunculin B in nocodazole-treated cells should lead to the scission of Golgi pairs and doubling of the Golgi elements. To test this possibility, we incubated cells with nocodazole in the presence of DMSO, cytochalasin B, or latrunculin B. Confocal microscopy analysis showed that nocodazole treatment dispersed the Golgi ribbon into ministacks; however, additional treatment with cytochalasin B did not further increase the number of Golgi elements. Similarly, depletion of Mena by siRNA did not affect the number of Golgi elements in nocodazole-treated cells (Supplemental Figure S5A). Under EM, the Golgi stacks in nocodazole-treated cells were shorter than in normal interphase cells and were always located adjacent to ER membranes, presumably ER exit sites. Neither Mena depletion nor cytochalasin B and latrunculin B treatment further affected the length and location of the Golgi ministacks (Supplemental Figure S5B). These results indicated that Golgi pairing is unlikely to be the mechanism of Mena and actin-mediated ribbon linking.
Mena and actin filament are required for Golgi fusion
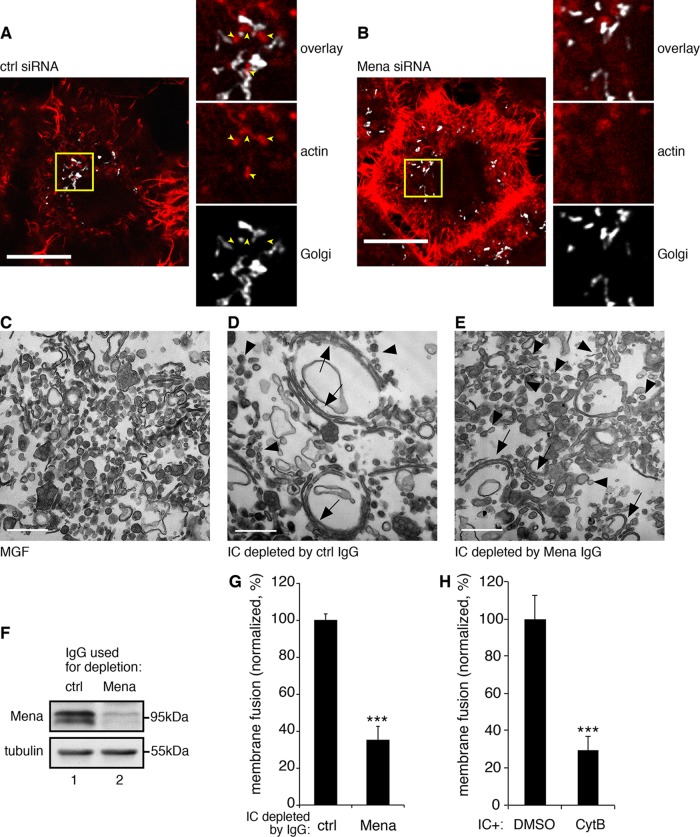
During the reformation of the Golgi ribbon after nocodazole removal, we often observed actin patches localized around and between Golgi elements in control cells (Figure 6A), possibly facilitating Golgi stacks to link with each other, whereas in Mena-depleted cells, these actin patches were rarely detectable (Figure 6B). Because Golgi ribbon linking requires membrane fusion activity, we determined whether Mena-mediated actin elongation facilitates Golgi membrane fusion, using a well-established in vitro Golgi reassembly assay (Tang et al., 2010a). We first treated purified Golgi membranes with mitotic HeLa cell cytosol to induce Golgi fragmentation. We reisolated the resulting mitotic Golgi fragments (MGFs) and further treated them with interphase cytosol to allow vesicles to fuse. We then measured Golgi membrane fusion activity by quantifying long cisternal membranes generated from vesicles. To determine whether Mena is required for Golgi membrane fusion, we immunodepleted Mena from interphase cytosol. When incubated with control immunoglobulin G (IgG)–treated cytosol, the mitotic Golgi fragments (Figure 6C) reassembled into long, stacked Golgi cisternae (Figure 6D). However, when Mena was depleted, which removed 85.5% of Mena from the cytosol, membrane fusion activity was reduced to 35.1 ± 7.7% of control cytosol (Figure 6, E–G). Similarly, depolymerizing actin by adding cytochalasin B into the reassembly reaction reduced the membrane fusion efficiency to 29.7 ± 7.1% relative to the DMSO control (Figure 6H). Thus Mena and actin are essential for Golgi membrane fusion.
FIGURE 6:
Mena regulates Golgi membrane fusion through F-actin elongation. (A, B) HeLa cells were transfected with control (A) or Mena (B) siRNA and incubated with nocodazole for 2 h. At 15 min after nocodazole removal, cells were stained for actin (red) and GRASP65 (white) and analyzed by confocal microscopy. Shown are single scans; parts of the images are enlarged to show the actin patches between Golgi membranes. Bars, 10 μm. (C) Mena was efficiently depleted from interphase cytosol, as shown by Western blot. (D–F) In vitro Golgi reassembly assay. Purified RLG membranes were disassembled by mitotic cytosol treatment, reisolated (D), and further incubated with interphase cytosol immunodepleted by control IgG (E) or by anti-Mena antibodies (F). Membranes were processed for EM and imaged. Bars, 500 nm. (G) Quantification of relative Golgi membrane fusion efficiency in E and F. ***p < 0.001. (H) Purified Golgi membranes that were first disassembled by mitotic cytosol treatment and then incubated with interphase cytosol in the presence of DMSO or 40 μM cytochalasin B (CytB). Membranes were processed for EM and quantified as in G.
Mena and elongated actin filament enhance GRASP65 oligomerization
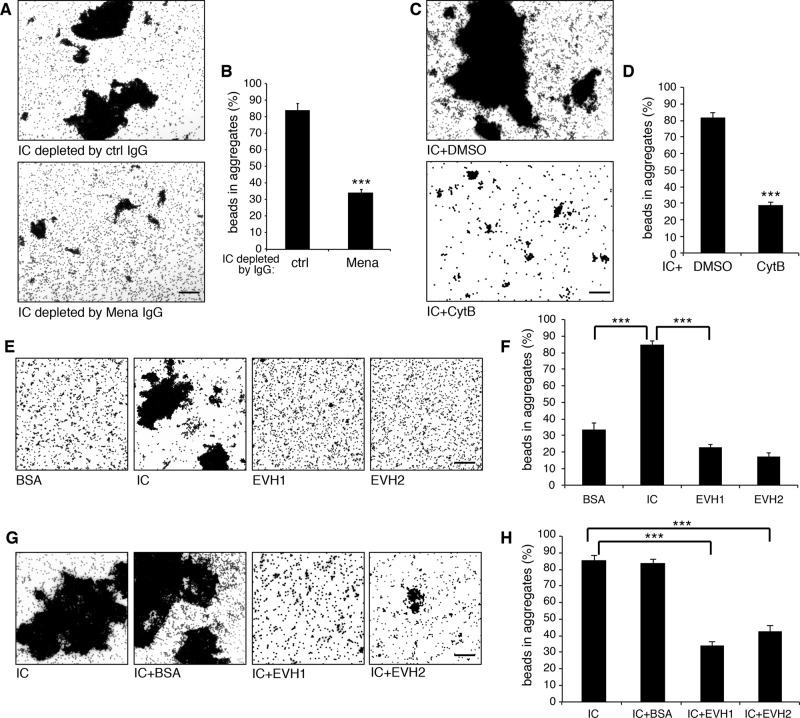
Because Mena directly interacts with GRASP65 and Mena promotes actin elongation to link Golgi stacks into a ribbon, we asked whether GRASP65 only functions as the membrane anchor for Mena or Mena and actin filaments also regulate GRASP65 oligomerization. This question was addressed using the bead aggregation assay, in which GRASP65 oligomerization can be directly visualized (Wang et al., 2003; 2005). As shown in Figure 7, A and B, GRASP65-coated beads formed large aggregates when treated with interphase cytosol, whereas depletion of Mena from the cytosol significantly reduced this activity, suggesting that Mena is at least one of the major components in the interphase cytosol that enhance GRASP65 oligomerization (Figure 7, A and B). Hyperpolymerization of GRASP65 requires actin polymerization, because adding cytochalasin B into the reaction significantly reduced GRASP65 bead aggregation compared with the DMSO control (Figure 7, C and D).
FIGURE 7:
Mena and F-actins enhance GRASP65 oligomerization. (A, B) GRASP65-coated beads were incubated with interphase cytosol (IC) immunodepleted by nonspecific or anti-Mena IgG. Beads were imaged by microscopy (A), and the percentage of beads in aggregates was quantified (B). (C, D) GRASP65-coated beads were incubated with IC in the presence of DMSO or 40 μM cytochalasin B and results quantified. (E, F) GRASP65-coated beads incubated with indicated purified proteins or IC were imaged and quantified. (G, H) GRASP65-coated beads incubated with interphase cytosol or interphase cytosol that was preincubated with BSA, Mena EVH1, domain or Mena EVH2 domain were imaged and quantified. Bars, 50 μm. ***p < 0.001.
Perhaps the best way to test how Mena and actin regulate GRASP65 oligomerization is to incubate GRASP65-coated beads with purified Mena and actin; however, efforts to express and purify full-length Mena failed, and we were able to purify only the fragments of Mena. Therefore we first determined whether Mena fragments could increase GRASP65 oligomerization. As shown in Figure 7, E and F, the EVH1 domain, which interacts with GRASP65 (Figure 3A) but does not promote actin polymerization due to the lack of the actin polymerization domain, did not increase GRASP65 bead aggregation. Similarly, the EVH2 domain, which promotes actin polymerization but does not bind to GRASP65, also did not enhance GRASP65 oligomerization. We then tested whether adding Mena fragments into interphase cytosol could inhibit GRASP65 oligomerization promoted by Mena. Indeed, adding purified EVH1 domain into the reaction, which competes with Mena in the cytosol for GRASP65 binding, significantly reduced the activity in the interphase cytosol in promoting GRASP65-coated bead aggregation. The EVH2 domain, known to compete for actin binding, had a similar effect. As a negative control, BSA had no effect on GRASP65 oligomerization (Figure 7, G and H). These results demonstrate that both GRASP65 interaction and actin elongation activity are required for Mena to enhance GRASP65 oligomerization and Golgi ribbon linking.
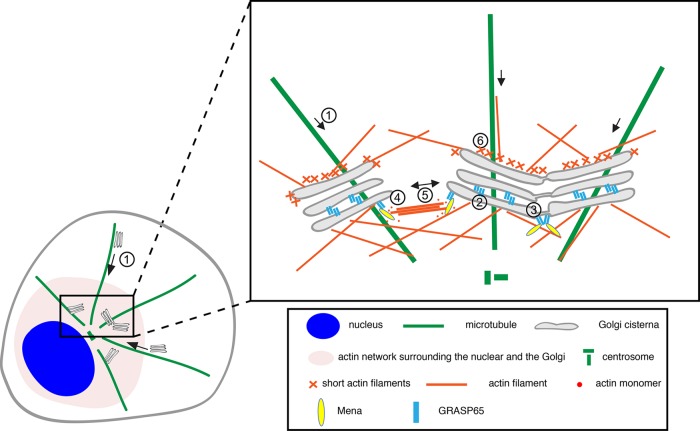
Taking the results together, we have identified Mena as a novel GRASP65-binding partner that plays an essential role in Golgi ribbon formation. This explains how GRASP65 plays a dual role in Golgi structure formation. When located between Golgi cisternae, GRASP65 oligomers directly tether adjacent cisternae into stacks. When located at the rims of Golgi stacks, GRASP65 recruits Mena onto the Golgi through a direct interaction and triggers local actin filament growth. Subsequently, the actin fibers cross-link GRASP65 and promote GRASP65 oligomerization. Both actin filaments and GRASP65 oligomers generate forces to link neighboring Golgi stacks into a ribbon (Figure 8).
FIGURE 8:
Model of Mena–GRASP65 interaction in Golgi ribbon linking. Golgi ministacks are transported to the cell center along microtubules (1). GRASP65 oligomers zipper Golgi cisternae into stacks (2) and tether nearby Golgi stacks into a ribbon (3). Ribbon formation initially requires Mena; GRASP65 recruits Mena to the Golgi, which promotes actin filament elongation (4). The actin fibers then link scattered Golgi stacks and pull them toward each other to form a ribbon (5). On both the cis and the trans faces of the stack, Golgi-associated actin filaments may facilitate membrane deformation for vesicle formation, fission and fusion, and short-range movements (6).
DISCUSSION
In this study, we identified the actin filament elongation factor Mena as a novel GRASP65-interacting protein. Mena directly interacts with GRASP65, and its depletion results in Golgi fragmentation. Of interest, VASP, another member of the Ena/VASP family of proteins, with an EVH1 domain highly similar to Mena, showed much lower affinity for GRASP65, indicating that the Mena–GRASP65 interaction is real and specific. Mena is recruited by GRASP65 to the Golgi membranes, where it promotes formation of local actin network or bundles, which enhance GRASP65 oligomerization and facilitate Golgi membrane tethering and fusion. Thus Mena–GRASP65 interaction couples actin polymerization, GRASP65 oligomerization, and Golgi ribbon linking.
How actin filaments facilitate membrane connection between Golgi stacks remains elusive, but previous studies suggested a few possibilities. First, actin filaments have been implicated in Golgi trafficking in many different ways (Egea et al., 2006). The WAHMM-Arp2/3 pathway functions in ER-to-Golgi anterograde transport (Campellone et al., 2008), whereas cdc42/N-WASP-Arp2/3 is required for Golgi-to-ER retrograde trafficking (Luna et al., 2002). Compared to the ER/Golgi interface, the coupling between actin and the fission of transport carriers at the trans-Golgi network (TGN) has been better studied. Both the cortactin/dynamin II/Arp2/3 and the Rho-mDia pathways promote actin nucleation and transport carrier formation at the TGN (Cao et al., 2005; Zilberman et al., 2011). Myosin and proteins like GOLPH3 also facilitate the fission of transport carriers at the TGN (Dippold et al., 2009; Miserey-Lenkei et al., 2010; Almeida et al., 2011). Because the Golgi morphology at the steady state is the result of a fine balance between membrane input and output, any significant imbalance of the flux will likely disrupt the Golgi structure (Wei and Seemann, 2010; Mironov and Beznoussenko, 2011). Suppressing ER-to-Golgi transport by inhibition of the WHAMM pathway resulted in Golgi fragmentation (Campellone et al., 2008), whereas reducing TGN export by inhibition of GOLPH3 or myosin resulted in a more condensed Golgi morphology (Dippold et al., 2009). Conversely, increasing Golgi-to-ER output by enhancing cdc42 activity (Dubois et al., 2005) or increasing TGN export through the Rho-mDia pathway both caused Golgi dispersion (Zilberman et al., 2011). However, in this study, our results showed that Mena is not required for membrane trafficking, suggesting that Mena maintains Golgi structure not through regulating trafficking.
Second, actin filaments might link Golgi stacks into pairs. In Drosophila S2 cells, actin depolymerization induces the scission of Golgi pairs at late G2 phase, which is mediated by the inactivation of actin nucleation–promoting factors WAVE/Scar/Abi (Kondylis et al., 2007). Similar Golgi pairs were observed in HeLa cells in the same study upon nocodazole treatment. In this study, we did not observe Golgi in pairs in our experimental conditions, such as after nocodazole treatment. Further incubation with latrunculin B or cytochalasin B or depletion of Mena had no effect on the size or number of Golgi stacks. Because we did not specifically select for G2 phase cells as in the previous report (Kondylis et al., 2007), it is possible that Golgi pairing only happens in G2 phase. Whether Mena and actin filaments are required for Golgi pairing in G2 phase needs further investigation.
Third, actin plays a critical role in vesicle priming and fusion at the plasma membrane (Bader et al., 2004). It is well accepted that vesicles derived from the TGN are transported along microtubule tracks to the cell cortex, where they are handed to actin, which pulls the vesicles toward the plasma membrane, an action required for priming the vesicles so that they are ready to fuse with the plasma membrane in a rapid and regulated manner (Gasman et al., 2004). The role of actin filaments in Golgi ribbon formation appears to be parallel to this function. Mena and actin filaments are required for fusion of Golgi membranes recovering from nocodazole treatment (Figure 5 and Supplemental Figure S4). Consistent with this observation, Mena and actin filaments are important for Golgi membrane fusion in the in vitro Golgi reassembly assay (Figure 6). In addition, incubation of purified Golgi stacks with purified actin results in clustering of Golgi membranes (Figure 6). These results suggest that actin filaments may pull Golgi stacks together to facilitate Golgi membrane fusion and ribbon formation. Therefore the cytoskeleton contributes to Golgi structure formation in a similar way as in exocytosis: first, microtubules take charge of the long-distance transport of Golgi stacks from cell periphery to the pericentriolar region, and then actin filaments modulated by the GRASP65–Mena interaction establish and maintain the integrity of the Golgi ribbon (Figure 8).
Our results on Mena are consistent with a recent study of its Drosophila homologue, Ena (Kannan et al., 2014). Both are localized on the cis-Golgi, and loss of function leads to Golgi fission, suggesting a conserved role for Mena and actin in Golgi organization between Drosophila and mammals. In addition, our study provided further information on how Mena and actin function to regulate Golgi structure. Mena is recruited to the Golgi membrane through the interaction with GRASP65 to promote actin polymerization for Golgi membrane fusion.
Another recent study showed that actin filaments are required for Golgi rigidity. In cells treated with cytochalasin D, an optically trapped bead could be pushed across the Golgi ribbon, whereas in nontreated cells, the bead was rejected out of the Golgi area (Guet et al., 2014). This result might suggest that actin filaments provide a rigid microenvironment surrounding the Golgi, or it might suggest that the rigidity of the Golgi reflects the intactness of the Golgi ribbon: when the Golgi stacks are unlinked, the optically trapped bead can be easily pushed across the Golgi through the gaps between the Golgi stacks. To summarize, actin and actin regulators are important for Golgi structure. At the cis-Golgi, Mena promotes actin filament elongation to maintain Golgi ribbon integrity.
GRASP65 was originally identified as a Golgi stacking factor (Barr et al., 1997; Shorter et al., 1999). Later reports suggested that GRASP65 might also play a role in Golgi ribbon linking because GRASP65 depletion caused Golgi fragmentation (Puthenveedu et al., 2006; Duran et al., 2008; Feinstein and Linstedt, 2008). These two models are not mutually exclusive, because both stacking and ribbon linking require GRASP65 to form trans-oligomers. Given the fact that GRASP65 is small and globular whereas the gaps between the stacks are large and heterogeneous, it is possible that other proteins might facilitate GRASPs in ribbon linking. Recently, a GRASP65-knockout mouse has been reported, with normal Golgi stacks but defects in ribbon linking (Veenendaal et al., 2014). One major concern of this mouse strain is that the targeting vector for knockout was inserted between exons 3 and 4 of the GRASP65 locus. The resulting mRNA, confirmed by real time quantitative PCR, contains exons 1–3 and encodes a 115-aa N-terminal fragment of GRASP65, including the membrane anchor, the entire PDZ1, which is sufficient to form oligomers but not the SPR domain, which contains phosphorylation and Mena-binding sites. Detection of this truncated protein was unsuccessful due to the lack of antibodies specific to this region (Veenendaal et al., 2014), but if produced, this fragment is sufficient for stacking but not ribbon linking due to the lack of Mena interaction.
In summary, we identified Mena as a novel protein that directly interacts with GRASP65 on the cis-Golgi and facilitates Golgi ribbon linking by enhancing local actin polymerization and GRASP65 oligomerization. Our study further revealed GRASP65 as a multifaceted protein in both Golgi stack and ribbon formation: the N-terminal GRASP domain (or the first PDZ domain) is responsible for membrane association and stacking by forming trans-oligomers (Tang et al., 2010b), whereas the C-terminal SPR domain is for both phosphorylation regulation during the cell cycle (Tang et al., 2012) and interaction with Mena and perhaps other proteins to link Golgi stacks into a ribbon.
MATERIALS AND METHODS
Reagents, plasmids, antibodies, and siRNA
All reagents were purchased from Sigma-Aldrich (St. Louis, MO), Thermo Fisher Scientific (Waltham, MA), Roche (Mannheim, Germany), EMD Millipore (Billerica, MA), and Calbiochem (Gibbstown, NJ), unless otherwise stated. Rhodamine–phalloidin was purchased from Life Technologies (Thermo Fisher Scientific). In all in vivo and in vitro assays, cytochalasin B and latrunculin B were used at concentrations of 40 and 0.5 μM, respectively. The following antibodies were used: monoclonal antibodies against Mena (a gift from Frank Gertler, MIT, Cambridge, MA), β-actin (AC-15; Sigma-Aldrich), α-tubulin (AA4.3-c; Developmental Studies Hybridoma Bank, Iowa City, IA), GST (B-14; Santa Cruz Biotechnology, Santa Cruz, CA), and histidine (His) tag (Proteintech, Chicago, IL); and polyclonal antibodies against human GRASP65 (Joachim Seemann, UT Southwestern, Dallas, TX), human GRASP55 (Proteintech), α–mannosidase II (Kelley Moreman, University of Georgia, Athens, GA), human TGN46 (AbD Serotec, Kidlington, UK), and syntaxin 5 (Graham Warren, Max F. Perutz Laboratories, Vienna, Austria).
Control siRNA (Silencer Select Negative Control #1 siRNA) was purchased from Applied Biosystems (Thermo Fisher Scientific). Mena-specific siRNA was synthesized by Sigma-Aldrich (sense sequence, GCCAUUCCUAAAGGGUUGAAGUACA) targeting to human Mena (5′-GCCATTCCTAAAGGGTTGAAGTACA-3′; Higashi et al., 2009). The β- and γ-actin–specific siRNAs were also synthesized by Sigma-Aldrich (sense sequences, AAUGAAGAUCAAGAUCAUUGC(TT) and AAGAGAUCGCCGCGCUGGUCA(TT)) targeting to human β-actin (5′-AATGAAGATCAAGATCATTGC-3′) and γ-actin (5′-AAGAGATCGCCGCGCTGGTCA-3′; Harborth et al., 2001). GRASP65-specific siRNA was Stealth siRNA oligo synthesized by Thermo Fisher Scientific (sense sequence, CCUGAAGGCACUACUGAAAGCCAAU) targeting to human GRASP65 sequence (5′-CCTGAAGGCACTACTGAAAGCCAAT-3′; Sutterlin et al., 2005; Tang et al., 2010b).
Constructs of GST- or His-tagged GRASP65 fragments or full length were prepared as described previously (Wang et al., 2005). GFP-Mena cDNA was excised from a construct of pMSCV-EGFP-MENA (a gift from Frank B. Gertler) and inserted into the pEGFP-C1 vector. RNAi-resistant mutant of Mena was generated by mutating the siRNA targeting sequence 5′-GCCATTCCTAAAGGGTTGAAGTACA-3′ to 5′-GCGATCCCAAATGGCTTAAAGTACA-3′. GFP-tagged Mena ΔFAB and ΔGAB mutants were constructed based on the RNAi-resistant form of GFP-Mena, with aa 411-429 and 362–37 deleted, respectively. Constructs of GST-tagged Mena EVH1 and EVH2 domain (in pGEX-2TK) were kindly provided by Frank B. Gertler. GFP-VASP construct was a gift from Maria Diakonova (University of Toledo, Toledo, OH). GFP-tagged GRASP65 mutants M1–M6 were generated by site-directed mutagenesis based on the template pEGFP-N1-rGRASP65 (Wang et al., 2005). All constructs were confirmed by DNA sequencing.
Bead aggregation assay
Bead aggregation assays were performed as previously described (Wang et al., 2003; 2005; Tang et al., 2010a). Dynabeads M-500 (or M-450) Subcellular was purchased from Invitrogen. For quantification of the aggregation efficiency, 10–20 random phase contrast digital images of each reaction were captured by Zeiss Observer Z1 epifluorescence microscope with a 10× lens. Images were analyzed with the MATLAB7.4 software to determine the surface area of objects, which was used to calculate the number of beads in the clusters. Aggregates were defined as those with ≥6 beads. From 10 to 20 random taken images were used to quantify the aggregation efficiency in each experiment.
To characterize the active components in the cytosol that enhance GRASP65 oligomerization, the aggregates of GRASP65-coated beads were washed with Tris-HCl 50 mM, pH 7.4, 0.2 M sucrose, 50 mM KCl, 20 mM β-glycerophosphate, 15 mM EGTA, 10 mM MgCl2, 2 mM ATP, 1 mM GTP, 1 mM glutathione, and protease inhibitors (MEB) buffer and incubated with purified Cdk1 and Plk at 37ºC for 60 min as previously described (Wang et al., 2003). The dispersed beads were then washed with KHM buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.0, 0.2 M sucrose, 60 mM KCl, 5 mM Mg(OAc)2, 2 mM ATP, 1 mM GTP, 1 mM glutathione, and protease inhibitors) and further incubated with either interphase cytosol or purified PP2A at 37°C for 60 min (Tang et al., 2010a). In other reactions, cytosol was heat inactivated at 56°C for 30 min or 95°C for 3 min or pretreated by adding 3 M KCl stock to a final concentration of 0.5 M and kept on ice for 30 min. These pretreated cytosols were then applied to the bead aggregation assay in parallel with nontreated cytosol. To determine the requirement of nucleotide, we tested 2 mM ATP or GTP or their analogues or 0.02 U/μl apyrase in the assay in the presence of interphase cytosol. To evaluate the function of F-actin and Mena in GRASP65 oligomerization, we added 40 μM cytochalasin B or 20 μg of EVH1 or EVH2 recombinant protein with interphase cytosol into the reaction.
Ammonium sulfate precipitation
Interphase cytosol prepared from HeLa S3 cells (Rabouille et al., 1995; Tang et al., 2010a) was first diluted to 1 mg/ml and precipitated with the indicated concentration (percentage saturation, such as 15%) of ammonium sulfate. Ammonium sulfate was slowly added at 4ºC with stirring for 60 min to reach equilibrium. After centrifugation in a bench-top minicentrifuge for 30 min at full speed and 4ºC, the ammonium sulfate concentration in the supernatant was further increased to a higher concentration (e.g., 30%) by supplementing additional ammonium sulfate powder. To prepare 15–30% ammonium sulfate precipitation, we first treated the cytosol with 15% ammonium sulfate and centrifuged it as described. The supernatant was added with ammonium sulfate salt to reach 30% and centrifuged again. This pellet is referred to as the 15–30% ammonium sulfate precipitate. The precipitants in each pellet were dissolved and dialyzed into KHM buffer. The dissolved proteins were analyzed by SDS–PAGE or used in other assays.
Velocity gradient
Protein samples were loaded onto a 15–35% glycerol gradient and then centrifuged in a VTi65 rotor at 65,000 rpm (372,645 × g) for 80 min at 4ºC. After centrifugation, the gradient was fractionated from the top to the bottom into 12 fractions. Proteins from each fraction were analyzed by SDS–PAGE or concentrated for further use.
Identification of GRASP65-interacting proteins
Interphase cytosol was diluted and fractionated with 15–30% ammonium sulfate precipitation as described. Proteins in the pellet were dissolved and dialyzed into KHM buffer. Dissolved proteins were incubated with BSA or His-GRASP65–coupled, CNBr-activated Sepharose 4B beads (Sigma-Aldrich) in the presence of an ATP-regenerating system (10 mM creatine phosphate, 1 mM ATP, 20 μg/ml creatine kinase, and 20 ng/ml cytochalasin B) in KHM buffer at 4ºC overnight. The beads were washed extensively. Bound proteins were boiled in SDS buffer and analyzed by SDS–PAGE. Specific bands that appeared in the lane of GRASP65 beads but not in the lane of BSA beads were excised and analyzed by mass spectrometry (Proteomics and Peptide Synthesis Core, University of Michigan) and searched against International Protein Index human databases as previously described (Chen et al., 2010, 2012).
Preparation of recombinant proteins
His-tagged GRASP65 and Mena EVH1 were expressed and purified on nickel (Qiagen, Hilden, Germany) agarose as previously described (Wang et al., 2003). GST-tagged GRASP65 fragments, as well as Mena EVH1 and EVH2, were expressed in BL21(DE3)Gold bacteria and purified on glutathione–Sepharose beads (GE Healthcare Life Sciences, Pittsburgh, PA).
In vitro binding assay
To confirm that Mena interacts with GRASP65 in vitro, proteins in 10 mg of interphase cytosol were fractioned by 15–30% ammonium sulfate precipitation and loaded onto 50 μg of BSA or MBP-GRASP65–coupled CNBr beads in the presence of an ATP-regenerating system in KHM buffer at 4ºC overnight. The beads were washed extensively, and the proteins enriched on the beads were boiled in SDS buffer and analyzed by SDS–PAGE and Western blot.
To map the interaction domains of GRASP65 and Mena, 20 μg of purified GST- or His-tagged EVH1 was incubated with 20 μg of BSA, His-GRASP65, His-GRASP65 (aa 1–201), or His-GRASP65 (aa 202–446) coupled to CNBr beads (preblocked with BSA before the incubation) in the presence of an ATP-regenerating system in KHM buffer at 4ºC overnight. The beads were washed extensively and then analyzed by SDS–PAGE and Western blot for GST or His tag.
To map the Mena interaction site on GRASP65, 5 μg of GST or GST-tagged GRASP65 fragments was prebound to glutathione beads (GE Healthcare Life Sciences) and further incubated with 6 mg of HeLa cell lysate. Proteins that were copurified with the beads were analyzed by SDS–PAGE and Western blot for Mena.
Coimmunoprecipitation
For coimmunoprecipitation of endogenous Mena and GRASP65, HeLa cells transfected with control or Mena-specific siRNA were scraped and resuspended in PBS, lysed in immunoprecipitation buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.2% NP40, with protease inhibitors) and cleared by centrifugation in a bench-top centrifuge at 4ºC and top speed for 10 min. Mena antibodies preloaded to protein G beads were added to the lysate and incubated overnight at 4ºC with gentle agitation. The HeLa cell lysates were precleared by incubation with 20 μl of protein G beads for 1 h at 4ºC and then incubated with antibody-prebound protein G beads at 4ºC overnight. After washing, the beads were analyzed by SDS–PAGE and Western blot.
For coimmunoprecipitation of exogenously expressed GFP-tagged Mena and GRASP65, GFP-tagged Mena, VASP, or GRASP65 WT and its mutants were transfected into HeLa cells. After 24 h, cells were collected and lysed into lysis buffer (20 mM HEPES, pH 7.4, 300 mM NaCl, 0.1% Triton X-100, and protease inhibitors). Lysates of equal amount of proteins were incubated with protein A beads precoupled with GFP antibodies at 4ºC overnight. After washing, the beads were analyzed by SDS–PAGE and Western blot.
Cell culture and microscopy
HeLa cells were routinely cultured in DMEM supplemented with 10% donor bovine serum (Life Technologies/Invitrogen), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). For knockdown experiments, HeLa cells were transfected using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s instructions. For Mena and actin depletion, cells were analyzed 48 h after transfection. For GRASP65 depletion, cells were analyzed 96 h after transfection to achieve a better knockdown efficiency. For protein expression, HeLa cells were transfected with indicated constructs using Lipofectamine 2000 (Invitrogen) and analyzed 24 h afterward. For the rescue of Mena knockdown with exogenously expressed Mena, cells were transfected first with a Mena-specific siRNA for 24 h and then by either a GFP cDNA or an RNAi-resistant mutant of Mena with a GFP tag for another 24 h. To rescue the Golgi in GRASP65-depleted cells with GRASP65 mutants, HeLa cells were first transfected with siRNA targeting to human GRASP65 for 72 h and then transfected with GFP or GFP-tagged rat GRASP65 WT or mutants for an additional 24 h. For VSV-G-ts045 transport assay, cells pretransfected with siRNA were exposed to adenovirus that were packed with VSV-G-ts045-GFP plasmid and incubated at 40.5°C overnight to allow VSV-G-ts045 to accumulate in the ER and then shifted to 32°C for indicated times to allow VSV-G transport (Xiang et al., 2013).
Immunofluorescence and electron microscopy were performed as previously described (Wang et al., 2005; Tang et al., 2010a). Images were taken with a Zeiss Observer Z1 epifluorescence microscope with a 63× oil lens. In interphase cells, fragmented Golgi was defined as disconnected or scattered dots. To quantify the percentage of cells with fragmented Golgi, >300 cells transfected with control or target specific siRNAs were counted. The results are presented as the mean ± SEM from three independent experiments.
To quantify the numbers of Golgi particles in each cell, cells that were treated with nocodazole (with or without washout) were fixed and stained for GRASP65. Images were obtained with a Leica SP5 confocal laser-scanning microscope using a 100× oil objective, captured using fixed parameters with 0.3-μm intervals at the z-axis, and then processed for maximum value projection. The number of Golgi particle in each cell was quantified by the Analyze particles function of ImageJ software (National Institutes of Health, Bethesda, MD).
For EM analysis, Golgi stacks were identified using morphological criteria and quantified using standard stereological techniques (Wang et al., 2005; Tang et al., 2010a). Golgi images were captured at 11,000× magnification. The longest cisterna was measured as the cisternal length of a Golgi stack using the ruler tool in Photoshop CS5. At least 20 cells were quantified in each experiment, and the results represent at least three independent experiments.
Immunodepletion of Mena from interphase cytosol
Interphase cytosol prepared from HeLa S3 cells (Rabouille et al., 1995; Tang et al., 2010a) was incubated with nonspecific control IgG or anti-Mena IgG and protein G–agarose beads at 4ºC overnight. The beads were pelleted and supernatant analyzed by immunoblotting for Mena. An equal amount of cytosol immunodepleted by control IgG or anti-Mena IgG was used in the bead aggregation assay and the Golgi disassembly and reassembly assay described next.
Golgi disassembly and reassembly assay and quantification
The Golgi disassembly assay was performed as described previously (Tang et al., 2010a). Briefly, purified Golgi membranes (20 μg) were mixed with 2 mg of mitotic cytosol, 1 mM GTP, and an ATP-regenerating system in MEB buffer in a final volume of 200 μl. After incubation for 60 min at 37°C, MGFs were isolated, and soluble proteins were removed by centrifugation (135,000 × g for 30 min in a TLA55 rotor) through a 0.4 M sucrose cushion in KHM buffer onto 6 μl of 2 M sucrose cushion. The membranes were resuspended in KHM buffer and either fixed or processed for EM (Tang et al., 2010a) or used in reassembly reactions. For Golgi reassembly, 20 μg of mitotic Golgi fragments was mixed with 400 μg of interphase cytosol (either control or Mena depleted as described) in the presence of an ATP regeneration system in a final volume of 30 μl in KHM buffer and incubated at 37°C for 60 min. The membranes were pelleted by centrifugation and processed for EM.
To quantify the reassembly of the Golgi, EM images of Golgi membranes were captured at 11,000× magnification. The percentage of membranes in cisternae or vesicles was determined by the intersection method (Misteli and Warren, 1994; Tang et al., 2010a). Cisternae were defined as long membrane profiles with a length greater than four times of their width, and the latter did not exceed 60 nm. Normal cisternae ranged from 20 to 30 nm in width and were longer than 200 nm. To quantify the relative Golgi regrowth rate, percentage of membranes in vesicles of MGFs was obtained first and designated A%. The percentages of membranes in vesicles of reassembled Golgi in control IgG-depleted interphase cytosol and Mena-depleted cytosol were obtained and designated B% and C%, respectively. The relative Golgi regrowth rate was calculated as (A% − C%)/(A% − B%) × 100%. At lease 10 random images were quantified in each set of experiments. All experiments were repeated at least three times.
Supplementary Material
Acknowledgments
We thank Maria Diakonova, Frank B. Gertler, Kelley Moremen, Joachim Seemann, and Graham Warren for antibodies and plasmids. We thank other members of the Wang lab for suggestions and reagents. This work was supported in part by the National Institutes of Health (Grants GM087364, GM105920, and GM112786), the American Cancer Society (Grant RGS-09-278-01-CSM), the Mizutani Foundation for Glycoscience, MCubed, and the Fastforward Protein Folding Disease Initiative of the University of Michigan to Y.W. D.T. was supported by a Rackham Predoctoral Fellowship from the University Michigan and granted a Chinese Government Award for Outstanding Self-financed Students Abroad by the China Scholarship Council.
Abbreviations used:
- EM
electron microscopy
- EVH
Enabled/vasodilator-stimulated phosphoprotein homology
- GRASP
Golgi reassembly stacking protein
- Mena
mammalian enabled
- Plk
polo-like kinase
- PP2A
protein phosphatase 2A
- TGN
trans-Golgi network
- VSV-G
vesicular stomatitis virus G protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0650) on November 4, 2015.
REFERENCES
- Almeida CG, Yamada A, Tenza D, Louvard D, Raposo G, Coudrier E. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat Cell Biol. 2011;13:779–789. doi: 10.1038/ncb2262. [DOI] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert C, Linstedt AD. Dual anchoring of the GRASP membrane tether promotes trans pairing. J Biol Chem. 2010;285:16294–16301. doi: 10.1074/jbc.M110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MF, Doussau F, Chasserot-Golaz S, Vitale N, Gasman S. Coupling actin and membrane dynamics during calcium-regulated exocytosis: a role for Rho and ARF GTPases. Biochim Biophys Acta. 2004;1742:37–49. doi: 10.1016/j.bbamcr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: structure, function and interactions. FEBS Lett. 2002;513:45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Braet F, De Zanger R, Jans D, Spector I, Wisse E. Microfilament-disrupting agent latrunculin A induces and increased number of fenestrae in rat liver sinusoidal endothelial cells: comparison with cytochalasin B. Hepatology. 1996;24:627–635. doi: 10.1053/jhep.1996.v24.pm0008781335. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TE, Small JV, Curth U, Dickinson RB, Faix J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownhill K, Wood L, Allan V. Molecular motors and the Golgi complex: staying put and moving through. Semin Cell Dev Biol. 2009;20:784–792. doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- Chen X, Andrews PC, Wang Y. Quantitative analysis of liver Golgi proteome in the cell cycle. Methods Mol Biol. 2012;909:125–140. doi: 10.1007/978-1-61779-959-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Simon ES, Xiang Y, Kachman M, Andrews PC, Wang Y. Quantitative proteomics analysis of cell cycle regulated Golgi disassembly and reassembly. J Biol Chem. 2010;285:7197–7207. doi: 10.1074/jbc.M109.047084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluett EB, Brown WJ. Adhesion of Golgi cisternae by proteinaceous interactions: intercisternal bridges as putative adhesive structures. J Cell Sci. 1992;103:773–784. doi: 10.1242/jcs.103.3.773. [DOI] [PubMed] [Google Scholar]
- Colon-Franco JM, Gomez TS, Billadeau DD. Dynamic remodeling of the actin cytoskeleton by FMNL1gamma is required for structural maintenance of the Golgi complex. J Cell Sci. 2011;124:3118–3126. doi: 10.1242/jcs.083725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois T, Paleotti O, Mironov AA, Fraisier V, Stradal TE, De Matteis MA, Franco M, Chavrier P. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–364. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Lazaro-Dieguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Egea G, Serra-Peinado C, Salcedo-Sicilia L, Gutierrez-Martinez E. Actin acting at the Golgi. Histochem Cell Biol. 2013;140:347–360. doi: 10.1007/s00418-013-1115-8. [DOI] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yu W, Li X, Lin S, Zhou Y, Hu J, Liu X. Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 proteins. J Biol Chem. 2013;288:28418–28427. doi: 10.1074/jbc.M113.478024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasman S, Chasserot-Golaz S, Malacombe M, Way M, Bader MF. Regulated exocytosis in neuroendocrine cells: a role for subplasmalemmal Cdc42/N-WASP-induced actin filaments. Mol Biol Cell. 2004;15:520–531. doi: 10.1091/mbc.E03-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Guet D, Mandal K, Pinot M, Hoffmann J, Abidine Y, Sigaut W, Bardin S, Schauer K, Goud B, Manneville JB. Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr Biol. 2014;24:1700–1711. doi: 10.1016/j.cub.2014.06.048. [DOI] [PubMed] [Google Scholar]
- Gurel PS, Hatch AL, Higgs HN. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr Biol. 2014;24:R660–R672. doi: 10.1016/j.cub.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Higashi M, Ishikawa C, Yu J, Toyoda A, Kawana H, Kurokawa K, Matsuda M, Kitagawa M, Harigaya K. Human Mena associates with Rac1 small GTPase in glioblastoma cell lines. PLoS One. 2009;4:e4765. doi: 10.1371/journal.pone.0004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Wang Y. Golgi defects enhance APP amyloidogenic processing in Alzheimer’s disease. Bioessays. 2015;37:240–247. doi: 10.1002/bies.201400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan R, Kuzina I, Wincovitch S, Nowotarski SH, Giniger E. The Abl/enabled signaling pathway regulates Golgi architecture in Drosophila photoreceptor neurons. Mol Biol Cell. 2014;25:2993–3005. doi: 10.1091/mbc.E14-02-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, van Nispen Tot Pannerden HE, Herpers B, Friggi-Grelin F, Rabouille C. The Golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lanzetti L. Actin in membrane trafficking. Curr Opin Cell Biol. 2007;19:453–458. doi: 10.1016/j.ceb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Lazaro-Dieguez F, Jimenez N, Barth H, Koster AJ, Renau-Piqueras J, Llopis JL, Burger KN, Egea G. Actin filaments are involved in the maintenance of Golgi cisternae morphology and intra-Golgi pH. Cell Motility Cytoskeleton. 2006;63:778–791. doi: 10.1002/cm.20161. [DOI] [PubMed] [Google Scholar]
- Loureiro JJ, Rubinson DA, Bear JE, Baltus GA, Kwiatkowski AV, Gertler FB. Critical roles of phosphorylation and actin binding motifs, but not the central proline-rich region, for Ena/vasodilator-stimulated phosphoprotein (VASP) function during cell migration. Mol Biol Cell. 2002;13:2533–2546. doi: 10.1091/mbc.E01-10-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M. Structural organization of the Golgi apparatus. Curr Opin Cell Biol. 2011;23:85–93. doi: 10.1016/j.ceb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Luna A, Matas OB, Martinez-Menarguez JA, Mato E, Duran JM, Ballesta J, Way M, Egea G. Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell. 2002;13:866–879. doi: 10.1091/mbc.01-12-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin AA. Dispersal of Golgi apparatus in nocodazole-treated fibroblasts is a kinesin-driven process. J Cell Sci. 1997;110:2495–2505. doi: 10.1242/jcs.110.19.2495. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV. Molecular mechanisms responsible for formation of Golgi ribbon. Histol Histopathol. 2011;26:117–133. doi: 10.14670/HH-26.117. [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J Pharmacol Sci. 2010;112:255–264. doi: 10.1254/jphs.09r03cr. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Ramabhadran V, Korobova F, Rahme GJ, Higgs HN. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22:4822–4833. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y. Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol. 2013;23:296–304. doi: 10.1016/j.tcb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010a;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Vielemeyer O, Perez F, Wang Y. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol Open. 2012;1:1204–1214. doi: 10.1242/bio.20122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in golgi cisternal stacking and cell cycle progression. Traffic. 2010b;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Truschel ST, Sengupta D, Foote A, Heroux A, Macbeth MR, Linstedt AD. Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J Biol Chem. 2011;286:20125–20129. doi: 10.1074/jbc.C111.245324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal T, Jarvela T, Grieve AG, van Es JH, Linstedt AD, Rabouille C. GRASP65 controls the cis Golgi integrity in vivo. Biol Open. 2014;3:431–443. doi: 10.1242/bio.20147757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J, Duran JM, Forlanelli E, Alleaume AM, Egorov M, Polishchuk R, Molina H, Malhotra V. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J Cell Biol. 2009;187:1055–1069. doi: 10.1083/jcb.200908040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi cisternal unstacking stimulates COPI vesicle budding and protein transport. PLoS One. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Unraveling the Golgi ribbon. Traffic. 2010;11:1391–1400. doi: 10.1111/j.1600-0854.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix D, Katoh T, Aoki K, Tiemeyer M, Wang Y. Regulation of cargo sorting and glycosylation by the Golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Linstedt AD. Golgi positioning. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005322. a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, Bershadsky A. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–2911. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.