Centromeres are embedded within heterochromatin but are transcriptionally active. Centromeric transcription and the centromere function of a human artificial chromosome resist repression mediated by nucleation of repressive marks H3K27me3 or H3K9me3 via tethering of EZH2 or the SET domain of Suv39h1, respectively.
Abstract
Centromeres are characterized by the centromere-specific H3 variant CENP-A, which is embedded in chromatin with a pattern characteristic of active transcription that is required for centromere identity. It is unclear how centromeres remain transcriptionally active despite being flanked by repressive pericentric heterochromatin. To further understand centrochromatin’s response to repressive signals, we nucleated a Polycomb-like chromatin state within the centromere of a human artificial chromosome (HAC) by tethering the methyltransferase EZH2. This led to deposition of the H3K27me3 mark and PRC1 repressor binding. Surprisingly, this state did not abolish HAC centromere function or transcription, and this apparent resistance was not observed on a noncentromeric locus, where transcription was silenced. Directly tethering the reader/repressor PRC1 bypassed this resistance, inactivating the centromere. We observed analogous responses when tethering the heterochromatin Editor Suv39h1-methyltransferase domain (centromere resistance) or reader HP1α (centromere inactivation), respectively. Our results reveal that the HAC centromere can resist repressive pathways driven by H3K9me3/H3K27me3 and may help to explain how centromeres are able to resist inactivation by flanking heterochromatin.
INTRODUCTION
Chromatin is the composite of proteins and nucleic acids that forms the chromosomes and regulates access to DNA. This regulation takes place largely through chemical modifications of DNA or the histones (termed “chromatin marks”) that can change the local electrostatic behavior and/or act as docking sites for secondary chromatin effectors (dubbed “readers” of marks; Allfrey et al., 1964; Bannister et al., 2001; Bannister and Kouzarides, 2011). Readers recruit downstream activities that establish characteristic chromatin states. Some marks can also interfere with the binding of readers, and histone variants can confer additional functional specificity (Talbert et al., 2012). It is believed that the combination of chromatin marks and effector readers at a given locus is responsible for determining and maintaining its chromatin state, ranging from transcriptionally active to poised or constitutively silenced (Strahl and Allis, 2000; Ernst and Kellis, 2010; Filion et al., 2010).
Experimental introduction of certain marks by chromatin editors into a given locus can cause specific downstream effectors to be subsequently recruited and induce a de novo change in overall chromatin state (Hansen et al., 2008; Kagansky et al., 2009). Some of these initial modifications are relatively stable, thus granting the locus a degree of “memory” that promotes continuous recruitment of downstream readers that functionally maintain that chromatin state (Zaidi et al., 2014; Audergon et al., 2015). These sequential chromatin pathways take the form of Editor → Mark → Reader → Chromatin state, although it should be noted that there is significant cross-talk and feedback between each step.
The centromere is a specialized chromosomal locus (Fukagawa and Earnshaw, 2014) that is the foundation for assembly of the kinetochore, a multiprotein superstructure that directs chromosome segregation during mitosis and meiosis (Cheeseman and Desai, 2008). Most centromeres are located in gene-poor loci containing tandemly repeated DNA (Fukagawa and Earnshaw, 2014). Of interest, specification of centromere location on the chromosome is not stringently dependent on DNA sequence (Voullaire et al., 1993; Sullivan and Schwartz, 1995; du Sart et al., 1997; Tyler-Smith et al., 1999). Instead, centromeres seem to be determined epigenetically (Earnshaw and Migeon, 1985; Earnshaw et al., 1989; Vafa and Sullivan, 1997; Warburton et al., 1997; Mendiburo et al., 2012), at least in part by the presence of CENP-A, a centromere-specific H3 variant (Earnshaw and Rothfield, 1985; Vafa and Sullivan, 1997; Warburton et al., 1997).
In many eukaryotes, the centromere and the pericentromeric region are widely enriched for chromatin marks characteristic of heterochromatin, including H3K9me3 (histone H3 lysine 9 trimethylation) and DNA methylation (Aagaard et al., 1999; Blower and Karpen, 2001; Guenatri et al., 2004; Partridge et al., 2000; Sullivan and Karpen, 2004). Pericentromeric heterochromatin has been suggested to play a role in sister chromatid cohesion (Bernard et al., 2001; Nonaka et al., 2002) during cell division.
Heterochromatin is typically associated with constitutive transcriptional repression that can spread across adjacent loci by progressive addition of H3K9me3 to nearby nucleosomes (Schultz, 1936; Bannister et al., 2001; Seum et al., 2001; Jost et al., 2012). Paradoxically, centromeres are actively transcribed in many organisms (reviewed in Scott, 2013; Fukagawa and Earnshaw, 2014). Human centromeric chromatin contains, in addition to CENP-A, marks such as H3K4me2 and H3K36me2 that correlate with active transcription (Sullivan and Karpen, 2004; Bergmann et al., 2011). It also recruits RNA polymerase II (Bergmann et al., 2012a; Chan et al., 2012) and produces transcripts (Saffery et al., 2003; Yan et al., 2005; Wong et al., 2007). This particular chromatin signature has been termed centrochromatin (Sullivan and Karpen, 2004). It is unknown how centromeres maintain this open/transcribed chromatin signature in close proximity to heterochromatin, particularly given that the underlying DNA across both these domains is the same repetitive sequence.
Recent advances in our understanding of centrochromatin have stemmed from using human artificial chromosome (HAC) technology and protein-tethering strategies (Nakano et al., 2008; Bergmann et al., 2012b; Kouprina et al., 2013). By generating HACs containing engineered centromeric α-satellite repeats that include TetO sites (henceforth referred to as alphoidTetO), it has been possible to specifically recruit TetR proteins fused to chromatin modifiers (editors) to the centromeric region of these chromosomes (Nakano et al., 2008).
Evidence from HAC-tethering studies has shown that local active transcription, or a chromatin signature compatible with it, may be required for centromere function in human cells despite the repressive nature of the surrounding (peri)centromeric heterochromatin. Tethering of the repressive heterochromatin editor Suv39h1 (Ohzeki et al., 2012), the repressive reader HP1α (Nakano et al., 2008), or the repressive scaffold KAP1 (Cardinale et al., 2009) caused loss of HAC centromeric transcription and inactivation of its centromere. Similar results were obtained by tethering LSD1 (Bergmann et al., 2011), which demethylates H3K4 and recruits other corepressors of transcription via CoREST (Lee et al., 2005).
Understanding how centrochromatin responds to diverse kinds of challenges to its transcriptional activity can help tease out key determinants that sustain its function. In this study, we probed the response of the HAC centromere to facultative heterochromatin mediated by proteins of the Polycomb group (Lewis, 1978; Di Croce and Helin, 2013; henceforth referred to as Polycomb chromatin). By comparatively generating H3K27me3 or H3K9me3 within the HAC centromere, we found that it may possess mechanisms that prevent repressive pathways producing these marks from fully translating into effective repression.
RESULTS
Diversity of chromatin signatures at human centromeres
To better understand the changes that nucleation of repressive chromatin could have on a human centromere, we first analyzed the natural distribution of several chromatin marks on human endogenous centromeres and the HAC.
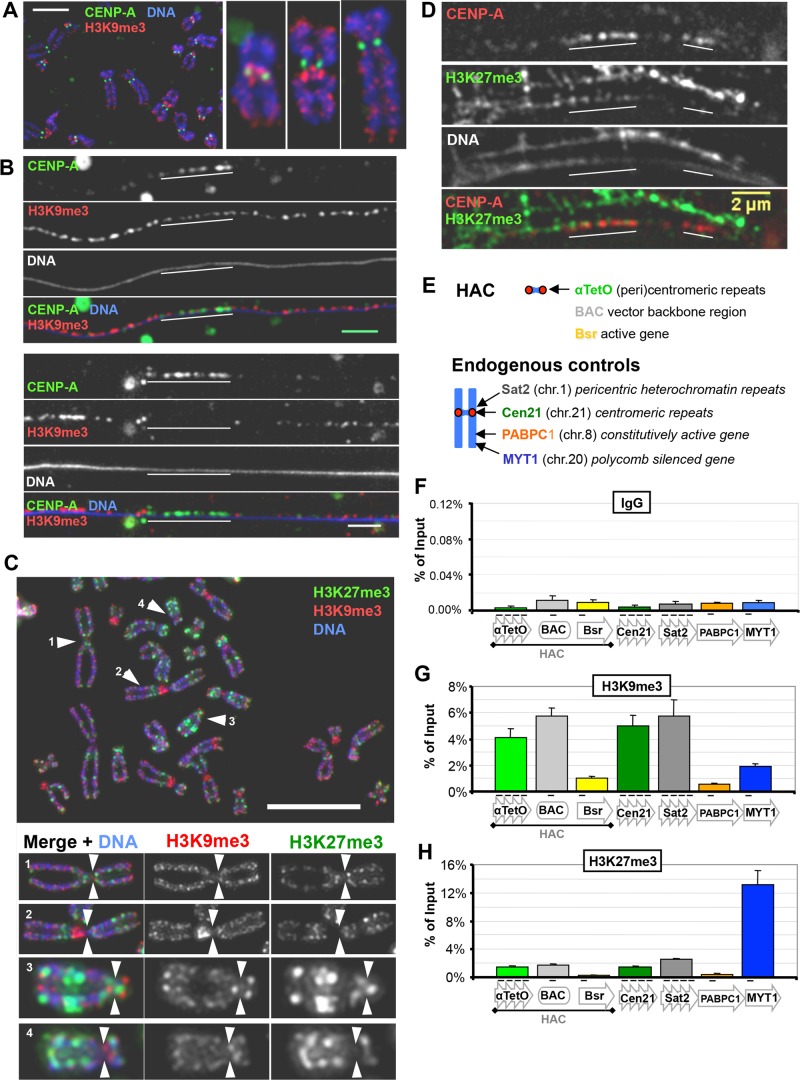
Pericentromeric regions of most metaphase chromosomes are enriched for the heterochromatic mark H3K9me3 (Figure 1A). Analysis of extended chromatin fibers of these chromosomes revealed that H3K9me3 enrichment mostly flanks the centromere core but is not entirely excluded from it (Figure 1B). The centromere core region, as delimited by the CENP-A signal, has been shown to be enriched with euchromatin-related marks (Sullivan and Karpen, 2004; Greaves et al., 2007; Bergmann et al., 2011). In agreement with this, our chromatin fiber analysis also revealed the presence of low levels of H2A.Z (a mark associated with enhancer loci and the 5′ region of active genes; Guillemette and Gaudreau, 2006) and H3K4me3 (a mark associated with the promoter and 5′ region of nonrepressed genes; Guenther et al., 2007) across this region (Supplemental Figure S1, A and B). This domain organization has been observed in humans and other metazoans, where clusters of CENP-A–containing nucleosomes are interspersed with H3-containing nucleosomes bearing marks associated with active transcription, whereas the surrounding pericentromeric nucleosomes are highly enriched for marks characteristic of heterochromatin (Blower and Karpen, 2001; Sullivan and Karpen, 2004; Lam et al., 2006). Together these data further emphasize the contrast in chromatin state between the pericentromere and the core CENP-A region.
FIGURE 1:
Distribution of repressive chromatin marks H3K9me3 and H3K27me3 at human (peri)centromeres. (A) H3K9me3 is enriched at pericentromeres, adjacent to CENP-A domains. Mitotic chromosomes from human HT1080 cells were stained with antibodies against H3K9me3 (red) and CENP-A (green). Example of a chromosome spread (scale bar, 5 μm). (B) As in A, but showing chromatin fibers from mitotic chromosomes. Examples of stretched chromatin fibers (scale bar, 2 μm). (C) H3K27me3 is enriched at some pericentromeres. Metaphase spreads of chromosomes from HeLa 1C7HAC cells were stained with antibodies against H3K9me3 (red) and H3K27me3 (green). Insets, individual chromosomes; arrows indicate the primary constriction (centromere). Scale bar, 5 μm. (D) H3K27me3 can be found adjacent to CENP-A domains. Mitotic stretched chromatin fibers of HeLa 1C7HAC cells, stained with antibodies against CENP-A (red) and H3K27me3 (green). Scale bar, 2 μm. (E–H) Chromatin analysis by ChIP shows that HAC and chromosome 21 (peri)centromeres are primarily heterochromatic and not enriched in Polycomb chromatin. ChIP analysis of HeLa 1C7HAC cells was performed using antibodies against H3K9me3 and H3K27me3, as shown in G and H, respectively. Mouse unspecific IgG was used as negative pull-down control for ChIP, as shown in F. Primer sets used are shown at the top, with Sat2 and MYT1 used as positive controls for bona fide heterochromatic and Polycomb loci, respectively. Mean of three independent experiments; error bars denote SEM.
Of interest, some pericentromeric regions did not exhibit obvious H3K9me3 enrichment (Figure 1A). Instead, they were enriched for the Polycomb mark H3K27me3 (Figure 1C). H3K27me3, like H3K9me3, can also occasionally be found close to CENP-A in extended chromatin fibers (Figure 1D). This variability between chromosomes with regard to their complement of α-satellite and other repetitive DNAs has long been known (Waye and Willard, 1987), and recent reports from the 1000 Genomes Project Consortium (2015) and others have revealed that extraordinary levels of variability extend into regions of unique-sequence DNA as well. Analysis of the most highly repetitive regions has lagged behind the rest of the genome, but new approaches to sequencing and data analysis are beginning to document more precisely the variability in these chromosome regions (Aldrup-Macdonald and Sullivan, 2014; Miga, 2015). In addition, enrichment of H3K27me3 at pericentromeres of the same chromosome is reported to vary between different cell types (Mravinac et al., 2009). We conclude that both active and repressive chromatin signatures of more than one type can occur in close proximity to the CENP-A domain.
We quantified the levels of these chromatin marks on the HAC by chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) in HeLa 1C7 cells. HeLa 1C7 cells carry one copy of the alphoidTetO HAC per cell (Cardinale et al., 2009; Bergmann et al., 2011; see Materials and Methods). The HAC alphoidTetO array and adjacent noncentromeric BAC regions are preferentially enriched with H3K9me3 (Figure 1, E and G) at levels similar to those found at the centromeric α-satellite repeats of chromosome 21 (Cen21) and the pericentromeric Sat2 repeat and have lower levels of H3K27me3 (Figure 1, E and H). Quantitation revealed that the levels of RNA polymerase II in its elongating state (Ser2ph) are higher on the HAC and at Cen21 than at the pericentromeric Sat2 locus (Supplemental Figure S1, C, E, and F). Thus alphoid arrays apparently possess a higher RNA polymerase II transcriptional activity than pericentromeres, which are predominantly heterochromatic. It is possible that other RNA polymerases might be involved in Sat2 transcription, but qPCR reveals that levels of Sat2 transcription are far below those of α-satellite transcription in 1C7 cells (see later discussion of Figure 4C). Of interest, levels of both elongating and initiating RNA polymerase II are higher on the alphoid HAC array than at Cen21 (Supplemental Figure S1, C, E, and F), suggesting that the HAC centromere is more transcriptionally active than endogenous centromeres.
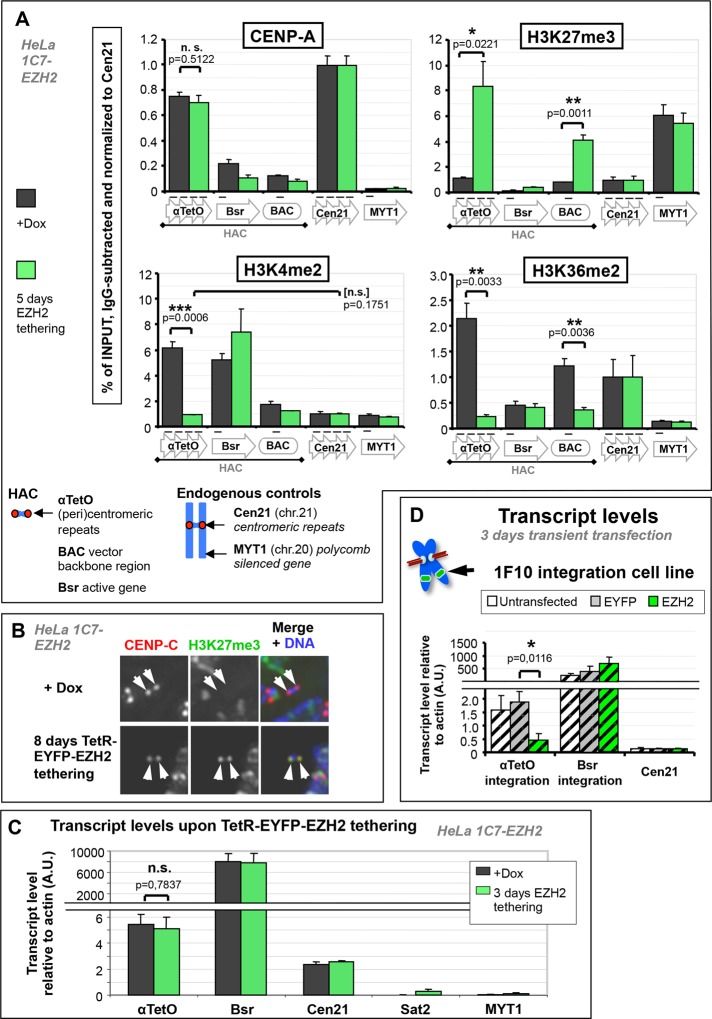
FIGURE 4:
EZH2 tethering severely reduces active transcriptional marks on the HAC alphoidTetO array but has no effect on HAC centromere transcription. (A) EZH2 tethering nucleates H3K27me3 and severely reduces HAC H3K4me2 and H3K36me2 but has no effect on its CENP-A levels. Chromatin analysis of 1C7-EZH2 cells by ChIP. Cells were washed free of doxycycline to allow TetR-EYFP-EZH2 to tether to the HAC and grown for 5 d before harvesting and processing for ChIP. Cells grown in doxycycline were processed in parallel. Antibodies against CENP-A, H3K27me3, H3K4me2, and H3K36me2 were used; mouse unspecific IgG was used as negative pull-down control. Values for each pull down were normalized to the signal of control locus Cen21 after subtraction of background signal (IgG). Primer sets used are shown below graphs. Mean of three independent experiments; error bars denote SEM. Two-tailed Student’s t test was used to evaluate significance of differences observed. (B) The HAC kinetochore-proximal region is enriched with H3K27me3 upon EZH2 tethering. Example of HAC H3K27me3 levels before and after TetR-EYFP-EZH2 tethering. Mitotic chromosome spreads of 1C7-EZH2 cells grown in presence or absence of doxycycline were stained with antibodies against CENP-C (red) and H3K27me3 (green). Arrows denote HAC chromatids. (C) EZH2 tethering does not affect levels of centromeric HAC transcripts. Quantification of transcripts from 1C7-EZH2 cells grown in presence or absence of doxycycline. Cells were grown with or without doxycycline for 3 d and harvested for RNA extraction. Primers against HAC alphoidTetO repeats or Bsr gene were used and against Cen21 as an endogenous centromere control. Transcript level for each locus was normalized to its genomic copy number (for comparison between repetitive regions) and further normalized to β-actin. Mean of three independent experiments; error bars denote SEM. Two-tailed Student’s t test was used to evaluate significance of differences observed. (D) A noncentromeric alphoidTetO array does not prevent silencing of alphoidTetO repeat transcription by EZH2-dependent repression. HeLa 1F10 cells were transiently transfected with either TetR-EYFP or TetR-EYFP-EZH2 for 3 d and subsequently harvested for transcript quantification as in C. Transcripts from untransfected cells were examined as negative controls for TetR-EYFP binding. Mean of three independent experiments; error bars denote SD. Two-tailed Student’s t test was used to evaluate significance of differences observed.
In summary, human centrochromatin has a chromatin signature correlating with active transcription, but the level of enrichment of particular marks can differ among centromeres of different chromosomes. We also observed heterogeneity of pericentromeres, which can be enriched with either heterochromatin or Polycomb-associated marks that encroach at lower levels into the active centromere.
EZH2 tethering to the HAC nucleates Polycomb chromatin
A key question is how the transcriptionally active state of centrochromatin is maintained within a repressive heterochromatic environment. We previously showed that targeting heterochromatin proteins to centrochromatin leads to its inactivation (Nakano et al., 2008; Cardinale et al., 2009). Given the link between centromere activity and transcription, we therefore sought to determine whether an alternative repressive chromatin state—Polycomb-mediated repressive chromatin—was similarly incompatible with centromere activity. Polycomb complexes achieve transcriptional repression through different means than classical heterochromatin repressors (by blocking RNA polymerase II and compacting chromatin) and also respond differently to the chromatin state of their target locus (Di Croce and Helin, 2013).
To nucleate Polycomb chromatin on the HAC, we transfected a plasmid expressing the fusion protein TetR–enhanced yellow fluorescent protein (EYFP)–EZH2 into HeLa 1C7 cells (Figure 2A). EZH2, the enzymatic subunit of the PRC2 complex, methylates histone H3 on K27 to generate H3K27me3 (Margueron et al., 2009), the chromatin mark for PRC1-complex binding (Stock et al., 2007; Eskeland et al., 2010). PRC1 binding induces chromatin compaction and generates H2AK119ub1, thereby blocking transcription elongation. The Polycomb canonical repressive pathway is therefore EZH2 → H3K27me3 → PRC1 → silent chromatin (Figure 2A′).
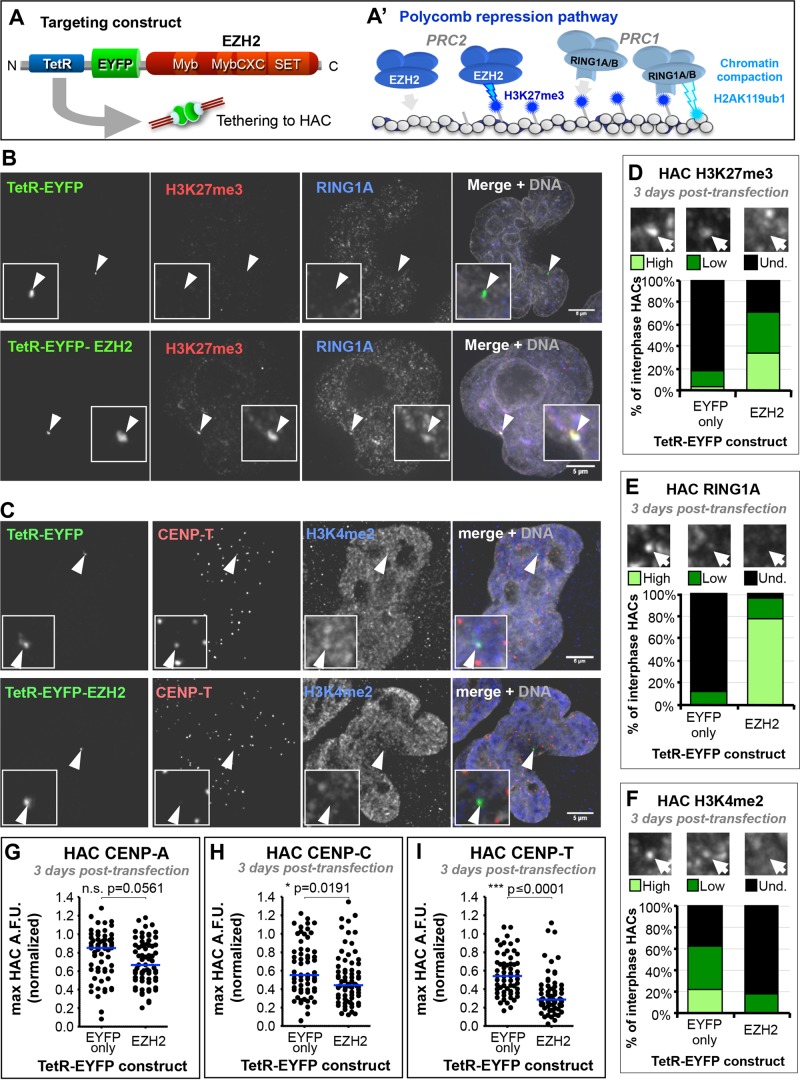
FIGURE 2:
EZH2 tethering to the HAC recapitulates the Polycomb-silencing pathway and reduces levels of centromere proteins. (A, A′) Strategy for inducing enrichment of the Polycomb chromatin state at the HAC. (A) TetR targeting construct used for this experiment. (A′) Canonical Polycomb-repressive pathway, by which PRC2-catalyzed H3K27me3 recruits PRC1, which enacts effective transcriptional repression. (B) EZH2 tethering to the HAC nucleates H3K27me3 and recruits PRC1 subunit RING1A. HeLa 1C7HAC cells were transiently transfected for 3 d with the TetR constructs shown (green), and processed for immunofluorescence using antibodies against H3K27me3 (red) and RING1A (blue). Arrows and insets highlight the HAC. Scale bar, 5 μm. (C) EZH2 tethering to the HAC slightly affects CCAN proteins and reduces HAC H3K4me2. Experimental conditions as in B, but stained with antibodies against CENP-T (red) and H3K4me2 (blue). (D) EZH2 tethering to the HAC enriches it for H3K27me3. Qualitative assessment of H3K27me3 HAC enrichment, as shown in B. Total of two independent experiments, ≥19 cells analyzed per experiment for each condition. (E) EZH2 tethering to the HAC enriches it for PRC1 subunit RING1A. Qualitative assessment of RING1A HAC enrichment, as shown in B. Total of two independent experiments, ≥20 cells analyzed per experiment for each condition. (F) EZH2 tethering effectively reduces the active transcription-related H3K4me2 mark at the HAC. Qualitative assessment of H3K4me2 HAC enrichment, as shown in B. Total of two independent experiments, ≥18 cells analyzed per experiment for each condition. (G) EZH2 tethering to the HAC has little effect on its CENP-A levels. Quantification of HAC centromere protein fluorescence levels in microscopy images after 3 d of TetR construct tethering, as shown in C. Total of three independent experiments, ≥18 cells analyzed per experiment. Mann–Whitney U statistical test was used to evaluate significance of differences observed. (H) EZH2 tethering to the HAC reduces the levels of CCAN component CENP-C. Conditions and quantification as described in G, using antibodies specific for CENP-C, with ≥19 cells per experiment. (I) EZH2 tethering to the HAC reduces the levels of CCAN component CENP-T. Conditions and quantification as described in G, using antibodies specific for CENP-T, with ≥18 cells per experiment.
Three days after transfection, TetR-EYFP-EZH2 binding to the HAC was associated with the appearance of H3K27me3 and recruitment of PRC1 subunit RING1A (Figure 2, B, D, and E, and Supplemental Figure S2, A and B). Both of these Polycomb-associated markers were absent when the TetR-EYFP control fusion protein was tethered to the HAC.
The synthetic Polycomb state appeared to be functional, as it induced a decrease in the HAC-associated signal of H3K4me2, a mark associated with open chromatin and transcription, compared with control TetR-EYFP-tethered HACs (Figure 2, C and F). TetR-EYFP-EZH2 tethering for 3 d also induced a decrease in levels of HAC centromere proteins CENP-C and CENP-T (Figure 2, C, H, and I, and Supplemental Figure S2B), which are part of the constitutive centromere-associated network (CCAN; Cheeseman and Desai, 2008; Perpelescu and Fukagawa, 2011). TetR-EYFP-EZH2 also caused a mild reduction of CENP-A levels on the HAC (Figure 2G and Supplemental Figure S2A).
We conclude that tethering of TetR-EYFP-EZH2 specifically initiates the Polycomb repressive pathway on the HAC, inducing changes in its centrochromatin state that can affect kinetochore assembly.
Long-term EZH2 tethering does not abolish HAC kinetochore assembly or mitotic segregation fidelity
To better characterize the effects of EZH2 tethering to the HAC, we generated a HeLa 1C7 cell line stably expressing TetR-EYFP-EZH2 (henceforth 1C7-EZH2). These cells were grown in the presence of doxycycline, which inhibits TetR binding to the TetO sites located on the HAC (Nakano et al., 2008), to prevent premature tethering of TetR-EYFP-EZH2.
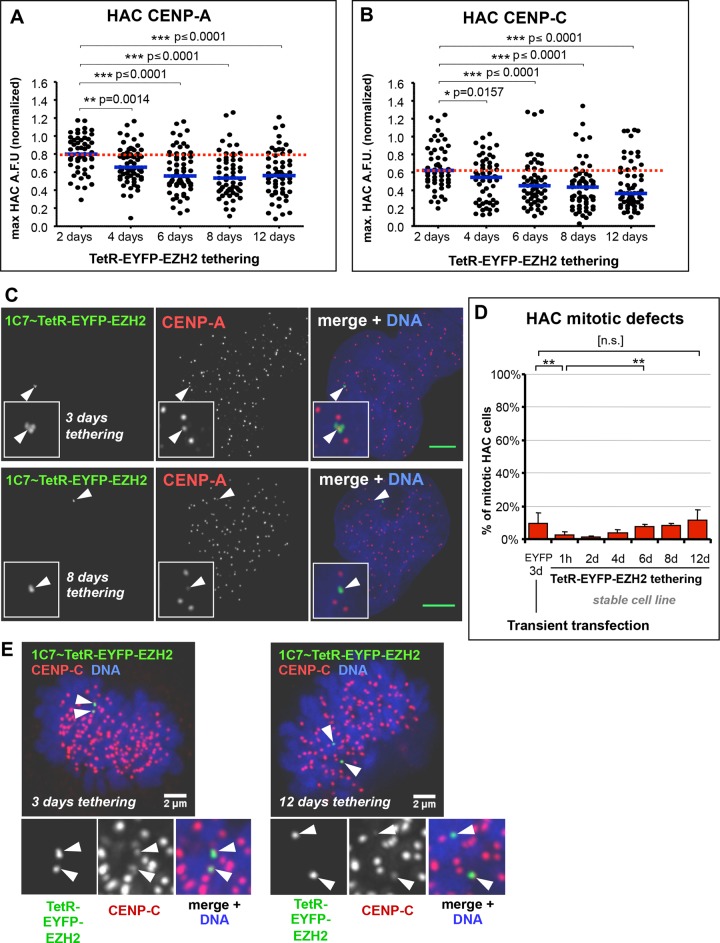
Long-term tethering of TetR-EYFP-EZH2 to the HAC resulted in ∼30% reduction of CENP-A and CENP-C levels after 6 d. Of interest, these levels stabilized and did not decrease further, remaining relatively constant up to 12 d of tethering (Figure 3, A–C). Errors in mitotic segregation of the HAC increased very little over the 12-d period and were never significantly higher than those observed for control HACs tethered with TetR-EYFP only (Figure 3, D and E).
FIGURE 3:
Long-term EZH2 tethering has little effect on HAC centromere proteins and mitotic fidelity. (A) Long-term EZH2 tethering in 1C7-EZH2 cells reduces but does not deplete HAC CENP-A. The 1C7-EZH2 cells were grown in the absence of doxycycline for a 12-d time course to allow TetR-EYFP-EZH2 to tether to the HAC, and at each time point indicated, samples were fixed and processed for immunofluorescence using antibodies against CENP-A. HAC centromere–specific fluorescence signals were quantified from microscopy images. Total of two independent experiments, ≥28 cells analyzed per each experiment for each time point. Red dotted line indicates level of the analyzed protein at the earliest time point. Mann–Whitney U statistical test was used to evaluate significance of differences observed. (B) Long-term EZH2 tethering in 1C7-EZH2 cells reduces but does not deplete HAC CENP-C. Conditions and quantification as described in A, using antibodies specific against CENP-C, with ≥24 cells per each experiment for each time point. (C) Persistence of the HAC centromere under EZH2 tethering. Microscopy analysis of CENP-A staining at 3 and 8 d of EZH2 tethering to the HAC, as described in A, using antibodies against CENP-A (red). Arrows and insets highlight the HAC. Scale bar, 5 μm. (D) Tethering of EZH2 to the HAC has little effect on HAC mitotic fidelity. Quantification of HAC mitotic defects in fixed cultures of 1C7-EZH2 cells at each time point described in A. A given HAC was scored as positive for mitotic defects if it did not congress in metaphase or lagged during anaphase or if chromatids missegregated during anaphase/telophase. Transient transfection of TetR-EYFP for 3 d into HeLa 1C7 cells was used as a negative control. Mean of three independent experiments, ≥40 cells each time point; error bars denote SD. Fisher’s exact test was used to evaluate significance of differences observed: EYFP 3 d vs. EZH2, **p = 0.0018; EZH2 1 h vs. 6 d, **p = 0.0011; EYFP 3 d vs. EZH2 12 d, n.s., p = 0.4167. (E) The HAC CCAN and mitotic function persists during long-term EZH2 tethering. Microscopy analysis of CENP-C staining in metaphase cells at 3 and 12 d after EZH2 tethering, as described in B, using antibodies against CENP-C (red). Arrows and insets highlight the HAC chromatids and associated CENP-C signal.
We next examined the effects of EZH2 tethering on the incorporation of CENP-A at centromeres using a SNAP-tag quench-chase-pulse assay (Jansen et al., 2007; Bodor et al., 2013). To do this, we cotransfected two plasmids expressing CENP-A:SNAP and TetR-EYFP-EZH2, respectively. In controls, we also cotransfected plasmids expressing CENP-A:SNAP and TetR-EYFP-LSD1. We previously showed that tethering of the LSD1 demethylase inhibits new CENP-A assembly (Bergmann et al., 2011).
Because CENP-A only assembles once during the cell cycle, in early G1, it is possible, by specifically labeling newly synthesized CENP-A:SNAP with fluorescent TMR-Star, to visualize only newly assembled CENP-A:SNAP at centromeres (Supplemental Figure S2C). In these experiments, by observing the first events of CENP-A assembly on the HAC after tethering, we found an inhibitory effect of LSD1, as reported previously (Bergmann et al., 2011), whereas EZH2 had little effect (Supplemental Figure S2, D and E).
Together these results indicate that, unlike previous experiments involving tethering of repressive complexes such as LSD1 (Bergmann et al., 2011), KAP1 (Cardinale et al., 2009) or Suv39h1 (Ohzeki et al., 2012), tethering of EZH2 has a remarkably mild effect on HAC centromere function.
EZH2 tethering to the HAC reduces transcription-related chromatin marks but not centromeric transcription
To understand the chromatin state induced by tethering EZH2 to the HAC centromere, we analyzed the levels of several chromatin marks on the HAC, as well as its transcriptional output, before and after tethering.
ChIP-qPCR analysis revealed no significant decrease of CENP-A levels on the HAC alphoidTetO array after 5 d of EZH2 tethering (Figure 4A). This contrasted with our microscopy observations, in which a limited (but significant) reduction of CENP-A was observed after TetR-EYFP-EZH2 tethering (compare Figures 3A and 4A). Because the primer pair used can only quantify CENP-A present at the alphoidTetO repeats, it is possible that microscopy may detect loss of CENP-A associated with other regions of the HAC that is preferentially lost after EZH2 tethering. We could indeed observe a low level of CENP-A on the Bsr gene, which decreased on TetR-EYFP-EZH2 tethering (Figure 4A).
Although it is formally possible that the levels of tethered EZH2 were insufficient to produce a full Polycomb response, examination of the downstream consequences of EZH2 tethering suggest that a full Polycomb-repressive state was produced. These included a large increase in H3K27me3 to levels comparable to or greater than those at the endogenous Polycomb-repressed gene MYT1 (Figure 4, A and B). We also noted an ∼10-fold increase in the downstream product of the Polycomb pathway, H2AK119ub1, which is produced by the PRC1 complex (Supplemental Figure S3B). Both were consistent with our microscopy results in transiently transfected cells (Figure 2, B, D, and E) and are consistent with a robust establishment of a Polycomb-like chromatin signature. Production of the Polycomb signature was accompanied by a sharp drop in the transcription-associated chromatin marks H3K4me2 and H3K36me2 on the HAC (Figure 4A). The low H3K4me2 level remaining on the HAC after EZH2 tethering was comparable to that found at the endogenous Cen21, unlike the more severe depletion previously observed after LSD1 tethering (Bergmann et al., 2011), and may explain why HAC centromere function appeared to suffer few ill effects (Figure 3). The changes appeared to be specific for the Polycomb pathway, as levels of HAC-associated H3K9me3 were unchanged (Supplemental Figure S3A).
An increase in H3K27me3 and H2AK119ub, together with a decrease in H3K36me2, was also observed on the adjacent BAC sequences on the HAC (Figure 4A and Supplemental Figure S3B), despite their being located ∼3 kb away from the α-satelliteTetO array and not possessing TetO-binding sites. The Polycomb-mediated repressive state has been shown to spread to adjacent loci (Chan et al., 1994; Kahn et al., 2006) via binding of the EZH2-containing PRC2 complex to H3K27me3 (its own catalytic product; Margueron et al., 2009). Together these results show that EZH2 can successfully establish a Polycomb-like chromatin state on the HAC alphoidTetO array.
Given these changes in the chromatin pattern, we were thus surprised when reverse transcription (RT) qPCR analysis revealed no significant effect on transcription from the HAC alphoidTetO array after 3 d of TetR-EYFP-EZH2 tethering (Figure 4C). Consistent with this, we confirmed by ChIP-qPCR that total levels of RNA polymerase II throughout the array did not change from their initial levels after EZH2 tethering (Supplemental Figure S3, C and C′). We observed that the Polycomb-repressed MYT1 gene had levels of transcription much lower than those observed for EZH2-tethered HACs (Figure 4C), indicating the extent to which endogenous Polycomb repression can reduce transcription. In addition, the pericentromeric Sat2 repeats were expressed at levels far lower than those of the centromeric repeats of both the HAC and Cen21 (Figure 4C), confirming that centromeres are indeed transcribed at levels greater than that of heterochromatin.
An important control for these experiments was to confirm that alphoidTetO sequences are not intrinsically resistant to Polycomb-induced silencing, for reasons unrelated to the presence of the HAC’s centromere. We therefore conducted TetR-EYFP-EZH2 targeting in the HeLa/HT1080 fusion cell line 1F10, which contains a noncentromeric α-satelliteTetO array integrated on a chromosomal arm (Supplemental Figure S4, A, C, and D). This array does not recruit significant levels of CENP-A and does not function as a centromere. Analysis of selected histone marks reveals that the noncentromeric alphoidTetO array is significantly more euchromatic than the HAC array (Supplemental Figure S4A), and its transcriptional output is greater than ∼20-fold higher (Supplemental Figure S4B).
After 3 d of transient transfection with plasmids expressing the TetR-EYFP fusion proteins, quantification of transcription by RT-qPCR revealed that EZH2 tethering to the noncentromeric integration array did indeed significantly reduce alphoidTetO transcription (Figure 4D), although not to the level observed at the endogenous MYT1 locus. Although it is possible that the HAC’s small size and lack of arms compared with a conventional chromosome might somehow interfere with effective establishment of repression, the total size of the HAC TetO array is ∼2.3 Mb, and TetR binding could potentially occur very densely: every 340 base pairs across the array (Nakano et al., 2008). Furthermore, we consistently observe that the TetR-EYFP signal corresponding to the noncentromeric integration in interphase 1F10 cells is noticeably smaller (and more difficult to detect) than that observed at the HAC in 1C7 cells. This suggests that the integrated array is actually smaller than the HAC array (a fact reported previously in Nakano et al. 2008).
In summary, the HAC centromere appeared to resist silencing induced by a Polycomb- repressive pathway initiated within it. Despite substantial reductions in transcription-related marks, alphoidTetO transcription in the context of centrochromatin was unaffected, whereas similar targeting of a euchromatic alphoidTetO array (integrated into a chromosome arm) did result in transcriptional silencing. These results suggest that the presence of a centromere on an otherwise identical DNA array can somehow prevent the Polycomb pathway from fully establishing its repressive target chromatin state.
Mitotic release of PRC1 from chromatin does not explain HAC centromere resistance to Polycomb-dependent repression
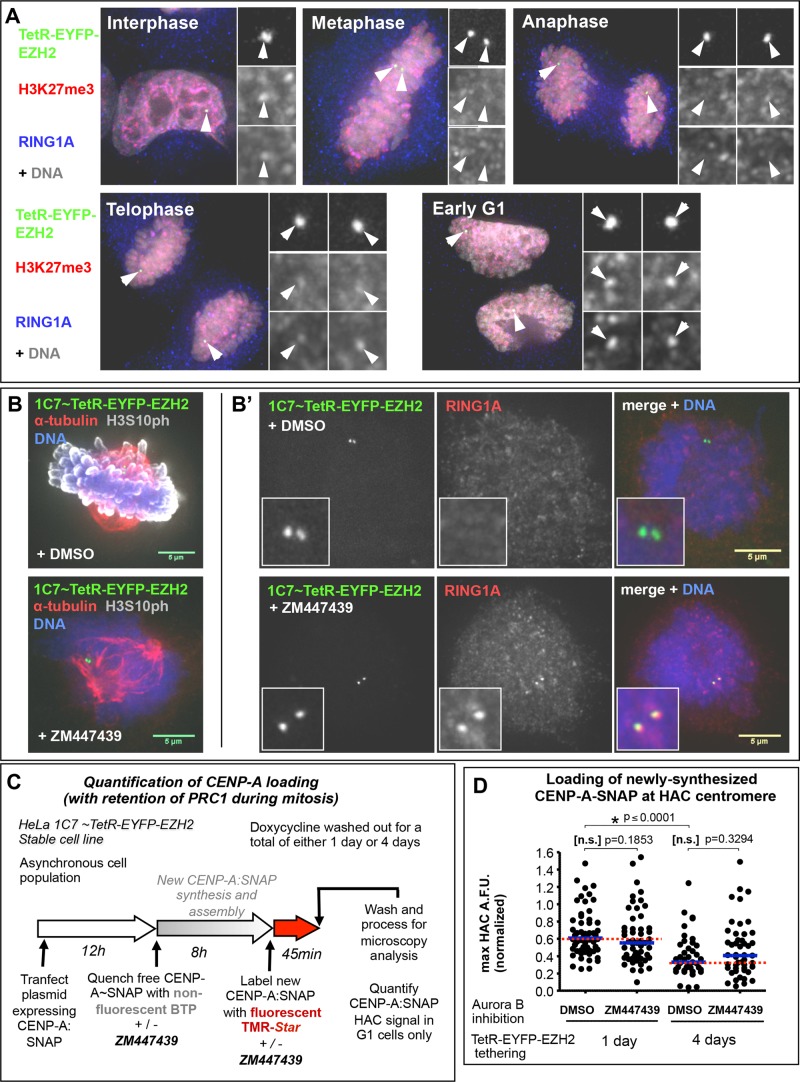
Cell cycle regulation events might account for this apparent resistance of centrochromatin to Polycomb-induced silencing. Human centromeres are transcribed during mitosis (Chan et al., 2012), and CENP-A assembly starts only at telophase (Jansen et al., 2007), continuing throughout G1 (Lagana et al., 2010). It has been reported that the PRC1 complex dissociates from chromosomes during mitosis, relocalizing again only after mitotic exit (Voncken et al., 1999). We confirmed this observation in HeLa 1C7 cells (Figure 5A). If the temporary absence of the PRC1 repressor complex coincides with the timing of centromeric transcription, the HAC centromere might use this “window of opportunity” to transcribe and thereby maintain its centromeric function in a transcriptionally repressive environment.
FIGURE 5:
Retention of PRC1 at HAC chromatin during mitosis is insufficient to affect CENP-A assembly. (A) PRC1 relocates away from chromatin during mitosis. Microscopy images of 1C7-EZH2 cells, after 4 d of EZH2 tethering (green), were analyzed for PRC1 signal at the HAC (as detected by its RING1A subunit) both at interphase and at various stages of mitosis. Antibodies against H3K27me3 (red) and RING1A (blue) were used for detection. Note that H3K27me3 signal is also shown as being reduced during mitosis, but the anti-H3K27me3 antibody used suffers from lack of epitope accessibility due to occlusion by the adjacent H3S28p modification present during mitosis (Hiroshi Kimura, personal communication). (B, B′) Inhibition of Aurora B activity leads to retention of PRC1 during mitosis at the EZH2-tethered HAC. Microscopy images of mitotic 1C7-EZH2 cells after 1 d of EZH2 tethering (green) in the presence or absence of Aurora B inhibitor ZM447439. (B) Confirmation of the inhibition of Aurora B kinase activity; antibodies against α-tubulin (red) and H3S10 phosphorylation (gray) were used. (B′) PRC1 is retained at the HAC during mitosis upon inhibition of Aurora B; antibodies against RING1A (red) were used to detect the PRC1 complex. Scale bar, 5 μm. (C) Experimental outline of procedure to analyze CENP-A assembly on EZH2-tethered HACs with or without PRC1 retention during mitosis (by inhibiting Aurora B activity). 1C7-EZH2 cells were grown in the absence of doxycycline, to allow for EZH2 tethering, for a total of 1 or 4 d before fixation. At 1 d before fixation, a plasmid expressing CENP-A-SNAP was transfected into cells, and ∼9 h before fixation, a quench-chase-pulse assay was performed to specifically label newly synthesized CENP-A-SNAP with TMR-Star. During the quench-chase-pulse period, cells were grown in parallel in medium supplemented either with dimethyl sulfoxide (DMSO) or ZM447439 (to inhibit Aurora B). G1 cells (those that loaded newly synthesized CENP-A onto centromeres during the assay timeline) were analyzed for the levels of CENP-A:SNAP∼TMR on the HAC. (D) PRC1 retention during mitosis at EZH2-tethered HACs has no visible effect on CENP-A assembly in the subsequent G1 phase. Quantification of C. Two independent experiments; ≥23 cells for each condition. Mann–Whitney U statistical test was used to evaluate significance of differences observed. A reduction in CENP-A assembly could be noted between controls of days 1 and 4; this loss of total CENP-A assembly is likely due to reduction in total centromere proteins (and consequently the assembly recruitment platform), in line with results in Figure 3A.
H3S28 phosphorylation, catalyzed by Aurora B, has been suggested to cause PRC1 release from H3K27me3 (Frangini et al., 2013), similar to the release of HP1 from H3K9me3 by phosphorylation of H3S10 during mitosis (Fischle et al., 2005; Hirota et al., 2005). We therefore hypothesized that inhibition of Aurora B function might prevent PRC1 release from chromatin and thus maintain its binding to the HAC during mitosis.
Addition of the Aurora B–specific inhibitor ZM447439 (Ditchfield et al., 2003) to 1C7-EZH2 cells substantially lowered H3S10 phosphorylation (Figure 5B), and PRC1 enrichment was maintained at the HAC during mitosis (Figure 5B’). To determine whether maintaining PRC1 at the HAC in this way interfered with CENP-A assembly in the subsequent G1 phase, we transiently transfected a plasmid expressing CENP-A:SNAP the day before the experiment and performed a SNAP-tag quench-chase-pulse assay (Figure 5C). We then quantified the amount of newly synthesized CENP-A incorporated at the HAC in the presence or absence of Aurora B inhibition. We observed no significant decrease in levels of newly incorporated CENP-A:SNAP after Aurora B inhibition (Figure 5D). This was true whether TetR-EYFP-EZH2 was tethered for 24 h (the first event of CENP-A loading after tethering) or for 4 d.
Thus the release of PRC1 during mitosis is not sufficient to explain the apparent resistance that HAC centromeric transcription shows against Polycomb silencing.
Direct tethering of PRC1, as opposed to its recruitment by H3K27me3, negatively affects HAC centromere function
Given that the HAC centromere seems to resist transcriptional repression by an EZH2-initiated Polycomb pathway despite being significantly enriched for downstream markers of Polycomb chromatin, we sought to understand the nature of this resistance and whether it was specific for the Polycomb pathway. It is unlikely that centromeric proteins form a physical barrier to effective Polycomb repression, as tethering of the reader HP1 or KAP1 and the editor Suv39h1 efficiently inactivated the HAC centromere (Nakano et al., 2008; Cardinale et al., 2009; Ohzeki et al., 2012). We hypothesized that the nature of centrochromatin, including the presence of CENP-A, might somehow block the Polycomb pathway from enacting effective repression at the centromere core (Figure 6A). This might occur because either H3K27me3 cannot be locally enriched in the immediate vicinity of CENP-A-containing nucleosomes or PRC1 is not properly recruited there despite the fact that both are efficiently enriched throughout the majority of the alphoidTetO array (as observed by microscopy and ChIP analysis). Alternatively, it could be that as-yet-unknown mechanisms specifically promoting the highly unusual mitotic transcription at centromeres (Chan et al., 2012) are resistant to repressive pathways that function during interphase.
FIGURE 6:
Repression-related H3 marks have little effect on the HAC centromere, but direct tethering of downstream repressor complexes induces severe HAC centromere defects. (A) Two-step pathways of the Polycomb and heterochromatin transcription repression systems: repression-related marks H3K9me3 and H3K27me3 are first generated on chromatin, which in turn recruit downstream repressor complexes to enact transcriptional silencing (directly or indirectly). (B) TetR-EYFP-BMI1 can recruit the PRC1 complex to the HAC. HeLa 1C7HAC cells were transiently transfected with the indicated TetR constructs for 1 d, fixed, and processed for immunofluorescence. Antibodies against RING1A were used as a surrogate to detect enrichment of PRC1. (C) Tethering of complexes that generate repressive H3 marks have only mild effects on HAC CENP-A levels, but direct tethering of downstream repressors causes pronounced loss of CENP-A. HeLa 1C7HAC cells were transfected for 3 d with the TetR constructs shown before fixation and processing for immunofluorescence. EZH2 and Suv39h1SET were used as catalysts of repressive H3 marks, and BMI1 and HP1α were used as downstream repressors, one each from the Polycomb and heterochromatin pathways, respectively. KAP-1 was used as a positive control for centromere inactivation. Antibodies against CENP-A (red) and H3K4me2 (blue) were used to analyze each construct’s effect on the HAC centromere and active transcription marks, respectively. (D) Quantification of HAC CENP-A fluorescence in microscopy images, as described in C. Mean of three independent experiments; ≥18 cells per condition in each experiment. Red dotted line indicates level of the analyzed protein in the EYFP-only negative control. The Mann–Whitney U test was used to evaluate the significance of differences observed. (E) Downstream transcription repressors induce much more pronounced defects in HAC mitotic fidelity than complexes that generate repressive H3 marks. HeLa 1C7HAC cells were transfected and processed as in C. Missegregation of HAC chromatids in transfected cells was scored whenever HAC EYFP signals segregated unequally in telophase-early G1 (confirmed by the presence of a midbody). Mean of three independent experiments, and ≥25 cells per condition in each experiment; error bars denote SD. Fisher’s exact test was used to evaluate significance of differences observed. (F) Decrease in HAC CENP-A levels induced by downstream repressors BMI1 and HP1α increases with longer tethering periods. HeLa 1C7HAC cells were transfected with the indicated TetR constructs for a long-term tethering of 5 d (see Materials and Methods). Antibodies against CENP-A were used to quantify its fluorescence levels at the HAC in microscopy images. Quantification, number of cells, and statistical analysis were performed as in D. (G) Prolonged tethering of downstream repressor BMI1 or HP1α increases the severity of HAC mitotic defects. HeLa 1C7HAC cells were transfected and processed as in F, and missegregation of HAC chromatids was scored as in E. Mean of three independent experiments, with ≥25 cells per condition in each experiment; error bars denote SD. Fisher’s exact test was used to evaluate significance of differences observed.
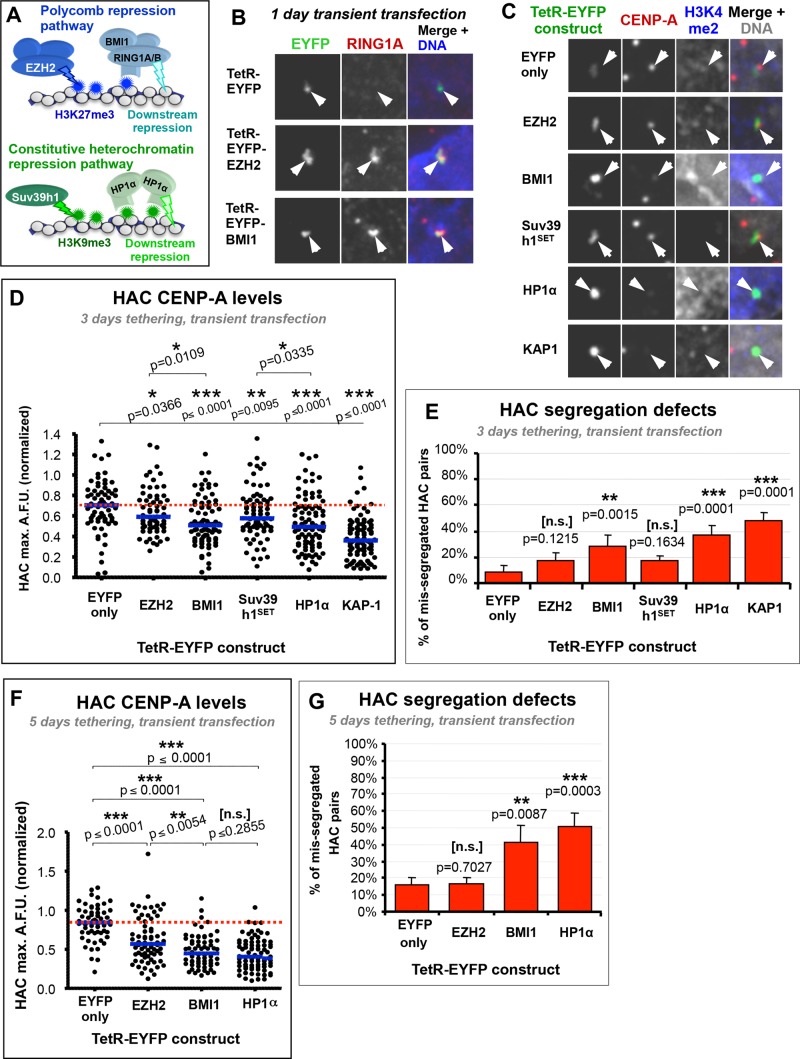
To bypass the need of H3K27me3 for PRC1 recruitment (Figure 6A), we tethered the PRC1 complex directly to the HAC. Tethering of TetR-EYFP-BMI1 (a subunit of the PRC1 complex) resulted in efficient recruitment of the RING1A subunit of PRC1 (Figure 6B and Supplemental Figure S5, A and B), and HAC H3K4me2 was reduced (Figure 6C). This suggested that a functional PRC1-repressive complex could be recruited using this approach. After 3 d of TetR-EYFP-BMI1 tethering, we observed a significant decrease in HAC CENP-A levels (Figure 6, C and D) and increased HAC segregation errors (up to ∼30%; Figure 6E) compared with control tethering of TetR-EYFP.
The HAC defects induced by tethering BMI1 were comparable to those induced by directly tethering HP1α and KAP1, which inactivate the HAC centromere (Nakano et al., 2008; Cardinale et al., 2009). Tethering of TetR-EYFP-EZH2 for the same duration had significantly milder effects, as before, despite also inducing recruitment of PRC1 (Figure 6B, Supplemental Figure S5B). We quantified the level of RING1A at HACs tethered with the aforementioned constructs and observed that BMI1 recruited more than twice the amount of RING1A than EZH2 compared with control TetR-EYFP (Supplemental Figure S5, A and B). This increased concentration of PRC1 on the HAC (as measured by presence of RING1A) brought about by BMI1 tethering might account for the increased severity of the phenotype relative to EZH2 tethering. It is possible that EZH2 tethering does not recruit sufficient PRC1 to the HAC to induce repression. However, this is less likely, as RING1A levels recruited to the HAC via EZH2 tethering are closer to those observed in other nuclear clusters of RING1A, whereas those recruited by BMI1 are much higher (Supplemental Figure S5B). This suggests that EZH2 tethering might recruit physiological levels of PRC1, whereas direct tethering of BMI1 might recruit levels above those normally observed physiologically, and those increased levels might overcome the normal resistance of centromeric transcription.
The effects of TetR-EYFP-BMI1 tethering on the HAC centromere were much stronger after 5 d. Tethering of BMI1 induced a significant decrease of HAC CENP-A levels (Figure 6F) and mitotic fidelity (Figure 6G) to a degree comparable to that induced by tethering HP1α (∼40 and 50% missegregated HACs, respectively) and more severe than that induced by tethering EZH2 (∼17% missegregation).
Although it is possible that direct constitutive binding of BMI1 via TetR exacerbates PRC1-dependent effects or might recruit a different variant of the PRC1 complex (Di Croce and Helin, 2013), the results presented here suggest that targeting the canonical endogenous Polycomb pathway to the HAC centromere via H3K27me3 is not on its own sufficient to silence centrochromatin transcription and centromeric function. Because direct targeting of a downstream repressor complex does inhibit centromere function, we conclude that centromeres are not intrinsically immune to Polycomb repression. This raises the interesting possibility that centrochromatin may locally modulate how the canonical Polycomb-repressive pathway translates into effective repression.
H3K9me3 nucleation on the HAC also does not abolish centromere function
Our observation that EZH2-mediated initiation of the Polycomb-repressive pathway was insufficient to abolish HAC centromere function, whereas tethering the H3K27me3 reader PRC1 overcame this resistance, led us to ask whether the same behavior might be true for H3K9me3 and HP1 (a reader of H3K9me3). The proteins that comprise the Polycomb-mediated as well as the constitutive heterochromatin repression pathways have similar domains and chromatin-targeting mechanisms. Both largely rely on readers (PRC1 or HP1) bearing chromodomains that bind to trimethylated ARKme3S motifs on the H3 tail (Jacobs and Khorasanizadeh, 2002; Min et al., 2003; Figure 6A). If the same mechanism that mediates the resistance of the HAC centromere to H3K27me3-dependent repression also applies to the H3K9me3 mark, this could help to explain how actively transcribing centromeres in human cells persist within transcriptionally repressed regions without becoming silenced. We therefore decided to study whether the HAC centromere was also resistant to H3K9me3-induced transcriptional repression initiated via the constitutive heterochromatin pathway.
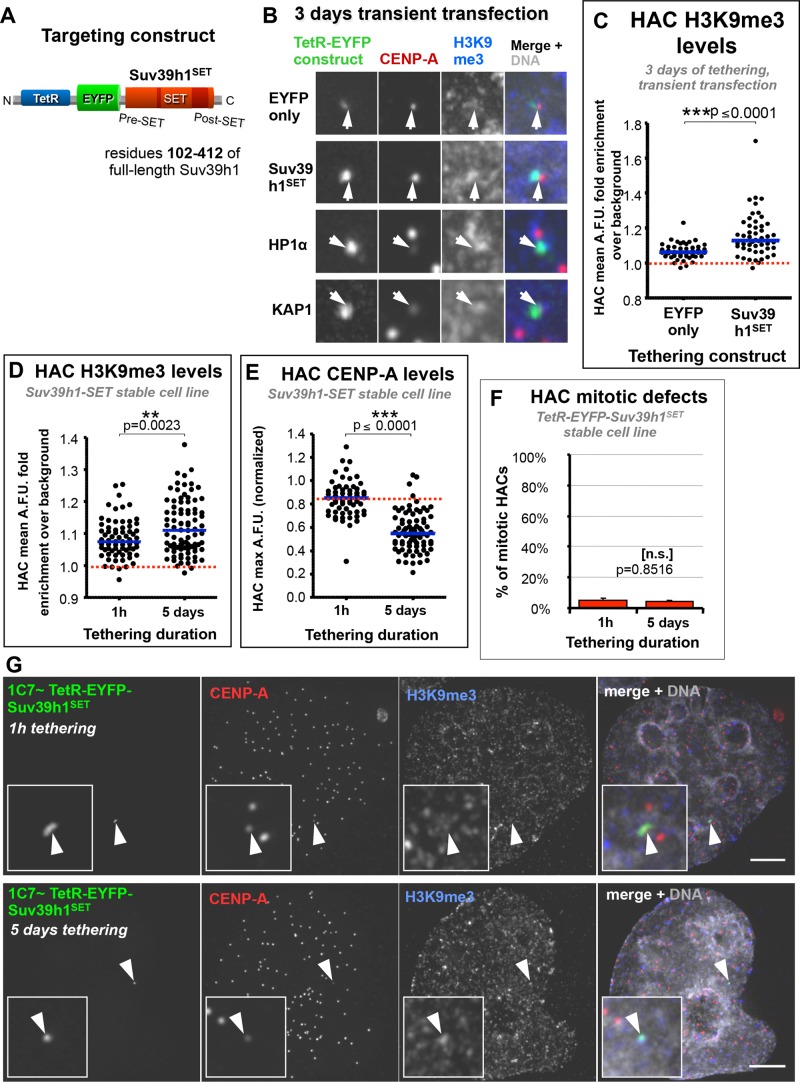
To establish the canonical pathway for constitutive heterochromatin (Suv39h1 → H3K9me3 → HP1 → silent chromatin) in HAC centrochromatin, we generated a construct containing the C-terminal H3K9 methyltransferase domain of hSuv39h1 (TetR-EYFP-Suv39h1SET; Figure 7A). This protein can nucleate H3K9me3 on chromatin (Figure 7, B and C) but lacks the H3K9me3-binding chromodomain and the interaction regions that directly recruit HDACs and HP1 in an H3K9me3-independent manner (Melcher et al., 2000; Jacobs and Khorasanizadeh, 2002; Vaute et al., 2002). In this way, we assessed H3K9me3-associated effects independently of direct HP1 recruitment by full-length Suv39h1, which induces centromere inactivation when tethered (Ohzeki et al., 2012).
FIGURE 7:
Tethering of Suv39h1SET to the HAC, which nucleates H3K9me3, has little effect on HAC centromere function. (A) Diagram of TetR-EYFP-Suv39h1SET construct, which contains a truncated version of Suv39h1 (amino acids 102–412), containing the enzyme’s functional SET domain, which catalyzes H3K9 trimethylation. (B) Suv39h1SET enriches the HAC for H3K9me3, with only mild effects on HAC CENP-A levels. HeLa 1C7HAC cells were transfected with the indicated TetR constructs as described in Figure 6, C–E. Antibodies against H3K9me3 (blue) and CENP-A (red) were used. (C) Tethering of TetR-EYFP-Suv39h1SET to the HAC increases its H3K9me3 levels. The indicated constructs were transfected into 1C7HAC cells for 3 d before being fixed and processed for immunofluorescence. Two independent experiments. Number of cells analyzed in each condition per experiment: EYFP only, ≥21; Suv39h1SET, ≥24. Red dotted line indicates mean level of local H3K9me3 background signal. Mann–Whitney U statistical test was used to evaluate significance of differences observed. (D) Long-term Suv39h1SET tethering to the HAC in 1C7-Suv39h1SET cells enriches it for H3K9me3. The 1C7-Suv39h1SET cells were grown in the absence of doxycycline for either 1 h or 5 d to allow TetR-EYFP-Suv39h1SET to tether to the HAC before fixation. Levels of H3K9me3 were assayed by fluorescence intensity from microscopy images in the HAC EYFP signal–delimited area. Total of three independent experiments. Number of cells analyzed: 1 h, ≥25; 5 d, ≥28. Mann–Whitney U statistical test was used to evaluate significance of differences observed. (E) Long-term Suv39h1SET tethering to the HAC reduces CENP-A at the HAC. The 1C7-Suv39h1SET cells were treated as described in D. HAC centromere-specific fluorescence signals were quantified from microscopy images. Total of three independent experiments. Number of cells analyzed in each condition per experiment: 1 h, ≥12; 5 d, ≥23. Mann–Whitney U statistical test was used to evaluate the significance of differences observed. (F) Long-term Suv39h1SET tethering to the HAC has little effect on its mitotic fidelity. 1C7 cells expressing TetR-EYFP-Suv39h1SET were treated as described in D. Any given HAC was scored as positive for mitotic defects if one of the following was true: it did not congress in metaphase, it lagged during anaphase, or chromatids missegregated during anaphase/telophase. Mean of three independent experiments, ≥100 cells per condition in each experiment; error bars denote SD. Fisher’s exact test was used to evaluate significant of differences observed. (G) Long-term Suv39h1SET tethering nucleates H3K9me3 at the HAC but has only a small effect on HAC CENP-A. The 1C7-Suv39h1SET cells were treated as described in D. Samples were fixed and processed for immunofluorescence using antibodies against H3K9me3 (blue) and CENP-A (red). Scale bar, 5 μm.
After 3 d of tethering after transient transfection, TetR-EYFP-Suv39h1SET caused a moderate reduction of CENP-A levels at the HAC, similar to that observed for TetR-EYFP-EZH2 but less severe than that observed for TetR-EYFP-HP1α (Figures 6, C and D, and 7B). As was the case for TetR-EYFP-EZH2, tethering of TetR-EYFP-Suv39h1SET for this period induced no significant increase in HAC mitotic defects (Figure 6E). In control experiments, TetR-EYFP-HP1α tethering to the HAC yielded the strong inhibition of centromere function observed in previous studies (Nakano et al., 2008; Cardinale et al., 2009) and similar to that induced by BMI1 (Figure 6, D and E). This similarity was particularly noticeable when BMI1 or HP1α was tethered to the HAC for 5 d (Figure 6, F and G).
To confirm these results, we generated a HeLa 1C7 cell pool stably expressing TetR-EYFP-Suv39h1SET in which ∼85% of the cells express the construct, albeit at different levels. The cells were maintained in doxycycline, which was then washed out of the growth medium for 5 d, allowing TetR-EYFP-Suv39h1SET to bind to the HAC. Between 1 h and 5 d of tethering, we observed a small but significant increase in HAC H3K9me3 by immunofluorescence (Figure 7, D and G). This relatively small increase may reflect the fact that the HAC already has H3K9me3 before tethering (Figure 1G), or that the nuclear background level of H3K9me3 is elevated in these cells due to overexpression of the ectopic construct (which was observed in transient transfections). When we analyzed CENP-A levels at the HAC of 1C7-Suv39h1SET cells, we detected ∼35% reduction after 5 d of tethering (Figure 7, E and G). Together with the previously observed reduction of HAC H3K4me2 (by transient transfection; Figure 6C), this suggests that the construct is functional and indeed affects HAC centrochromatin, shifting its signature into a less euchromatic state. Remarkably, tethering of Suv39h1SET for this time period caused no measurable increase in HAC segregation defects (Figure 7F).
In summary, our results indicate that the HAC centromere retains functionality despite local deposition of the repressive mark H3K9me3, although it could be that the moderate levels of H3K9me3 generated by this Suv39h1SET construct are sufficient to reduce CENP-A levels but not to perturb HAC segregation. Because recruitment of heterochromatin readers (in this case, downstream repressors) causes robust centromere inactivation, this suggests that although human centromeres are not specifically insensitive to heterochromatin repression, they are able to locally resist repressive pathways initiated by H3 marks.
DISCUSSION
Previous studies of centrochromatin in HACs and at endogenous human centromeres pointed to deleterious effects of transcriptional repression on centromere function. They relied on tethering of heterochromatin factors such as the editor Suv39h1 (Ohzeki et al., 2012), the reader HP1 (Nakano et al., 2008), strong multirepressor scaffolds like KAP1 (Cardinale et al., 2009), and editors such as LSD1 that remove transcription-related marks (Bergmann et al., 2011) or conversely by opposing repression via targeting of acetyltransferases (such as PCAF or p300) (Ohzeki et al., 2012) or deacetylase inhibitors (such as trichostatin A; Nakano et al., 2003). How is it, therefore, that natural human centromeres are embedded within constitutively heterochromatic regions, without losing transcriptional activity?
The present study suggests that, considering the normal cycle of Editor→ Mark → Reader → Chromatin environment, centrochromatin appears able to influence how locally enriched marks eventually translate into silent chromatin states.
Centromeres resist Polycomb repression mediated by H3K27me3
Tethering the Polycomb PRC2 editor EZH2 to the HAC recapitulated Polycomb-like chromatin and reduced the levels of active transcription marks. Surprisingly, this did not inactivate centromere transcription despite reducing levels of centromere proteins. This agrees with a recent report that human centromeres can remain functional with reduced CENP-A levels (Bodor et al., 2014). Because EZH2 tethering to a noncentromeric α-satelliteTetO array effectively represses local transcription, we conclude that the HAC centromere may harbor mechanisms for resisting Polycomb-mediated transcriptional repression. This could occur by preventing the binding of repressive readers to their target marks. However, this is unlikely, since we showed that RING1 can bind centromeric H3K27me3 nucleated by tethered EZH2. Alternatively, the concentration of repressive marks within CENP-A chromatin could be insufficient to achieve a full-blown repressive response, or the specialized centromeric chromatin containing both CENP-A and associated CCAN proteins and/or specialized characteristics of transcription during mitosis might actively oppose the silencing functions of readers bound to repressive marks.
HAC centromere resistance to Polycomb-mediated repression is bypassed by direct tethering of the downstream repressor PRC1
Direct tethering of the PRC1 reader BMI1 disrupted the HAC centromere, whereas its recruitment by EZH2-induced H3K27me3 did not. Direct constitutive tethering of elevated levels of PRC1, possibly with altered binding dynamics not examined here (J. Ruppert and W. C. Earnshaw, unpublished data), might induce an exacerbated repression not seen with normal recruitment via EZH2 tethering. This suggests that although PRC1 might be recruited to the HAC centromere core by H3K27me3, its binding dynamics and/or stability might be altered, or, possibly, a less active version of the PRC1 complex version is recruited. Alternatively, centrochromatin might fail to achieve a required threshold of H3K27me3 enrichment either due to lower H3 density (due to replacement by CENP-A) or because either the centromere or another aspect of specialized mitotic transcription somehow inhibits EZH2 activity locally. This could lead to lower PRC1 recruitment specifically near CENP-A but not on the remainder of the alphoidTetO array, a difference that is difficult to discriminate by immunofluorescence or ChIP. EZH2 activity can depend on local chromatin context (Tie et al., 2009; Schmitges et al., 2011; Yuan et al., 2011), and in one study, EZH2 tethered to active chromatin was unable to reduce local H3K4me2 and histone acetylation (Rush et al., 2009).
Regardless which of these explanations is correct, our results point to a previously unreported ability of centromeres to resist local transcriptional silencing.
Resistance to H3 repression–related marks H3K9me3 and H3K27me3 might be a conserved feature of human centromeres
Because PRC1 and HP1 have analogous chromatin-targeting mechanisms, we explored whether centrochromatin showed a similar resistance to the endogenous heterochromatin-repressive pathway. Indeed, we observed a similar response of the HAC centromere to Suv39h1SET as for EZH2. Long-term H3K9me3 nucleation in a cell line expressing Suv39h1SET confirmed that centromeres can resist inactivation by the repressive pathway involving H3K9me3, similar to H3K27me3.
We showed that whereas enforced repression through tethering of silencing readers BMI1 or HP1 can inactivate centrochromatin, silencing through nucleation of the canonical pathways is ineffective. The H3K27me3 and H3K9me3 repressive pathways share many similarities in chromatin-binding mechanisms (Beisel and Paro, 2011), and both can spread across chromatin domains (Seum et al., 2001; Kahn et al., 2006) and might even functionally interact with each other (Sewalt et al., 2002; Yamamoto et al., 2004; Vasanthi et al., 2013). Thus it is possible that centromeres use a similar mechanism to cope with marks from either repressive pathway.
Microscopy of stretched chromatin fibers of endogenous centromeres showed a difference in chromatin signature between the CENP-A–containing core and the surrounding pericentromere. Specifically, we detected marks characteristic of nonsilenced promoters (H3K4me3 and H2A.Z) and only low levels of H3K9me3 colocalizing within the centromere core, as have others (Sullivan and Karpen, 2004; Lam et al., 2006). Similar distributions of an attenuated heterochromatic signature across the centromere core were also observed in Neurospora crassa (Smith et al., 2011) and mouse centromeres (Lehnertz et al., 2003; Guenatri et al., 2004). Thus a variety of evidence suggests that an active mechanism may prevent H3K9me3/H3K27me3 spreading into the centromere core.
One hypothesis is that the semieuchromatic signature and/or active transcription at centromeres promote resistance against enrichment of repressive H3 marks. Both H3K9me3 and H3K27me3 are highly depleted at transcription start sites and 5′ regions of active genes (Barski et al., 2007; Mikkelsen et al., 2007). Higher histone turnover, generally associated with 5′ regions of transcribing chromatin (Mito et al., 2005; Nakayama et al., 2007), has been suggested to counteract repressive chromatin states by removing histones and any repressive marks that they carry (Aygun et al., 2013), thus forming a barrier against spreading of repressive chromatin (Lee et al., 2007; Wen et al., 2010; Klose et al., 2013). This may occur at centromeres as well (Figure 8A), because canonical nucleosomes within centrochromatin are enriched for the H3.3 variant (Dunleavy et al., 2011), which replaces replication-deposited H3.1 in a cotranscriptional manner (Ahmad and Henikoff, 2002; Elsaesser et al., 2010). Consistent with this, recently identified centromere-associated proteins Eaf6/CENP-28, MSL1v1/CENP-36, and PHF2/CENP-35 (Ohta et al., 2010) have been reported to be subunits of transcription initiation complexes, normally present at promoters. PHF2/CENP-35 binds H3K4me3 and demethylates H3K9 (Wen et al., 2010; Baba et al., 2011; Figure 8B).
FIGURE 8:
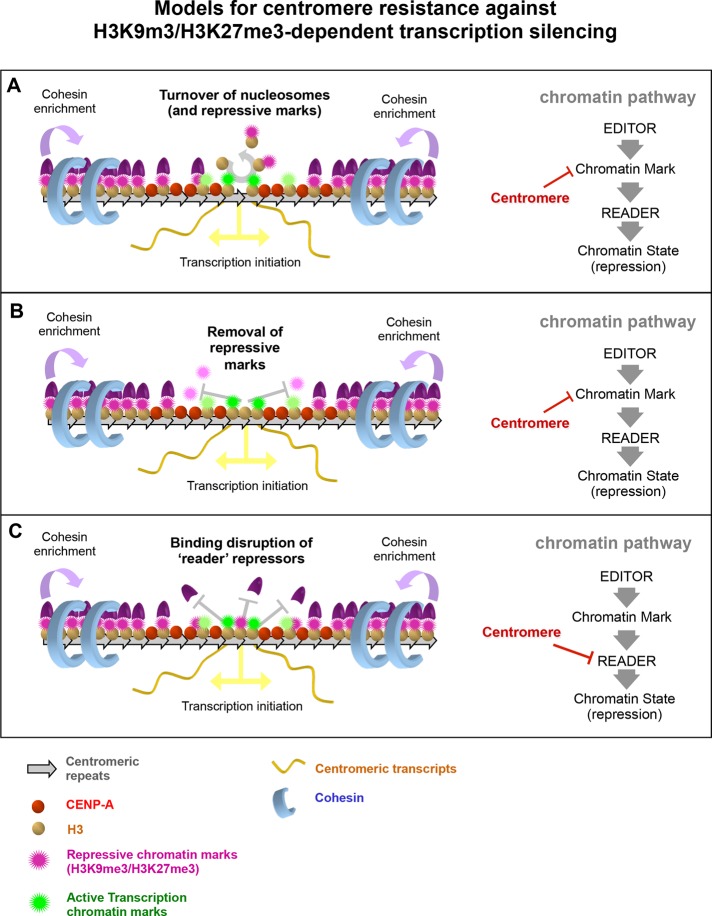
Models for centromere resistance against H3K9m3/H3K27me3-dependent transcription silencing. The ability of eukaryote centromeres to resist local transcription silencing potentially induced by repressive chromatin marks such as H3K9me3 or H3K27me3 might may be derived from one or more properties. Our proposals for how this might occur and where in a repressive pathway the centromere may be interfering. (A) Turnover of nucleosomes within the centromere core may evict repressive chromatin marks as a consequence, thus decreasing their local concentration and potentially preventing spreading of the repressive state. (B) Direct removal of repressive chromatin marks, possibly by centromere-localized demethylases, could clear the repressive chromatin state locally at the centromere core. As in A, it could help prevent spreading of the repressed state as well. (C) Modulation of the binding of readers of repressive chromatin marks, without preventing the spreading of the repressive marks into the centromere core, can potentially alleviate transcriptional silencing promoted by those repressor readers.
Alternatively, nucleosomes within centrochromatin may be compatible with H3 repressive marks, but an unknown centromeric component or some specialized aspect of transcription during mitosis might modulate the binding of repressive readers such as PRC1 or HP1, thus alleviating repression in a way that prevents a limited but sufficient level of transcription (Figure 8C).
Perspectives
Accumulating evidence suggests that active transcription within centrochromatin is critical for chromosome segregation. This raises the question of how it is that centromeres, which in many eukaryotes are flanked by large regions of pericentromeric heterochromatin, are able to resist “invasion” by such heterochromatin, given its known propensity to spread and silence transcription of nearby loci. Our experiments suggest that centromeres can specifically resist two types of canonical repressive pathways that establish silent chromatin states. This may be key to their maintenance and activity in the vicinity of transcriptionally silent heterochromatin and sets the stage for future avenues of experimentation regarding the compatibility of centromere transcription and heterochromatin.
MATERIALS AND METHODS
Cell culture
Human cells were grown in DMEM (+l-glutamine, +pyruvate) supplemented with 10% (vol/vol) fetal bovine serum and 100 U/ml penicillin G and 100 μg/ml streptomycin sulfate (all Life Technologies, Carlsbad, CA). Cells were grown at 37°C in humidified atmosphere containing 5% CO2. Cell harvesting was performed by washing in prewarmed Dulbecco’s phosphate-buffered saline (PBS) and using TrypLE Express (both Life Technologies) to release adherent cells before passage into fresh culture vessels or downstream applications. All cell lines containing an α-satelliteTetO array (1C7 HAC cells of 1F10 integration cells) were maintained in the presence of 4 μg/ml blasticidin S (Life Technologies), selectable via the Bsr gene copy adjacent to the α-satelliteTetO locus (HAC or integration).
HAC-containing HeLa 1C7 cells are described in Cardinale et al. (2009) and are the product of polyethylene glycol–mediated cell fusion between HeLa and HAC-containing HT1080 Ab2.2.18.21 cells (Nakano et al., 2008). HeLa 1F10 cells were generated in the same manner, from HeLa cells fused with integration-containing HT1080 Ab2.5.30 cells. The HT1080 Ab2.5.30 cell line was generated as described in Nakano et al. (2008) as part of a HAC generation assay but contains a noncentromeric α-satelliteTetO array integrated into a chromosome arm instead of an independent ectopic artificial chromosome.
Plasmid expression constructs
The coding sequence of full-length EZH2 was amplified from HeLa cDNA by PCR and cloned into tYIP vector (Cardinale et al., 2009), generating the TetR-EYFP-EZH2 construct. The coding sequence of SUV39H1 from amino acids 102–412 was amplified similarly and cloned into tYIP vector as well, generating the TetR-EYFP-Suv39h1SET construct. The tYIP vector (and derived constructs) expresses TetR–EYFP-(cloned protein) and the puromycin resistance gene (separated by a synthetic intron and an internal ribosome entry site [IRES] motif) from a cytomegalovirus promoter. The coding sequence of full-length BMI1 was amplified similarly and cloned into pJETY3-TetR-EYFP vector (Ohzeki et al., 2012). This generated the T-Y-BMI1 vector, which expresses TetR-EYFP-BMI1 and the hygromycin resistance gene (separated by an IRES motif) from an EF1 promoter. The plasmids TetR-EYFP (Nakano et al., 2008), TYIP-LSD1 (Bergmann et al., 2011), TetR-EYFP-HP1α (Nakano et al., 2008), and TetR-EYFP-KAP1 (Cardinale et al., 2009) have been described.
Immunofluorescence staining
Cells were fixed in 2,5% paraformaldehyde (PFA)/PBS for 5 min at room temperature, quenched in 125 mM glycine for 5 min, and subsequently processed for indirect immunofluorescence using standard protocols. Primary antibodies used are described later, and fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA). DNA was counterstained with 1 μg/ml Hoechst 333342 (Sigma-Aldrich, St. Louis, MO) in PBS and samples mounted onto glass slides with ProLong Gold (Life Technologies).
Staining of unfixed metaphase spreads and stretched chromatin fibers
Preparation and staining of unfixed mitotic chromosomes was essentially performed as described in Keohane et al. (1996). This protocol generates both spreads of metaphase chromosomes and stretched chromatin fibers. Mitotic cells from cultures arrested in prometaphase for 2 h in 100 ng/ml Colcemid (KaryoMax; Life Technologies) were collected by shake-off and incubated in 75 mM KCl for 10 min. Cells were cytospun at 1800 rpm for 10 min onto glass slides using a Cytospin3 (Thermo Fisher Scientific, Houston, TX) and incubated in KCM buffer (10 mM Tris, pH 8.0, 120 mM KCl, 20 mM NaCl, 0.5 mM EDTA, 0.1% Triton X-100) for 10 min. Samples were then labeled with primary and secondary antibodies (diluted in 1% bovine serum albumin in KCM buffer), fixed in 4% PFA (in KCM), stained with Hoechst 333342, and mounted in ProLong.
Antibodies
The following antibodies were used: normal mouse immunoglobulin G (IgG; Merck Millipore, Billerica, MA), mouse anti–CENP-A (A1), rabbit anti–CENP-C (R554), rat anti–CENP-T (r42F10; a kind gift from Kinya Yoda, Division of Biological Science, Nagoya University, Nagoya, Japan [deceased]), mouse anti-H3K27me3 (1E7), mouse anti-H3K27ac (9E2H10, for ChIP), rabbit anti-H3K4me2 (07-030, for immunofluorescence [IF]; Merck Millipore), mouse anti-H3K4me2 (27A6, for ChIP only), mouse anti-H3K36me2 (2C3), rabbit anti-H3K9me3 (07-523, for IF; Merck Millipore), mouse anti-H3K9me3 (2F3, for ChIP), rabbit anti-H3K9ac (07-352, for IF; Merck Millipore), mouse anti-H2AK119ub1 (cl.E6C5; Merck Millipore), rabbit anti-H2A.Z (07-594; Merck Millipore), and rabbit anti-RING1A (ASA3; a kind gift from Paul Freemont, Section of Structural Biology, Imperial College London, London, UK).
Microscopy, cytological analysis, and fluorescence quantification
Images were acquired on a DeltaVision Core system (Applied Precision, Issaquah, WA) using an inverted Olympus IX-71 stand with an Olympus UPlanSApo ×100 oil immersion objective (numerical aperture 1.4) and an InsightSSI light source. The camera (CoolSnap HQ, Photometrics, Tucson, AZ), shutter, and stage were controlled through SoftWoRx (Applied Precision). Z-sections were collected with a spacing of 0.2 mm, and images were projected and analyzed in ImageJ (National Institutes of Health, Bethesda, MD). When required, image stacks were first deconvolved in SoftWoRx.
All fluorescence signal quantification was performed on maximum intensity projections of nondeconvolved microscopy images acquired at a 1 × 1 binning using identical exposure conditions for each experimental subset. Fluorescence intensity is displayed as arbitrary fluorescence units. Cells displaying more than one HAC were quantified for only one of them, determined randomly.
To quantify centromeric proteins in interphase cells, an ImageJ macro (HAC & CRaQ), adapted from that of Bodor et al. (2013), was used to assess HAC centromere protein levels relative to those of endogenous centromeres. Briefly, the maximum signal intensity (of a given centromere protein staining) associated with the HAC was measured and the local nuclear background signal subtracted. The same measurement procedure was applied to endogenous centromeres and automatically detected by the macro, and the HAC-associated signal was normalized against the mean signal of all those centromeres.
To quantify HAC-associated signals for chromatin marks or chromatin proteins, maximum intensity projections of five Z-planes centered on the HAC, were used. An area thresholded to the EYFP HAC signal was used: the mean signal within the HAC area was quantified, and the mean of three local nuclear background areas was subtracted from that.
SNAP-tag assays to assess CENP-A assembly
Visualization and quantification of newly synthesized CENP-A assembled at centromeres were performed as described in Bodor et al. (2014). Briefly, cells were first transfected with a plasmid encoding CENP-A:SNAP, which, when expressed, is incorporated into centromeres, functioning as endogenous CENP-A (Bodor et al., 2014). Afterward, free SNAP-tag on CENP-A:SNAP was permanently labeled with nonfluorescent BTP (New England Biolabs, Ipswich, MA), thus quenching any existing centromere-localized CENP-A:SNAP from any future labeling. After an ∼8- to 11-h chase period (depending on experiment) to allow new CENP-A:SNAP to be synthesized and assembled onto centromeres (as cells exit from mitosis into G1), this pool of CENP-A (which bears free SNAP-tag) is then labeled with fluorescent TMR-Star (New England Biolabs) and can visualized by fluorescence microscopy.
ChIP-qPCR experiments
ChIP experiments were performed using a protocol adapted and modified from Kimura et al. (2008). A minimum of 5 × 106 cells were used for each separate ChIP experiment, cross-linked in 1% formaldehyde (Sigma-Aldrich) for 5 min at room temperature. Cross-linked chromatin was snap-frozen before shearing by sonication in a Bioruptor sonicator (Diagenode, Liège, Belgium). One fifth of input chromatin material was used for each immunoprecipitation, carried out with anti-mouse IgG Dynabeads M-280 (Life Technologies). Antibodies used were described earlier.
To quantify IP DNA, qPCR was performed on input and IP samples using a SYBR Green master mix (Roche, Penzberg, Germany). The primers used are described in Table 1. For each experiment and primer pair, standard curves were prepared from input, and percentage of recovered IP material was calculated relative to it to account for differential primer efficiency.
TABLE 1:
Primer sequences for ChIP and RT-qPCR.
| Primer name | Sequence |
|---|---|
| alphoidTetO fw | CCACTCCCTATCAGTGATAGAGAA |
| alphoidTetO rv | TCGACTTCTGTTTAGTTCTGTGCG |
| TetO_rv 2 (for RT-qPCR) | GTTAAACTCAGTCGTCACCAAGAG |
| Bsr fw | CAGGAGAAATCATTTCGGCAGTAC |
| Bsr rv | TCCATTCGAAACTGCACTACCA |
| Cen21 fw | GTCTACCTTTTATTTGAATTCCCG |
| Cen21 rv | AGGGAATGTCTTCCCATAAAAACT |
| Sat2 fw | TCGCATAGAATCGAATGGAA |
| Sat2 rv | GCATTCGAGTCCGTGGA |
| PABPC1_5′UTR_fw | CGGCGGTTAGTGCTGAGAGTG |
| PABPC1_5′UTR_rv | GGTCGGTCTCGGCTGCTTCACC |
| PABPC1-10kb_fw | CTAGCATCCGTGGGCCAAGAG |
| PABPC1-10kb_rv | CTCTTCCCCAACCCCAGCAAAAT |
| MYT1+40kb_fw | ACGAGGGCTATGGTGTGGACAG |
| MYT1+40kb_rv | GGGCGATGAATCTCGTCCTG |
| BAC_fw | GATTATCACAGTTTATTACTCTGAATTGGCTATC |
| BAC_rv | AGCGCAAGAAGAAATATCCACCG |
| Actin_fw | GCCGGGACCTGACTGACTAC |
| Actin_rv | AGGCTGGAAGAGTGCCTCAG |
fw, forward; rv, reverse.
Transcript quantification by RT-qPCR analysis
Total RNA was extracted and purified using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. Reverse transcription was performed with the Transcriptor High Fidelity cDNA Synthesis Kit (Roche) on purified RNA, using random hexamer primers, in a T3000 thermocycler (Biometra, Göttingen, Germany). qPCR was performed in a LightCycler 480 (Roche) using a SYBR Green master mix (Sigma-Aldrich); the primers used are described in Table 1. For each plate and primer pair used, a serial dilution of the relevant template DNA was included to determine a standard curve and account for differential reaction efficiencies. The specificity of reactions was validated by product melting curve analysis. Reaction crossing points were determined using the second derivative maximum algorithm in the LightCycler 480 software.
Supplementary Material
Acknowledgments
We thank Irina Stancheva, Patrick Heun, Philipp Voigt, and Nadine Vincenten for critical discussions and feedback on the manuscript. We thank Alexander Kagansky for the design and construction of the TetR-EYFP-Suv39h1SET expression construct. This work was funded by the Wellcome Trust, of which W.C.E. is a Principal Research Fellow (Grant 107022). The Wellcome Trust Centre for Cell Biology is supported by core grants 077707 and 092076. Additional experiments were supported by Ministry of Education, Culture, Sports, Science and Technology of Japan KAKENHI Grants 23247030 and 23114008 and the Kazusa DNA Research Institute Foundation (H.M.); grants-in-aid from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.K.); and the intramural research program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (V.L.). N.M.C.M. was funded by a PhD studentship from the Fundação para a Ciência e a Tecnologia (SFRH/BD/62092/2009), and J.H.B. was funded by a PhD studentship from the Wellcome Trust (080458).
Abbreviations used:
- CCAN
constitutive centromere-associated network
- ChIP
chromatin immunoprecipitation
- HAC
human artificial chromosome
- H3Knme3
histone H3 trimethylated on lysine n.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0605) on November 12, 2015.
REFERENCES
- 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–38. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Aldrup-Macdonald ME, Sullivan BA. The past, present, and future of human centromere genomics. Genes (Basel) 2014;5:33–50. doi: 10.3390/genes5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348:132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, NI M, Meyer CA, Igarashi K, Kanno J, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13:668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, Earnshaw WC. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci Suppl. 2012a;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Martins NM, Larionov V, Masumoto H, Earnshaw WC. HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosome Res. 2012b;20:505–519. doi: 10.1007/s10577-012-9293-0. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. Elife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Valente L, Mata J, Black BE, Jansen LE. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, C H, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting bubR1, Mad2 and CENP-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon B. A family of centromere proteins is absent from the latent centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, Van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, De Castro IJ, Kerkhoven RM, Bussemaker HJ, Van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Frangini A, Sjoberg M, Roman-Trufero M, Dharmalingam G, Haberle V, Bartke T, Lenhard B, Malumbres M, Vidal M, Dillon N. The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol Cell. 2013;51:647–661. doi: 10.1016/j.molcel.2013.08.022. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost KL, Bertulat B, Cardoso MC. Heterochromatin and gene positioning: inside, outside, any side. Chromosoma. 2012;121:555–563. doi: 10.1007/s00412-012-0389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- Keohane A, O’neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–30. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling—an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Earnshaw WC, Masumoto H, Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell Mol Life Sci. 2013;70:1135–1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, de Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C., Cooch N., Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, de Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2012;334:686–990. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Miga KH. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res. 2015;23:421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA. Histone modifications within the human X centromere region. PLoS One. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Okamoto Y, Ohzeki J, Masumoto H. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci. 2003;116:4021–4034. doi: 10.1242/jcs.00697. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Ohta S, Bukowski-Wills JC, Sanchez-Pulido L, Alves Fde L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Rush M, Appanah R, Lee S, Lam LL, Goyal P, Lorincz MC. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4:404–414. doi: 10.4161/epi.4.6.9392. [DOI] [PubMed] [Google Scholar]
- Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH. Transcription within a functional human centromere. Mol Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schultz J. Variegation in Drosophila and the inert chromosome regions. Proc Natl Acad Sci USA. 1936;22:27–33. doi: 10.1073/pnas.22.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC. Transcription and ncRNAs: at the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. 2013;21:643–651. doi: 10.1007/s10577-013-9387-3. [DOI] [PubMed] [Google Scholar]
- Seum C, Delattre M, Spierer A, Spierer P. Ectopic HP1 promotes chromosome loops and variegated silencing in Drosophila. EMBO J. 2001;20:812–818. doi: 10.1093/emboj/20.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt RG, Lachner M, Vargas M, Hamer KM, den Blaauwen JL, Hendrix T, Melcher M, Schweizer D, Jenuwein T, Otte AP. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol Cell Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. Chromatin containing CENP-A and a-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Vasanthi D, Nagabhushan A, Matharu NK, Mishra RK. A functionally conserved Polycomb response element from mouse HoxD complex responds to heterochromatin factors. Sci Rep. 2013;3:3011. doi: 10.1038/srep03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, Van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112:4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectible alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere. Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke C, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Nucleotide sequence heterogeneity of alpha satellite DNA: a survey of alphoid sequences from different human chromosomes. Nuclei Acids Res. 1987;15:7549–69. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, LI J, Song T, Lu M, Kan PY, Lee MG, Sha B, Shi X. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J Biol Chem. 2004;279:401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Grandy RA, Lopez-Camacho C, Montecino M, Van Wijnen AJ, Lian JB, Stein JL, Stein GS. Bookmarking target genes in mitosis: a shared epigenetic trait of phenotypic transcription factors and oncogenes. Cancer Res. 2014;74:420–425. doi: 10.1158/0008-5472.CAN-13-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.