Mutations in CCDC11 cause aberrant placement of internal organs and congenital heart disease in humans. Ccdc11 is a novel component of centriolar satellites and plays a critical role in motile and sensory ciliogenesis. The results implicate centriolar satellites in the pathology of left–right patterning and heart disease.
Abstract
The establishment of left–right (L-R) asymmetry in vertebrates is dependent on the sensory and motile functions of cilia during embryogenesis. Mutations in CCDC11 disrupt L-R asymmetry and cause congenital heart disease in humans, yet the molecular and cellular functions of the protein remain unknown. Here we demonstrate that Ccdc11 is a novel component of centriolar satellites—cytoplasmic granules that serve as recruitment sites for proteins destined for the centrosome and cilium. Ccdc11 interacts with core components of satellites, and its loss disrupts the subcellular organization of satellite proteins and perturbs primary cilium assembly. Ccdc11 colocalizes with satellite proteins in human multiciliated tracheal epithelia, and its loss inhibits motile ciliogenesis. Similarly, depletion of CCDC11 in Xenopus embryos causes defective assembly and motility of cilia in multiciliated epidermal cells. To determine the role of CCDC11 during vertebrate development, we generated mutant alleles in zebrafish. Loss of CCDC11 leads to defective ciliogenesis in the pronephros and within the Kupffer’s vesicle and results in aberrant L-R axis determination. Our results highlight a critical role for Ccdc11 in the assembly and function of motile cilia and implicate centriolar satellite–associated proteins as a new class of proteins in the pathology of L-R patterning and congenital heart disease.
INTRODUCTION
Cilia are highly conserved microtubule-based organelles found on the surface of nearly all human cells and serve a wide variety of essential functions. Primary (nonmotile) cilia are found on the majority of cells and play important chemosensory and mechanosensory roles that are fundamental for embryonic development and adult organ homeostasis (Nigg and Raff, 2009; Duldulao et al., 2010; Bettencourt-Dias et al., 2011; Nachury, 2014). Motile cilia are present on distinct cell types in the upper and lower respiratory tract, brain ventricles, and reproductive organs to provide specialized functions. For example, the coordinated cilia-based fluid movement across the surface of multiciliated epithelial cells of the airways is responsible for the major host-defense mechanism of mucociliary clearance (Brooks and Wallingford, 2014; Horani et al., 2014; Tilley et al., 2015). Moreover, motile cilia function in early embryogenesis to create flow across left–right (L-R) organizers (termed the embryonic node) in vertebrates, which is necessary for induction of an asymmetric gene expression cascade that determines L-R organ asymmetry (Norris, 2012; Babu and Roy, 2013; Yoshiba and Hamada, 2014). Both primary and motile cilia are composed of an axoneme containing nine outer doublet microtubules that extend from basal bodies (also called centrioles). Motile cilia of multiciliated cells contain an additional two central microtubules (9 + 2 axonemal arrangement), whereas motile monocilia in the embryonic node lack the central microtubule pair (9 + 0 arrangement). Axonemal microtubule-attached dynein-arm motors, radial spokes, and nexin–dynein regulatory complexes generate the force for ciliary beating.
Defects in the assembly or function of cilia lead to a host of human disease syndromes collectively termed ciliopathies. Mutations have been described that result in abnormal function of primary or motile cilia, with surprisingly little overlap in the disease phenotypes. Primary cilia syndromes have multiple clinical features, including renal dysplasia, polydactyly, retinal dystrophy, and neurocognitive impairment, among others (Powles-Glover, 2014; Valente et al., 2014; Wheway et al., 2014). Defects in motile cilia result in phenotypes that include respiratory tract dysfunction, infertility, and abnormal L-R placement of internal organs (situs). The classic inherited disorder of motile cilia dysfunction is primary cilia dyskinesia (PCD), an autosomal recessive disorder characterized by recurrent respiratory tract infections and irreversible, destructive airway disease (Kennedy and Ostrowski, 2006; Knowles et al., 2013; Kurkowiak et al., 2015). Of note, PCD manifestations also include laterality defects in ∼50% of affected individuals. Moreover, defects in both immotile and motile cilia in the embryonic node during early development result in aberrant L-R situs determination, including situs inversus totalis and a spectrum of heterotaxy syndromes, which are often associated with congenital heart disease (Nonaka et al., 1998; Olbrich et al., 2002; Brueckner, 2007; Nakhleh et al., 2012; Li et al., 2015).
The complex genetics of PCD and L-R asymmetry is rapidly unfolding, and the identification of causative mutations in patients has led to a better understanding of the molecular mechanisms that contribute to the observed ciliary defects. To date, 30 genes have been associated with PCD, 18 of which also affect L-R axis organization (reviewed in Horani et al., 2014; Kurkowiak et al., 2015; Werner et al., 2015). These 30 genes can be classified into four major categories according to protein localization and the ciliary structure affected: 1) those encoding components of the outer and inner axonemal dynein arms, 2) genes for proteins implicated in organization of the central pair microtubules, radial spokes, and the nexin–dynein regulatory complex, 3) genes encoding proteins that are part of the cytoplasmic machinery involved in the preassembly and targeting of ciliary motor proteins, and 4) genes encoding proteins that play distinct roles in specifying the assembly and orientation of basal bodies in rare syndromic forms of PCD (Horani et al., 2014; Kurkowiak et al., 2015; Werner et al., 2015). The list of mutated genes that cause L-R asymmetry defects not associated with PCD is much shorter and includes the X-linked transcription factor ZIC3 (Ware et al., 2004), the transforming growth factor β family members Nodal (Mohapatra et al., 2009) and Lefty1 and Lefty2 (Kosaki et al., 1999a), and the activin receptor, ACVR2B (Kosaki et al., 1999b). Although much progress in gene identification of PCD and L-R asymmetry has been achieved, it is estimated that the known causative genes account for only ∼65% of the cases (Ferkol and Leigh, 2012; Knowles et al., 2013; Kurkowiak et al., 2015). Thus our understanding of the critical components of ciliary assembly and function implicated in these disease syndromes remains incomplete.
Recently, recessive mutations in CCDC11 (coiled-coil domain containing 11) were discovered in patients with situs anomalies (Perles et al., 2012). The authors identified homozygous splice-site mutations in two affected brothers from consanguineous parents, whereas the remaining five siblings and the parents were heterozygous for this mutation. One of the affected siblings displayed phenotypes consistent with heterotaxy syndrome and/or PCD, including abdominal situs anomalies and congenital heart defects, whereas the other sibling presented with situs inversus totalis with an otherwise normal cardiac anatomy and function (Perles et al., 2012). Another recent study described homozygous loss-of-function mutations in CCDC11 in a patient with situs inversus totalis and mild respiratory defects but no other discernible ciliopathy phenotypes (Narasimhan et al., 2015). Previous studies reported elevated expression of CCDC11 during differentiation of multiciliated tracheal epithelial cells in mice (Hoh et al., 2012), human airway epithelia (Ross et al., 2007), and the multiciliated epidermal cells of Xenopus embryos (Hayes et al., 2007). In addition, Ccdc11 was identified in a proteomic screen of purified centrioles from bovine sperm cells (Firat-Karalar et al., 2014). Collectively these findings suggest a potentially critical role for CCDC11 in regulating ciliary assembly and/or function, yet the cellular role(s) of Ccdc11 remain unknown. Thus we aimed to characterize the molecular functions of Ccdc11 in cells with either immotile or motile cilia.
Here we demonstrate that Ccdc11 is a novel component of centriolar satellites—nonmembranous cytoplasmic structures that concentrate around the centrosome and play critical roles in trafficking proteins to and from the centrosome and primary cilium. We show that depletion of CCDC11 disrupts the subcellular organization of satellites and perturbs primary and motile cilium assembly in vitro. Moreover, ciliary assembly and function were interrupted in CCDC11-deficient Xenopus embryos and zebrafish in vivo. Finally, we observed defective L-R axis formation in the CCDC11-deficient zebrafish, consistent with the human disease phenotype. Collectively our results uncover a critical role for Ccdc11 in the assembly and function of motile and primary cilia and implicate centriolar satellite–associated proteins as a new category of proteins in the pathology of PCD and aberrant L-R patterning.
RESULTS
Ccdc11 is a novel component of centriolar satellites
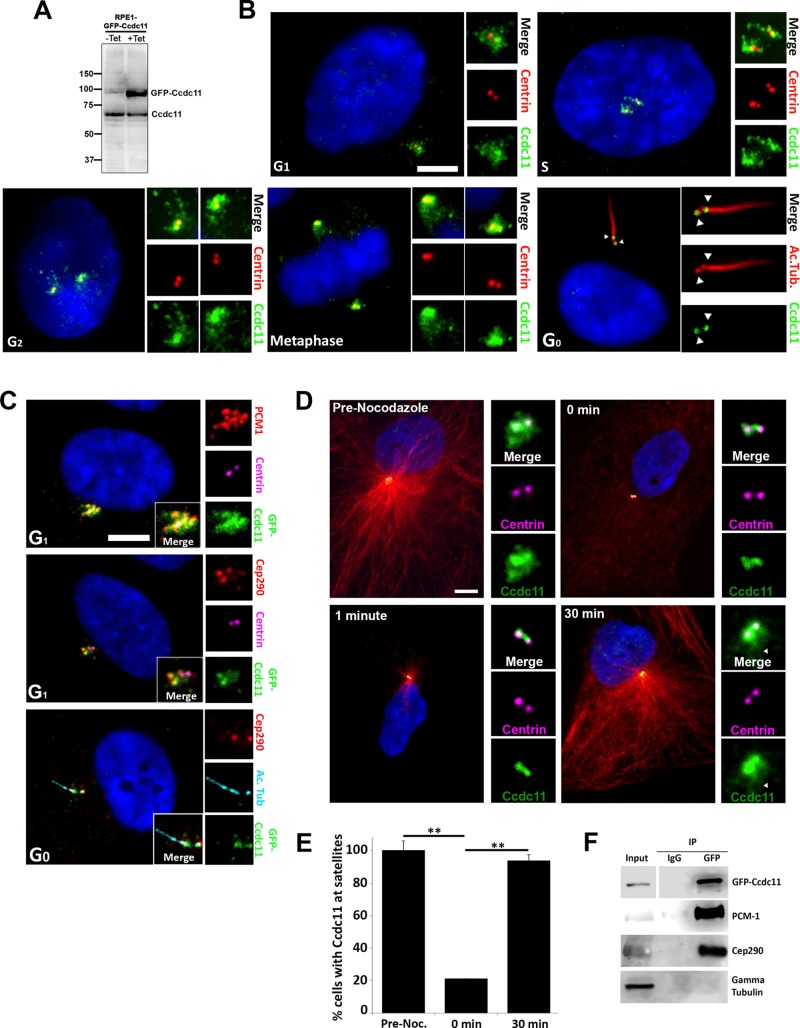
Ccdc11 is a protein of 514 amino acids with a predicted molecular weight of ∼62 kDa containing three putative coiled-coil domains. It is conserved in organisms containing centrioles/basal bodies and cilia ranging from humans to Chlamydomonas (Supplemental Figure S1, A–C). To characterize the localization and function of Ccdc11 at the cellular level, we generated polyclonal antibodies directed against human Ccdc11. The antibody recognized a protein of expected size in immunoblots of human telomerase–immortalized retinal pigment epithelial cells (hTERT-RPE-1; hereafter referred to as RPE-1; Figure 1A). The antibody also recognized the green fluorescent protein (GFP)–tagged fusion protein in RPE-1 cells stably expressing tetracycline-inducible GFP-Ccdc11 (RPE::GFP-Ccdc11; Figure 1A). Next we investigated the localization of endogenous Ccdc11 at various stages of the cell division cycle. Ccdc11 was present on both centrioles in G1 cells and was also distributed in small aggregates that surrounded the two centrioles (Figure 1B). This localization pattern is highly reminiscent of centriolar satellites, which are 70- to 100-nm cytoplasmic granules that concentrate around the centrioles/centrosome and are involved in trafficking of proteins to and from the centrosome (Kubo et al., 1999; Dammermann and Merdes, 2002).
FIGURE 1:
Ccdc11 is a component of centriolar satellites. (A) Immunoblot of lysates from tetracycline-inducible GFP-Ccdc11–expressing RPE-1 stable cell line (RPE::GFP-Ccdc11) probed with anti-Ccdc11 antibody. Numbers on the left indicate molecular mass of markers in kilodaltons. (B) Localization of endogenous Ccdc11 throughout the cell cycle. Asynchronously growing MEF cells were fixed and stained for Ccdc11 (green), centrin to identify centrioles (red), acetylated tubulin to mark cilia (red), and DNA (4′,6-diamidino-2-phenylindole [DAPI]; blue). The centriolar satellite distribution of Ccdc11 is conserved through G1, S phase, G2, and mitosis. After serum starvation to induce ciliogenesis, the satellite pool of Ccdc11 is lost, and the protein is enriched at the transition zone (distal end of each centriole, arrowheads). Insets, magnified images of the centrosome region. Scale bar, 5 μm. (C) RPE::GFP-Ccdc11 cells immunostained with antibodies against GFP (green), PCM-1 (red), or Cep290 (red), as well as centrin (magenta) or acetylated tubulin (light blue) to mark centrioles and cilia, respectively. Ccdc11 colocalizes with PCM-1 and Cep290 at satellites in G1 and with Cep290 at the transition zone in G0. Scale bar, 5 μm. (D, E) RPE-1 cells were treated with nocodazole for 1 h to depolymerize microtubules and then allowed to recover for up to 30 min. Samples were fixed at the indicated times and stained for Ccdc11 (green), centrioles (centrin, magenta), microtubules (α-tubulin, red), and DNA (DAPI, blue). Ccdc11 localization to centriolar satellites is microtubule dependent, but the centriolar pool of Ccdc11 does not require microtubules. Scale bar, 10 μm. Arrowhead indicates the satellite pool of Ccdc11. We scored 300 cells for each sample, from three independent experiments; **p < 0.05. (F) Ccdc11 interacts with satellite proteins PCM-1 and Cep290. Immunoprecipitation was performed on extracts from RPE::GFP-Ccdc11 cells using anti-GFP antibody or control IgG and then probed for GFP and endogenous PCM-1, Cep290, and γ-tubulin (negative control).
To investigate further the satellite-like staining pattern of Ccdc11, we costained RPE::GFP-Ccdc11 cells with antibodies against PCM-1, a core component of centriolar satellites essential for the proper organization of satellites around the centrioles (Kubo et al., 1999; Kubo and Tsukita, 2003; Dammermann and Merdes, 2002; Lopes et al., 2011). GFP-Ccdc11 signal overlapped with PCM-1 in G1 (Figure 1C and Supplemental Figure S2A). Furthermore, GFP-Ccdc11 colocalized with another component of satellites, Cep290, a protein that is also mutated in patients with ciliopathies (Figure 1C and Supplemental Figure S2A; Chang et al., 2006; Valente et al., 2006; Kim et al., 2008). The centriolar satellite–staining pattern of both endogenous Ccdc11 and GFP-Ccdc11 persisted during centriole duplication and growth in S and G2 (Figure 1B and Supplemental Figure S2A). A subset of satellite proteins (e.g., PCM-1, BBS4) disperse in mitosis, such that in metaphase/anaphase, they show little point of focus (Balczon et al., 1994; Dammermann and Merdes, 2002; Kim et al., 2004). In contrast, other centriolar satellite proteins (e.g., Cep290, Ofd1) redistribute onto spindle poles in mitosis (Lopes et al., 2011). We found that Ccdc11 remains enriched on the spindle poles in mitosis, with slight staining evident on spindle microtubules (Figure 1B and Supplemental Figure S2A). To assess the localization of Ccdc11 upon assembly of the primary cilium, we serum starved cells for 48 h to induce ciliogenesis. The satellite pool of Ccdc11 was reduced, but a portion of the protein remained associated with the transition zones at the distal end of each centriole (Figure 1B), an area at the base of cilia involved in regulating trafficking into the cilium (Czarnecki and Shah, 2012). Costaining with anti-Cep290, which relocalizes from satellites to the transition zone upon ciliogenesis (Garcia-Gonzalo et al., 2011; Klinger et al., 2014), confirmed this localization pattern (Figure 1C).
Centriolar satellite organization around the centrosome is dependent on an intact microtubule array and the microtubule-dependent dynein motor (Dammermann and Merdes, 2002; Kim et al., 2004; Kodani et al., 2010). Disrupting the microtubule network or interfering with microtubule-dependent transport leads to dispersal of a subset of satellite proteins, including PCM-1 and Cep290 (Dammermann and Merdes, 2002; Kim et al., 2004, 2008). We therefore sought to determine the localization pattern of Ccdc11 in RPE-1 cells after depolymerization of the microtubule network. Incubation of cells with nocodazole resulted in dispersal of the satellite pool of Ccdc11, consistent with redistribution of centriolar satellites into the cytoplasm (Figure 1, D and E). A fraction of Ccdc11 remained associated with the centrioles and became more obvious to detect. Microtubule regrowth after nocodazole washout led to reaccumulation of the satellite Ccdc11 aggregates (Figure 1D), consistent with refocusing of centriolar satellites around the centrosome (Dammermann and Merdes, 2002; Kim et al., 2004, 2008). Finally, we sought to determine whether Ccdc11 interacts with satellite proteins in vivo. GFP-Ccdc11 was affinity purified from asynchronously growing RPE::GFP-Ccdc11 cells by immunoprecipitation with anti-GFP antibodies (Supplemental Figure S2B) and the protein complexes analyzed using mass spectrometry as previously described (MacCoss et al., 2002; Tabb et al., 2002). As predicted, both PCM-1 and Cep290 specifically coimmunoprecipitated with GFP-Ccdc11 (Figure 1F). Collectively these data demonstrate that Ccdc11 behaves as a bona fide centriolar satellite protein and is present in a complex with core components of satellites in vivo.
Depletion of Ccdc11 causes dispersal of satellites and disrupts primary cilium assembly
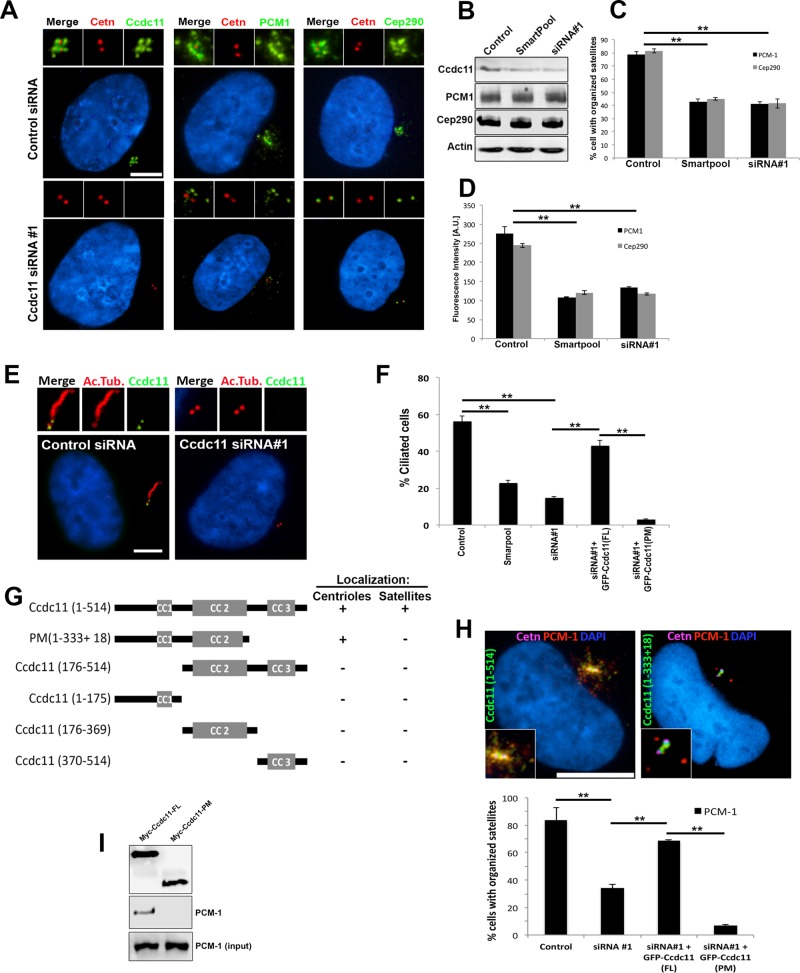
To address the function of Ccdc11, we used small interfering RNA (siRNA) oligonucleotides to deplete the endogenous protein. RPE-1 cells were transfected either with a pool of four siRNAs or a single siRNA targeting CCDC11, which resulted in 65 and 78% reductions in Ccdc11 protein level, respectively (Figure 2, A and B). It is intriguing that depletion of Ccdc11 resulted in loss of PCM-1 and Cep290 signal at centriolar satellites, with slight Cep290 staining still evident at centrioles (Figure 2, A, C, and D). Immunoblot analysis of endogenous PCM-1 and Cep290 in Ccdc11-depleted cells showed no change in the level of either protein, which suggests that the loss of PCM-1 and Cep290 signal was not due to lower expression or degradation of those proteins (Figure 2B). We interpret these data to suggest that Ccdc11 is essential for the integrity and organization of PCM-1 and centriolar satellites, but in their absence, CEP290 may still be targeted to centrioles. In support of this hypothesis, siRNA-mediated depletion of PCM-1 caused loss of CEP290 from satellites but not the centrioles (Supplemental Figure S3, A and B), as previously reported (Kim et al., 2008; Stowe et al., 2012). In addition, depletion of PCM-1 resulted in dispersal of Ccdc11 from satellites but not centrioles (Supplemental Figure S3, A and B), further highlighting the codependence of the two proteins in regulating centriolar satellite organization. We then assessed the consequences of Ccdc11 depletion on primary cilium assembly, since centriolar satellite organization and function are known to be essential for ciliogenesis (reviewed in Tollenaere et al., 2015). RPE-1 cells were transfected with either control or CCDC11 siRNAs and then serum starved for 48 h to induce ciliogenesis. Knockdown of Ccdc11 resulted in a significant reduction in the percentage of cells that assembled primary cilia (Figure 2, E and F), similar to depletion of PCM-1 (Supplemental Figure S3, C and D). Of importance, coexpression of GFP-tagged, siRNA-resistant Ccdc11 rescued the satellite association of PCM-1 and Cep290 (Figure 2H and unpublished data), as well as primary cilium assembly (Figure 2F), highlighting the specificity of the satellite dispersal and ciliary assembly phenotypes. We conclude that Ccdc11 is involved in maintaining centriolar satellite organization and is essential for primary ciliogenesis.
FIGURE 2:
Depletion of Ccdc11 causes dispersal of centriolar satellites and disrupts primary cilium assembly. (A) RPE-1 cells transfected with either nontargeting control siRNA or siRNA against Ccdc11. Cells were fixed and immunostained with antibodies against Ccdc11, PCM-1, or Cep290 (green). Cells were also stained for centrioles (centrin, red) and nuclei (DAPI, blue). Knockdown of Ccdc11 results in loss of satellites surrounding the centrioles. Insets, magnified images of the centrosome region. Scale bar, 10 μm. (B) Immunoblot analysis of extracts from RPE-1 cells transfected with control or Ccdc11 siRNA oligonucleotides and probed with antibodies targeting Ccdc11, PCM-1, Cep290, and actin (as a loading control). Depletion of Ccdc11 does not affect the expression levels of endogenous PCM-1 or Cep290. (C, D) siRNA-mediated depletion of Ccdc11 causes a significant reduction in the percentage of cells with organized satellites. Total fluorescence intensities (D) for PCM-1 and Cep290 were determined within a 2-μm circle centered on the centrosome in cells treated with the indicated siRNAs. The histogram presents the mean total intensities relative to that in the control siRNA-treated samples. We scored 450 cells for each sample from three independent experiments; **p < 0.05. (E, F) Control and Ccdc11-depleted RPE-1 cells were serum starved, fixed, and immunostained for Ccdc11 (green) and acetylated tubulin (red) to label primary cilia. Depletion of Ccdc11 significantly reduced the percentage of cells with cilia, which is rescued by cotransfection with siRNA-resistant, full-length GFP-Ccdc11 but not with the truncated protein GFP-Ccdc11(PM). We scored 300 cells for each sample from three independent experiments; **p < 0.05. Scale bar, 10 μm. (G) Schematic of Ccdc11 deletion constructs and summary of their localizations to centrioles and/or satellites. Colocalization was determined by analyzing RPE-1 cells transfected with Myc-tagged Ccdc11 deletion constructs and immunostaining of fixed cells with anti-Myc and antibodies targeting centrin, PCM-1, and Cep290. CC, coiled-coil domains. (H) Ccdc11-depleted RPE-1 cells were transfected with either Myc-Ccdc11(FL) or Myc-Ccdc11(PM), fixed, and immunostained with antibodies against Myc, centrin, PCM-1, and nuclei (DAPI). Scale bar, 10 μm. We scored 300 cells for each sample from three independent experiments; **p < 0.05. (I) Immunoprecipitation using anti-Myc antibody was performed on extracts of RPE-1 cells expressing either Myc-Ccdc11(FL) or Myc-Ccdc11(PM). Immunoprecipitates were probed for Myc and endogenous PCM-1.
To identify functionally important domains of Ccdc11, we generated a series of deletion mutants (Figure 2G). These mutant forms of Ccdc11 were expressed at low levels in RPE-1 cells as Myc-tagged fusion proteins, and their localization was determined by immunofluorescence using anti-Myc antibodies. Of note, we engineered a deletion construct that mimics the truncated protein described in patients with mutations in CCDC11 (Perles et al., 2012). This mutation causes a frameshift that introduces 18 amino acids and a premature stop codon, resulting in loss of the third coiled-coil domain (Figure 2G; PM(1-333+18) [Patient Mutation; which includes the additional 18 amino acids present in patients with a frameshift mutation in CCDC11]). Loss of this C-terminal coiled-coil fragment disrupted localization of Myc-Ccdc11 to satellites but not to centrioles (Figure 2G and Supplemental Figure S2C). Myc-Ccdc11 truncations lacking the other coiled-coil domains failed to localize to satellites or centrioles and displayed a diffuse cytoplasmic distribution (Figure 2G and Supplemental Figure S2C). It is intriguing that overexpression of Myc-Ccdc11 PM(1-333+18) acted in a dominant-negative manner, resulting in dispersal of centriolar satellites and aberrant ciliogenesis (Supplemental Figure S2D). These results indicate that the C-terminal coiled-coil domain is necessary but not sufficient for localization of Ccdc11 to satellites and is a critical domain for regulating the organization of satellites structures around the centrosome. Consistent with this theory, expression of Myc-Ccdc11 PM(1-333+18) in Ccdc11-depleted cells did not rescue the centriolar satellite localization of PCM-1 (Figure 2H) or primary cilium assembly (Figure 2F) compared with the full-length protein. Finally, we tested whether this coiled-coil domain is involved in mediating the interaction between Ccdc11 and PCM-1 at satellites. We expressed either full-length Myc-Ccdc11 or Myc-Ccdc11 PM(1-333+18) in RPE-1 cells and immunoprecipitated the proteins using anti-Myc antibodies. Whereas full-length Ccdc11 was able to coimmunoprecipitate endogenous PCM-1, loss of the third coiled-coil domain disrupted this interaction (Figure 2I). Together these data indicate that Ccdc11 is required for maintaining the localization of PCM-1 and Cep290 at satellites, likely mediated via its third coiled-coil domain.
Loss of Ccdc11 disrupts motile cilia assembly and function
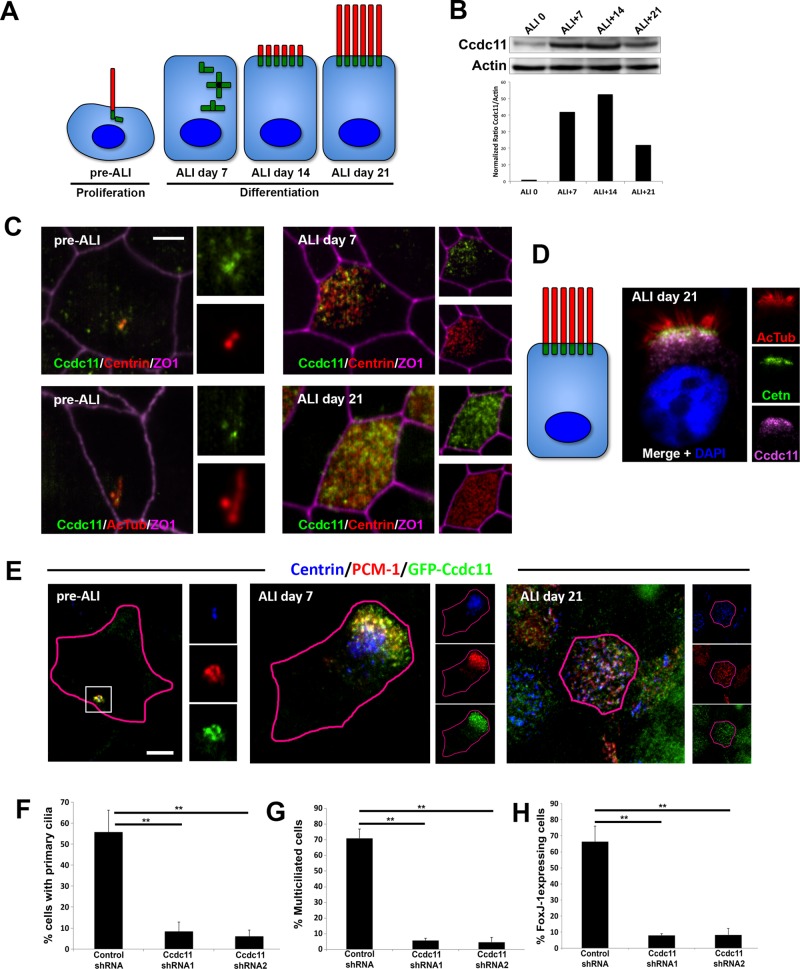
Next we sought to determine the function of Ccdc11 in cells with motile cilia using a human tracheobronchial epithelial cell (hTEC) culture system. This model provides an opportunity to study motile ciliogenesis in mammalian cells that produce hundreds of centrioles and cilia during differentiation (Figure 3A). Tracheobronchial progenitor cells were obtained from the Airway Epithelial Cell Core at Washington University (St. Louis, MO) and cultured as previously described (You et al., 2002; You and Brody, 2013; Mahjoub et al., 2010). Immunoblots of hTEC lysates showed increasing levels of Ccdc11 protein during differentiation (Figure 3B), which correlates with the previously described up-regulation in mRNA levels (Ross et al., 2007; Hoh et al., 2012). Immunofluorescence staining of hTEC cultures showed Ccdc11 localization to centriolar satellites in proliferating cells before differentiation (Figure 3C; pre–air/liquid interface [ALI]), consistent with its satellite localization in RPE-1 cells. Centriologenesis begins ∼7 d after shifting to an ALI in these cultures, as indicated by the appearance of dense clusters of centrin-positive spots (Figure 3C). The Ccdc11 signal showed significant overlap with centrin at this early stage of centriole assembly. The fully assembled centrioles then disperse throughout the cytoplasm, dock at the apical surface, and initiate the formation of cilia (Figure 3A). Ccdc11 remained in the vicinity of the centrioles throughout the course of ciliogenesis (Figure 3C). More specifically, the majority of Ccdc11 signal was present in a layer just beneath the centrioles (Figure 3D). This is somewhat analogous to how satellites appear in cells with a single centrosome, in which the majority of the satellite proteins (e.g., PCM-1, Cep290, or Ccdc11) are present in aggregates in a region neighboring the centrioles. To determine whether the localization pattern of Ccdc11 was consistent with satellite protein localization, we infected hTEC cells with a lentiviral construct expressing GFP-Ccdc11. Cells were fixed at various stages of differentiation and stained with antibodies to detect GFP-Ccdc11, PCM-1, and centrioles (centrin). As predicted, GFP-Ccdc11 colocalized with PCM-1 at all stages of hTEC differentiation (Figure 3E).
FIGURE 3:
Ccdc11 is required for motile cilia formation in human multiciliated epithelial cells. (A) Schematic of the differentiation steps of cultured multiciliated respiratory epithelia. The key phases of the culture are depicted in cartoon form and include the progenitor phase (pre-ALI), centriologenesis (ALI day 7), initiation of ciliogenesis (ALI day 14), and mature multiciliated cells (ALI day 21). (B) Expression levels of endogenous Ccdc11 in hTECs. Cells were grown on Transwell permeable filter supports, and ALI was established 2 d after cells reached confluence. Cells were harvested on the indicated days, and lysates were analyzed by immunoblotting with antibodies against Ccdc11 and actin. Graph shows quantification of Ccdc11 levels normalized to actin for each stage. (C, D) Localization of endogenous Ccdc11 in hTEC at different stages of differentiation, viewed en face (C) or longitudinally (D). Progenitor cells were grown until confluent, and differentiation was induced by creating ALI. Samples were fixed on the indicated days and stained with antibodies against Ccdc11, centrin, acetylated tubulin, and ZO1 (to mark cell boundaries). Scale bar, 5 μm. (E) Colocalization of GFP-Ccdc11 and PCM-1 in hTEC. Progenitor cells were infected with lentivirus expressing GFP-Ccdc11 and grown until confluent, and differentiation was induced by creating ALI. Samples were fixed on the indicated days and stained with antibodies against GFP, centrin, and PCM-1. Scale bar, 5 μm. (F–H) Quantification of ciliogenesis defects upon loss of Ccdc11 in hTEC. shRNA-mediated depletion of Ccdc11 results in significant reduction of primary cilium assembly in pre-ALI progenitor cells (F), reduced number of multiciliated cells at ALI day 21 (G), and decreased number of cells expressing FoxJ1 at ALI day 21 (H). We scored 300 cells for each sample from three independent experiments; **p < 0.05.
To test the function of Ccdc11 in hTECs, we infected progenitor cells with short hairpin RNA (shRNA)–expressing, puromycin-resistant lentivirus 6 d before establishing ALI and selected infected cells with puromycin as previously described (Horani et al., 2013). Cells were fixed and assayed once the progenitors reached confluence (pre-ALI) and then on ALI days 7–21 to monitor differentiation using centrin (to mark centrioles) and acetylated tubulin (to mark cilia). As expected, hTEC progenitors expressing control shRNA assembled a primary cilium pre-ALI (Figure 3F and Supplemental Figure S4A) and underwent centriologenesis and motile ciliogenesis (Figure 3G and Supplemental Figure S4B). In contrast, hTEC progenitors expressing two different shRNAs targeting Ccdc11 displayed dispersal of satellites consistent with our observations in RPE-1 cells and similarly failed to assemble a primary cilium (Figure 3F and Supplemental Figure S4A). Moreover, depletion of Ccdc11 in hTECs resulted in a decrease in the fraction of multiciliated cells at ALI day 21 (Figure 3G and Supplemental Figure S4B). To determine whether the decrease in multiciliated cells was due to a block in centriologenesis or in the differentiation pathway of those cells, we stained shRNA-expressing cells with antibodies against FoxJ1. This forkhead family transcription factor is expressed in cells that become committed to the multiciliated cell fate (Tichelaar et al., 1999; You et al., 2004). There was a marked decrease in the percentage of FoxJ1 expressing hTECs at ALI day 21 upon depletion of Ccdc11 (Figure 3H), which correlated with the decrease in the fraction of multiciliated cells in the population (Figure 3G). We interpret these results as Ccdc11 playing a critical role in the ciliogenesis program of multiciliated epithelia. However, it was not clear whether this phenotype was due to Ccdc11-dependent function(s) during centriologenesis and/or ciliogenesis or a consequence of satellite dispersal and loss of primary cilium assembly that were evident in pre-ALI progenitor cells.
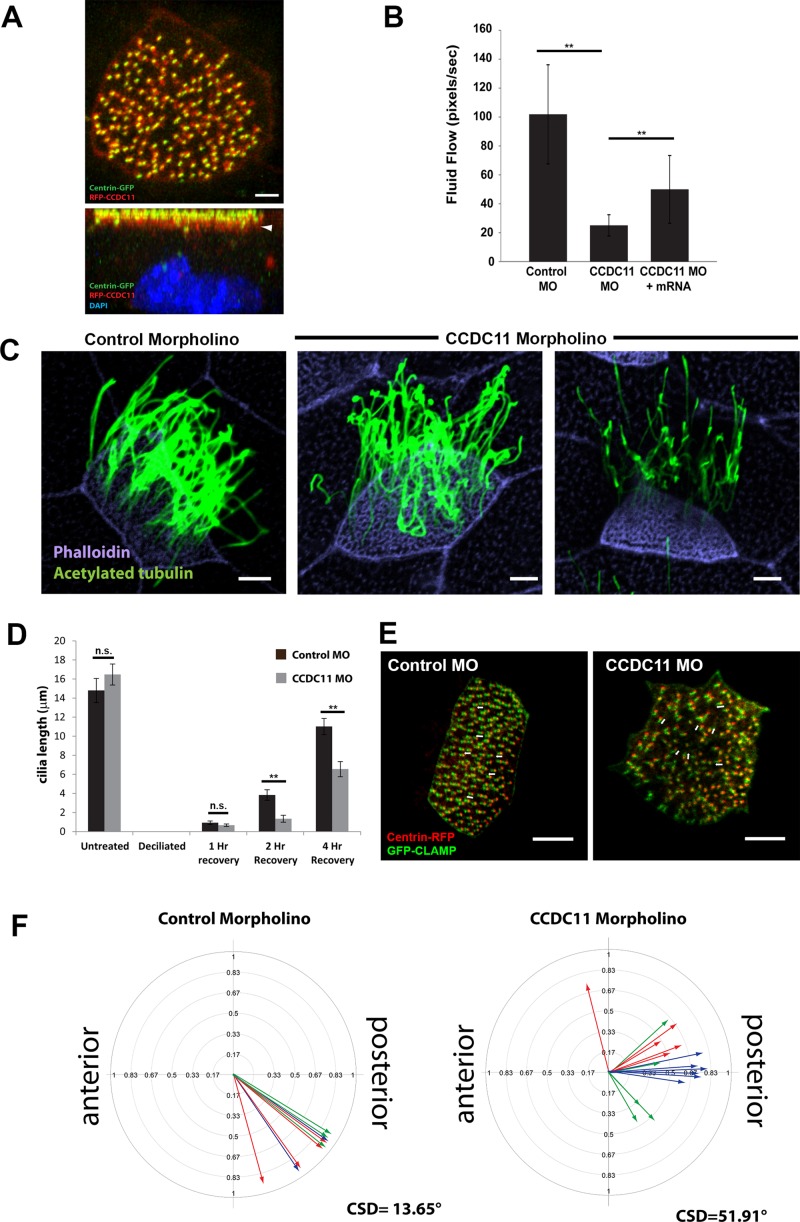
Because depletion of CCDC11 in hTECs disrupted the assembly of primary cilia and blocked the differentiation of multiciliated cells, we sought to examine the function of Ccdc11 in the motile ciliated epithelia of the Xenopus skin. Similar to tracheal epithelial cells, this tissue contains multiciliated cells that assemble hundreds of motile cilia responsible for generating directed fluid flow over the surface of the embryo. However, these cells do not form primary cilia, thereby allowing us to analyze directly the role of the protein in motile cilium assembly and function. First, we injected mRNA encoding a red fluorescent protein (RFP)–tagged Xenopus homologue of CCDC11 (Supplemental Figure S1; Xl.12748) and found that it localized to the centrioles/basal body region at the apical surface of cells (Figure 4A). The majority of RFP-Ccdc11 signal was present in a layer just beneath the centrioles (Figure 4A), similar to the localization of Ccdc11 in mature hTECs (Figure 3D). We then assessed the function of CCDC11 in Xenopus by injecting a morpholino oligo (MO) targeting CCDC11 to deplete the protein. We analyzed ciliary function by measuring fluid flow over the surface of embryos generated by the beating of motile cilia, using live-cell imaging of fluorescent microbeads as previously described (Werner et al., 2011; Werner and Mitchell, 2013). Remarkably, CCDC11 morphant embryos displayed a significant decrease in their ability to generate directed fluid flow as compared with embryos injected with control morpholino (Figure 4B and Supplemental Videos S1 and S2). Of importance, this decrease in flow was rescued by the addition of MO-resistant mRNA encoding CCDC11 (Figure 4B), indicating that the phenotype is specific to the loss of CCDC11. Given the localization of RFP-Ccdc11 to centrioles, we asked whether it had a role in centriole biogenesis but observed no difference in centriole number (unpublished data). However, there appeared to be a decrease in the number of cilia in CCDC11 morphant cells, which suggests a defect in ciliogenesis (Figure 4C). To address more accurately the ciliogenesis phenotype, we quantified cilia length in cells at different time points after deciliation and found that CCDC11 morphant cells regrew cilia significantly more slowly than did control morphants (Figure 4D). We interpret these results as indicating that CCDC11 has an important role in ciliogenesis of multiciliated cells in vivo. The somewhat mild ciliogenesis phenotype did not fully explain the significant loss in fluid flow seen in morphant embryos. Therefore we analyzed the orientation and polarity of cilia in the multiciliated cells, since defective ciliary orientation can also disrupt directed fluid flow (Mitchell et al., 2007, 2009). Control and CCDC11 morphant embryos were coinjected with GFP-CLAMP and centrin4-RFP to mark the positions of the centriolar rootlet structures and centrioles, respectively (Figure 4E). Centriole orientation was scored manually by measuring the angle of the rootlets relative to the anterior–posterior axis of the embryo (Mitchell et al., 2007, 2009; Park et al., 2008). CCDC11 morphant cells failed to polarize their centrioles (and thus cilia) to the extent of control cells (Figure 4F). Collectively these data indicate that loss of CCDC11 in Xenopus results in defective motile cilia assembly, morphology, and function in multiciliated cells.
FIGURE 4:
Ccdc11 is essential for motile cilia assembly and function in Xenopus. (A) Localization of RFP-Ccdc11 (red) in a multiciliated cell from Xenopus embryo shows accumulation near centrioles at the apical surface, marked with centrin-GFP (green). Arrow indicates the majority of Ccdc11 signal being present in a layer just beneath the centrioles. Scale bar, 5 μm. (B) Measurement of fluid flow across the surface of Xenopus embryos injected with a control MO (n = 12 embryos), CCDC11 MO (n = 10), or CCDC11 MO + CCDC11 mRNA (n = 9). Rates of fluid flow were recorded by visualizing the movement of fluorescent microbeads over the surface of the embryos (see Supplemental Videos S1 and S2). **p < 0.05. (C) Multiciliated cells from stage 28 embryos injected with control or CCDC11 MO. Cilia were stained with antibodies against acetylated tubulin (green) and actin stained with phalloidin to mark cell boundaries (purple). The abundance and morphology of cilia appear abnormal in CCDC11 morphant embryos. Scale bar, 5μm. (D) Quantification of ciliary regrowth after deciliation. Cilia were stained using antibodies against acetylated tubulin, and length was measured in untreated cells and in cells at 1, 2, and 4 h after chemical deciliation (28 cells taken from five embryos). **p < 0.05. (E, F) Quantification of cilia orientation and polarity in multiciliated cells. Embryos were injected with either control or CCDC11 MO, and orientation of basal bodies was determined using immunofluorescence staining of centrin-RFP and GFP-CLAMP (see Materials and Methods). White arrows in E show examples of the axis of basal body-rootlet orientation. Control MO, 10 cells from three embryos represented by color; CCDC11 MO, 16 cells from three embryos. CSD, circular SD.
Loss of function of CCDC11 causes defective ciliogenesis and L-R asymmetry in zebrafish
To investigate the biological role of Ccdc11 during vertebrate embryonic development, we used targeted mutagenesis to disrupt the gene in zebrafish. The zebrafish has emerged as a powerful vertebrate model in which to query developmental defects associated with ciliary dysfunction (Serluca et al., 2009; Jaffe et al., 2010; Austin-Tse et al., 2013). Because the embryos and larvae contain multiple different ciliated cell types, zebrafish has been used to model both sensory and motile cilia functions. A single homologue of Ccdc11 is present in the zebrafish genome (Zebrafish Model Organism Database [ZFIN] ID ZDB-Gene-060503-508), with 69% similarity and identical domain organization (with three coiled coils) to human Ccdc11 (Supplemental Figure S1). We designed a pair of transcription activator–like effector nucleases (TALENs; Boch et al., 2009; Moscou and Bogdanove, 2009) targeting the second exon in the CCDC11 gene (Supplemental Figure S5, B and C; described in detail in Materials and Methods). Sequencing of PCR products encompassing the target sequence from TALEN-injected embryos identified a number of independent strains with deletions that result in premature stop codons, two of which were used in the subsequent experiments (Δ5bp and Δ2bp; Supplemental Figure S5D). Reverse transcription PCR (RT-PCR) analysis of cDNA (synthesized from mRNA) showed loss of CCDC11 transcript in mutant embryos, likely due to degradation of the endogenous transcript (Supplemental Figure S5E).
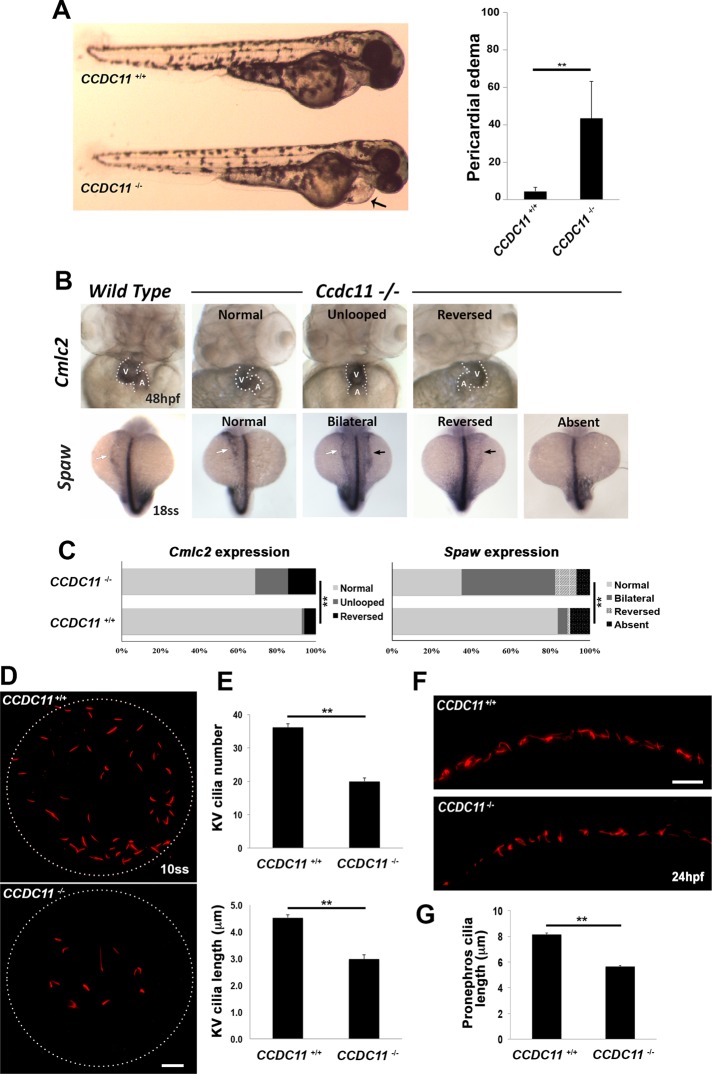
Zebrafish cilia mutants display pleiotropic phenotypes, including pericardial edema, hydrocephalus, dilated/cystic kidneys, and aberrant L-R asymmetry. We observed several prominent phenotypic defects in the CCDC11−/− mutants that are consistent with a role in ciliary assembly and function. At 2 d postfertilization (dpf), mutants exhibited pericardial edema, whereas wild-type and heterozygous CCDC11+/− embryos did not display an altered phenotype (Figure 5A). Because CCDC11 mutation causes situs abnormalities in humans (Perles et al., 2012), we sought to examine the specification of L-R asymmetry of the zebrafish body plan and patterning of asymmetrically placed organs such as the heart. In situ hybridization for southpaw (spaw), an early marker of L-R asymmetry (Long et al., 2003), showed normal spaw expression in the left-side lateral plate mesoderm in most of the control embryos at the 18-somite stage (Figure 5B). In CCDC11−/− mutants, spaw expression was more random and was detected in the right-side lateral plate mesoderm (reversed) or bilaterally or was absent (Figure 5, B and C). We also examined the expression of cardiac myosin light chain 2 (cmlc2), a marker for the orientation of heart looping (Yelon et al., 1999). Normal heart looping, with left placement of the ventricle and right placement of the atrium, was observed in the majority of wild-type embryos, whereas in CCDC11−/− mutants, heart looping was reversed or the heart failed to loop at all (Figure 5, B and C).
FIGURE 5:
Loss-of-function of CCDC11 causes defective ciliogenesis and L-R asymmetry in zebrafish. (A) CCDC11 mutant embryos display pericardial edema at 2 dpf. We scored 117 (wild-type) and 163 (mutant) embryos. **p < 0.05. (B) Whole-mount in situ hybridization showing the expression of laterality markers in wild-type and CCDC11 mutants. Cmlc2 expression shows the heart tube looping to the left in control embryos at 48 hpf, whereas looping was reversed or absent in CCDC11 mutant embryos (A, atrium; V, ventricle). Spaw is expressed in the left lateral plate mesoderm in control embryos at the 18-somite stage (white arrow), whereas its expression is bilateral (black and white arrows), reversed (black arrow), or absent in a significant number of CCDC11 mutants. (C) Quantification of laterality defects in CCDC11 mutant embryos. For Cmlc2 analysis, 89 embryos (WT) and 167 embryos (CCDC11 mutants) were scored. For Spaw analysis, 154 embryos (WT) and 231 embryos (CCDC11 mutants) were scored. **p < 0.05. (D) Immunofluorescence images of cilia in the KV of wild-type and CCDC11 mutant embryos. Embryos were fixed at the 8- 10-somite stages and stained with anti–acetylated α-tubulin. Scale bar, 10 μm. (E) Quantification of cilia abundance and length in KV. There was a significant reduction in cilia number and length in CCDC11 mutants compared with control embryos. Fifteen embryos from each genotype. **p < 0.05. (F) Immunofluorescence staining of cilia in the posterior segment of the pronephros from wild-type and CCDC11 mutant embryos at 24 hpf stained with anti–acetylated tubulin. Scale bar, 10 μm. (G) Quantification of cilia length in the pronephros. Fifteen embryos from each genotype. **p < 0.05.
To determine whether these phenotypic defects were due to impaired ciliogenesis upon loss of CCDC11, we immunolabeled control and mutant embryos with antibodies against acetylated tubulin to mark cilia. We began by examining the abundance and length of cilia in the Kupffer’s vesicle (KV), the transient ciliated organ of asymmetry in the zebrafish embryo that initiates L-R development of the brain, heart, and gut (Essner et al., 2005). Cilia number in KV of 8- to 10-somite-stage CCDC11−/− mutant embryos was significantly reduced compared to control embryos (Figure 4, D and E). Moreover, there was a reduction in length of the remaining cilia in KV of CCDC11−/− mutants (Figure 5E). Cilia in the zebrafish pronephros are also motile and drive fluid flow through the kidneys. Immunolabeling of cilia in pronephros of control 3-dpf zebrafish showed densely populated cilia that were long and extended into the lumen of the kidney tubule (Figure 5F). Of interest, the pronephric cilia of CCDC11−/− mutants appeared less dense, and the existing cilia were significantly shortened (Figure 5, F and G). Together these data suggest that CCDC11 plays a central role in regulating cilia assembly and function in zebrafish in vivo, similar to what we observed for primary and motile ciliated cells in vitro. Moreover, loss of Ccdc11 in zebrafish recapitulates the L-R patterning and heart defects observed in humans.
DISCUSSION
Mutations in CCDC11 have recently been reported in patients with laterality defects and heart disease (Perles et al., 2012; Narasimhan et al., 2015), which is often caused by aberrant assembly and function of cilia. To gain insights into the mechanistic role of Ccdc11, we characterized the function of the protein in ciliated cells in vitro and in vivo. We discovered that Ccdc11 is a component of centriolar satellites, and its localization throughout the cell cycle is consistent with what has been described for other satellite proteins. Immunoprecipitation of Ccdc11 identified a robust interaction with PCM-1, which is viewed as a fundamental component and often used to identify these structures (Balczon et al., 1994; Kubo et al., 1999; Kubo and Tsukita, 2003; Dammermann and Merdes, 2002). We also found an interaction with Cep290, which is present in a complex with PCM-1 in satellites and is essential for ciliogenesis (Kim et al., 2008).
Depletion of Ccdc11 resulted in dispersal of PCM-1 and Cep290 from the region surrounding the centrioles and disrupted primary cilium assembly. This is consistent with previous studies showing that dispersal of PCM-1 (and thus satellites) perturbs the formation of primary cilia. Moreover, deletion of the third coiled-coil domain, as seen in patients with mutations in CCDC11, abrogated its interaction with PCM-1 and also disrupted satellite organization and ciliogenesis. How would loss of Ccdc11 affect the organization of satellites around the centrioles? Satellites depend on an intact microtubule network and require a direct interaction with dynein/dynactin motor proteins for correct localization and cargo delivery (Kubo et al., 1999; Kim et al., 2004, 2008). This physical coupling between the satellites and microtubule-associated motor proteins can be mediated by several satellite proteins, such as PCM-1, Cep290, BBS4, and Par6α (Kim et al., 2004, 2008; Kodani et al., 2010). Thus one interesting possibility is that Ccdc11 similarly helps to tether PCM-1 and/or Cep290 to dynein/dynactin motors or the cytoplasmic microtubule array. Collectively our results indicate that Ccdc11 is a bona fide component of satellites, is necessary for the organization of PCM-1 and Cep290 at centriolar satellites, and is essential for primary ciliogenesis.
We also demonstrate that depletion of Ccdc11 disrupts motile ciliogenesis in multiciliated human tracheal epithelial cells, Xenopus multiciliated epidermal cells, and cilia in the zebrafish KV and pronephros. This is consistent with the recent finding by Narasimhan and colleagues (2015) that morpholino-mediated knockdown of Ccdc11 in zebrafish disrupts motile cilia function in the KV, spinal canal, and pronephros. Although they did not observe any overt changes in ciliary assembly, this may be partly explained by potentially incomplete depletion of the protein with morpholinos or the presence of maternally inherited Ccdc11 in the embryo. Nevertheless, these results collectively point toward a critical role for Ccdc11 in regulating motile cilia assembly and function. It is important to note that, in addition to primary ciliated cells, many satellite proteins are also expressed in cells with motile cilia. Indeed, the presence of satellite-like, electron-dense “fibrous granules” was long ago observed around the numerous assembling basal bodies in motile ciliated epithelia (Steinman, 1968; Anderson and Brenner, 1971; Dirksen, 1991).
Remarkably, most of the functional studies on satellite proteins have been performed in cells with immotile cilia, and little is known about the composition and function of these complexes in motile ciliated cells. This is likely due to the fact that mutations in a number of satellite proteins (e.g., Cep290, BBS4, OFD1) are linked to ciliopathy disease syndromes that display clinical features typically associated with immotile cilia defects, such as renal dysplasia, retinal dystrophy, mental retardation, and limb malformations (Feather et al., 1997; Mykytyn et al., 2001; Katsanis, 2004; Chang et al., 2006; Valente et al., 2006; Saal et al., 2010). However, these ciliopathy syndromes are heterogeneous disorders and may involve defects associated with motile cilia dysfunction, such as infertility, hydrocephalus, and respiratory/airway malfunction. For example, some patients harboring mutations in Cep290 show a decrease in the fraction of multiciliated respiratory epithelial cells, and the remaining cells contain shorter cilia and display abnormal ciliary motility (Papon et al., 2010). Such findings, along with our experiments demonstrating the importance of Ccdc11 in regulating motile cilia assembly in vitro and in vivo, highlight the critical role that satellite proteins may play in motile ciliogenesis. What would the function of satellite proteins such as Ccdc11 be in motile ciliated cells? We conceptualize that these protein complexes are involved in ferrying ciliary cargo proteins to the hundreds of basal bodies in multiciliated cells, analogous to their trafficking functions in primary ciliated cells. The localization of Ccdc11, together with PCM-1, in the vicinity of basal bodies during hTEC differentiation lends support to this theory. The complete functional proteome of satellites in multiciliated cells remains elusive, and we still do not understand how their composition, localization, and function are regulated during differentiation. To address such questions, methods to analyze satellite assembly and dynamics in multiciliated cells have to be further developed.
To the best of our knowledge, Ccdc11 is the first centriolar satellite component identified whose mutation specifically disrupts motile cilia function in humans without any overt phenotypes characteristic of defects in primary cilia, making it unique among the cohort of satellite proteins. Because Ccdc11 depletion disrupts primary ciliogenesis in vitro, one outstanding question is why these patients do not present with defects associated with primary cilia. One possibility is the existence of redundant mechanism(s) that compensate for its absence in tissues with immotile cilia, and there is precedence for such compensatory mechanisms in centrosome and ciliary biology. One such example is Azi1, a centriolar satellite protein that was shown to be essential for primary cilium formation in vitro by a number of independent groups (Graser et al., 2007; Staples et al., 2012; Hall et al., 2013). Remarkably, Azi1-null mice displayed none of the gross abnormalities commonly associated with primary cilia dysfunction (Hall et al., 2013). The difference in phenotypes observed after knockdown of Azi1 in vitro (referred to as “acute” loss of the protein) versus permanent deletion of the gene in vivo (referred to as “chronic’’ loss) is indicative of a compensation mechanism that, in essence, rescues the ciliary assembly defects caused by loss of Azi1 (Hall et al., 2013). Other examples of compensation derive from studies of human mutations in genes required for centrosome assembly and duplication, for which the predominant phenotype appears to be microcephaly (Kumar et al., 2009; Guernsey et al., 2010; Hussain et al., 2012). These last findings suggest that, during mammalian development, most tissues can compensate and overcome mutations in centrosome duplication proteins, except for neural progenitor cells, which appear especially sensitive to these perturbations. These types of functional compensations highlight the complexity and challenge in understanding cilia assembly and function in vivo, but a mechanistic understanding of the underlying compensation process is crucial and may ultimately be leveraged for therapy.
MATERIALS AND METHODS
Cell culture and media
Mammalian cell lines used in this study were hTERT-immortalized human RPE cells (RPE-1), mouse embryonic fibroblasts (MEFs), and human embryonic kidney 293T cells (HEK293T). Cells were grown in DMEM (Corning, Corning, NY) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin/streptomycin solution (Life Technologies, Carlsbad, CA). For serum-starvation experiments, cells were incubated for 24–48 h in DMEM supplemented with 0.5% FBS. Human tracheobronchial epithelial cells were obtained from the Airway Epithelia Cell Core at Washington University, as approved by the institutional review committees at Washington University School of Medicine. hTEC cultures were established as previously described (You et al., 2002; You and Brody, 2013; Mahjoub et al., 2010). Briefly, human airway epithelial cells were isolated from trachea and mainstem bronchi of lungs donated for transplantation. After isolation, hTECs were expanded on tissue culture dishes that were coated with 50 μg/ml type I rat-tail collagen. To initiate cultures, freshly isolated passage 1 or 2 hTECs were seeded on collagen-coated membranes (Transwell; Corning) at a density of 1 × 105 cells/cm2. The medium used for initiation of cell culture was hTEC/Plus supplemented with retinoic acid and human epidermal growth factor (You et al., 2002; You and Brody, 2013; Mahjoub et al., 2010). When cells achieved confluence, an ALI was established, and the medium in the lower compartment was replaced with one lacking serum, to promote epithelial cell differentiation. Cells were cultured at 37°C with 5% CO2, and medium was replaced every 2 d. All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. All media were supplemented with 100 U/mL penicillin, 100 mg/ml streptomycin, and 0.25 mg/ml Fungizone (all obtained from Life Technologies).
Plasmids, siRNA, and transfections
A full-length cDNA clone of human Ccdc11 was obtained from GE Healthcare Dharmacon (Lafayette, CO; clone ID 1573). The Ccdc11 open reading frame (ORF; 1545 base pairs encoding 514 amino acids) was amplified by PCR and subcloned into pEGFP-C1 vector (Clontech, Mountain View, CA) within unique BspEI and BamHI sites in the multiple cloning site. This vector contains an N-terminally located enhanced GFP gene, resulting in a GFP-Ccdc11 fusion protein. For lentiviral-based expression, the GFP-Ccdc11 cassette was PCR amplified and subcloned into pLVX-Puro or pLVX-Puro-TetOn vectors (Clontech) at unique XhoI and EcoRI sites. Alternatively, the full-length (FL) Ccdc11 ORF was cloned into a pCMV vector containing a single N-terminal Myc tag. For generation of truncated versions of Ccdc11, PCR amplification of the desired Ccdc11 cDNA sequences was performed to yield fragments that encode amino acids 1–175, 176–369, 176–514, 350–514, and 370–514 and PM(1–333+18) (Perles et al., 2012). All PCR products and constructs were verified by sequencing. Cells were subsequently transfected with 1–2 μg of the appropriate plasmid DNA using 10 μl of Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. A stable cell line with constitutive expression of recombinant GFP-Ccdc11 protein was generated by infecting RPE-1 cells with either pLVX-Puro-GFP-Ccdc11 or pLVX-Puro-TetOn-GFP-Ccdc11 lentiviral constructs. Infected cells were selected using 2 μg/ml puromycin, single colonies were isolated, and individual clones were analyzed for expression of GFP-Ccdc11 using immunoblotting and immunofluorescence.
For experiments using siRNA-mediated depletion of Ccdc11, a pool of four predesigned synthetic oligonucleotides targeting four unique sequences within the human Ccdc11 gene was obtained from GE Healthcare Dharmacon (SMARTpool ON-TARGETplus, L-016630-02-0005). The individual siRNA sequences were 5′-GUAACAACGCACAGAUUAA-3′, 5′-CCGUACAGCGGGAGGUUAA-3′, 5′-CAUUGAAGAAAGACGAAAU-3′, and 5′-GCUCCGUGUUGAAUUGUUA-3′. In addition, a high potency 21-mer siRNA oligonucleotide (referred to as siRNA#1) targeting a region within a coding strand of human Ccdc11 gene was obtained from Life Technologies (Silencer Select, 4392420, ID s47871, 5′-GAGACGAACAGGACUUGAAtt-3′). For siRNA-mediated depletion of PCM-1, a single ON-TARGETplus oligo (5′-UCA GCU UCG UGA UUC UCA GTT-3′) was used. A Silencer Select Negative Control siRNA (4390844; Life Technologies) was used as control. To deplete Ccdc11, RPE-1 cells were seeded at 20–30% confluence and transfected with 50 nM each siRNA oligonucleotide and Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol. Loss of Ccdc11 protein was observed 72 h after siRNA transfection by immunofluorescence and Western blot. Ciliogenesis was induced at 72 h postdepletion by shifting cells from high (10%) to low (0.5%) serum–containing medium for 24–48 h. Ciliogenesis rescue experiments were performed using an siRNA-resistant GFP-Ccdc11 or Myc-Ccdc11 construct, generated by PCR to introduce noncoding nucleotide changes in the siRNA#1-targeted region (nucleotides 911–929; 5′-AGGGATGAGCAAGATTTAAA-3′). A similar strategy was used to generate the siRNA-resistant truncated version of Ccdc11 covering amino acids 1–333 + 18. PCR products were cloned into pEGFP-C1 vector with unique BspEI and PstI sites, yielding recombinant GFP-siRNA–resistant Ccdc11(FL) or GFP-siRNA–resistant Ccdc11(PM). Each plasmid (2 μg) was transfected into RPE-1 cells 48–72 h after siRNA#1 delivery.
Lentivirus production and cell infection
We obtained two lentivirus-based shRNA constructs targeting human Ccdc11 (shRNA#1: nucleotides 789–810, 5′-CGCACAGATTAAACATGAGAA-3′; and shRNA#2: nucleotides 778–799, 5′-GAAAGTAACAACGCACAGATT-3′), as well as a control shRNA–expressing plasmid, from the RNAi Consortium (TRC) collection available at the Washington University School of Medicine DNA Core. Lentivirus was produced by cotransfection of HEK293T cells with the appropriate transfer and lentiviral helper plasmids (pCMVDR8.74 packaging vector and pMD2.VSVG envelope vector), using the calcium phosphate coprecipitation method. Medium was exchanged 18 h after transfection, and lentiviral supernatant was harvested after another 48 h. The lentiviral supernatants were concentrated 100- to 500-fold by ultracentrifugation at 20°C for 180 min at 50,000 × g. For infection of RPE-1, cells were seeded onto 24-well tissue culture plates the day before infection. The medium was removed the next day and replaced by medium containing lentivirus at a multiplicity of infection value of 1. Virus containing–medium was removed 24 h after infection. To infect hTECs, cells were seeded on Transwell filters in the presence of shRNA-expressing lentivirus, and infected cells were selected with puromycin for 4–6 d until they reached confluence. ALI was established as described, and samples were harvested on the indicated days.
Antibody generation and immunofluorescence microscopy
A polyclonal Ccdc11 antibody was raised in rabbits using a synthetic peptide against human Ccdc11 protein QGAEHHLERIRRSH (amino acids 31–44) and affinity purified using the manufacturer’s methods (GenScript, Piscataway, NJ). This antibody was used for immunofluorescence detection of endogenous Ccdc11 at a dilution of 1:250. In parallel, we generated a polyclonal Ccdc11 antibody from guinea pig. Briefly, recombinant Ccdc11-hexahistidine protein (amino acids 1–250) was expressed in bacteria and purified on a nickel–nitriloacetic acid agarose column following the manufacturer’s protocol (Qiagen, Valencia, CA). We used 1 mg of antigen to inject guinea pigs (Rockland Immunochemicals, Limerick, PA), and antibody was affinity purified from sera using the immobilized antigen. This antibody worked well for detection of Ccdc11 on immunoblots of whole-cell lysates and was used at a dilution of 1:2500. For indirect immunofluorescence, RPE-1 cells were grown on glass coverslips (Electron Microscopy Sciences, Hatfield, PA) coated with 1 mg/ml poly-l-lysine (Sigma-Aldrich). Cultured cells or hTECs were rinsed twice with phosphate-buffered saline (PBS) and fixed in either 100% ice-cold methanol at −20°C for 10 min or 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS at room temperature for 10 min, depending on antigen. For hTEC samples, filters were excised from their plastic supports and cut into quarters to provide multiple equivalent samples for parallel staining. Cells were permeabilized by washing three times with 0.1% Triton X-100 in PBS (PBS-T) and blocked for 1 h at room temperature with PBS-T supplemented with 3% bovine serum albumin (BSA). Cells were incubated with primary antibodies for 1 h at room temperature, followed by three washes for 5 min each in PBS-T. All antibodies were diluted in 0.1% Triton X-100 in PBS. Primary antibodies used in this study were as follows: mouse anti–acetylated tubulin (clone 6-11b-1; 1:10,000; Sigma-Aldrich), mouse anti-centrin (clone 20H5; 1:2000; EMD Millipore, Billerica, MA), rabbit anti-PCM1 (clone H-262, 1:3000; Santa Cruz Biotechnology, Dallas, TX), rabbit anti-Cep290 (1:1000; Abcam, Cambridge, MA), mouse anti-Myc (clone 4A6, 1:500; EMD Millipore), chicken anti-GFP (1:500; Life Technologies), mouse anti-FoxJ1 (1:3000; gift from S. Brody, Washington University School of Medicine), mouse anti–α-tubulin (clone DM1A, 1:4000; Sigma-Aldrich), and rat anti–ZO-1 (R40.76, 1:1000; Santa Cruz Biotechnology). Alexa Fluor dye–conjugated secondary antibodies (Life Sciences) were used at a dilution of 1:500 at room temperature for 1 h. Coverslips were mounted on glass slides using Mowiol mounting medium containing n-propyl gallate (Sigma-Aldrich). Images were captured using a Nikon Eclipse Ti-E inverted confocal microscope equipped with a 60× (1.4 numerical aperture [NA]) or 100× (1.45 NA) Plan Fluor oil immersion objective lens (Nikon, Melville, NY).
Analysis of centriolar satellites and ciliogenesis
To induce primary cilium formation after siRNA-mediated depletion of Ccdc11, cells were arrested in G0 by incubation in DMEM containing 0.5% FBS for 24–48 h. At least 100 cells were scored for the presence or absence of cilia by immunofluorescence in three independent experiments. For localization of Ccdc11 in the absence of microtubules, cells grown on coverslips were incubated in medium containing 0.1 μg/ml nocodazole (Sigma-Aldrich) for 1 h at 37°C. Coverslips were washed three times, incubated with normal growth medium to allow microtubule regrowth, and then fixed at 0, 1, 5, and 30 min after nocodazole treatment, using ice-cold methanol. Centriolar satellite dispersal was quantified by counting the percentage of cells with intact PCM-1 and Cep290 at satellites. In parallel, dispersal was determined by measuring total fluorescence intensity of PCM-1 or Cep290 within a 2-μm circular region of interest (ROI) around the centrosome, using Nikon Elements AR 4.20 Software. Background fluorescence values were measured using an ROI of identical size in the near proximity of each centrosome and subtracted from the centrosomal measurements. More than 150 cells were scored per experiment, and the average values were calculated from three independent experiments. For analysis of ciliogenesis in Ccdc11-depleted hTEC, cells infected with shRNA-expressing lentivirus were fixed before differentiation (pre-ALI) and at 7, 14, and 21 d post-ALI. Samples were stained with antibodies to mark Ccdc11, centrioles, and cilia as described. Primary cilium assembly was determined by counting the percentage of cells that formed a cilium at ALI day 0. The fraction of multiciliated cells was quantified by counting the percentage of cells in the total population that underwent centriologenesis (determined by centrin staining), assembled cilia (anti–acetylated tubulin), and were fated to become multiciliated (Foxj1 staining). The average numbers were calculated by counting 100 cells each from three independent experiments.
Cell extracts, immunoprecipitation, and Western blots
To obtain whole-cell extracts for immunoblotting, cells were washed in PBS and lysed by the addition of SDS sample buffer. Proteins were resolved on 6, 8, 10, 12, or 4–15% gradient SDS–PAGE gels, depending on protein size. For immunoprecipitation of GFP- or Myc-Ccdc11, RPE-1 cells stably expressing GFP-Ccdc11 or Myc-Ccdc11 at near-endogenous levels were grown on twenty 150-mm cell culture dishes to ∼90% confluence, washed in PBS, and trypsinized for 5 min in a 37°C incubator. After centrifugation, the cellular pellet (∼1000-1500 μl packed cell volume) was lysed in ice-cold lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 150 mM NaCl, and 10% glycerol) containing protease inhibitors (Mammalian Protease Arrest; G Biosciences, St. Louis, MO), phosphatase inhibitors (PhosSTOP; Roche, Indianapolis, IN), 10 μg/ml cytochalasin D (Sigma-Aldrich), and 1% NP-40 (Sigma-Aldrich) for 30 min on ice. Lysates were cleared by centrifugation at 3000 × g for 15 min and then at 50,000 × g for 30 min in an OPTIMA LE-80K ultracentrifuge using an SW 41-Ti rotor (Beckman Coulter, Pasadena, CA). Protein concentration of the clarified cellular extract was determined using a Pierce 660-nm Protein Assay kit (Pierce, Rockford, IL). Equal amounts of the clarified cellular extract (∼1 mg/ml) were mixed with 150 μl of affinity resin composed of the following antibodies: 55 μg of goat anti-GFP (Rockland), 55 μg of control goat immunoglobulin G (IgG; Santa Cruz Biotechnology), 20 μg of rabbit anti-Myc (Sigma-Aldrich), or 20 μg of control rabbit IgG (Santa Cruz Biotechnology). All antibodies were covalently coupled to Protein G or Protein A Magnetic Beads via imidoester cross-linker (DMP; Pierce) per manufacturer’s protocol. Samples were incubated for 2 h at 4°C with rotation. The beads were magnetically pelleted and washed five times with ice-cold lysis buffer for 5 min each. Protein complexes were eluted in 240 μl of lysis buffer containing SDS sample buffer and prepared for electrophoresis by boiling at 95°C for 5 min. For silver staining, the Silver Stain Plus kit (Bio-Rad, Hercules, CA) was used following manufacturer’s instructions. Antibodies used for immunoblotting include rabbit anti-GFP (1:2000; Hatch et al., 2010), mouse anti-Myc (clone 4A6, 1:1000; EMD Millipore), anti-Ccdc11 (1:2500), mouse anti–γ-tubulin (clone GTU88, 1:5000; Sigma-Aldrich), rabbit anti-PCM-1 (clone H-262, 1:1000; Santa Cruz Biotechnology), rabbit anti-Cep290 (1:1000; Bethyl Laboratories, Montgomery, TX), and mouse anti-actin (clone AC-40, 1:3000; Sigma-Aldrich). Secondary antibodies were horseradish peroxidase (HRP)–conjugated mouse anti-rabbit HRP (1:10,000; Sigma-Aldrich), donkey anti-mouse HRP (1:10,000; Jackson ImmunoResearch, West Grove, PA), and donkey anti–guinea pig HRP (1:10,000; Jackson ImmunoResearch). Immunoblots were imaged using WesternSure PREMIUM chemiluminescent substrate (LI-COR, Lincoln, NE) and imaged on a digital C-DiGit Blot Scanner (LI-COR).
MudPIT analysis of immunoprecipitated complexes were performed from protein pellets dissolved in buffer (8 M urea, 100 mM Tris, pH 8.5), reduced with Tris[2-carboxyethyl]-phosphine hydrochloride, and alklyated with chloroacetamide. After dilution of urea to 2 M, proteins were digested with trypsin. Digested peptides were analyzed by multidimensional chromatography coupled with tandem mass spectrometry using an LTQ-Orbitrap mass spectrometer. Multidimensional chromatography was performed online with three salt steps (MacCoss et al., 2002). Tandem mass spectra were collected in a data-dependent manner with up to 10 ms2 scans performed for each initial scan (m/z = 300–2000). The search program Prolucid was used to match data to a human protein database. Peptide identifications were filtered using the DTASelect program (Tabb et al., 2002).
Analysis of Ccdc11 in Xenopus
All Xenopus experiments were performed using previously described techniques (Werner and Mitchell, 2013). Briefly, Xenopus embryos were acquired by in vitro fertilization using standard protocols (Sive et al., 1998) approved by the Northwestern University (Evanston, IL) Institutional Animal Care and Use Committee; Xenopus CCDC11 (Xl.12748) was subcloned into pCS2-RFP vector using EcoRI (forward primer CGAATTCATGCTGTATAGTCAAAGAAGCCG) and XhoI (reverse primer GCCTCGAGTTGTGGGAGCCAGTCCTTAG). Fluorescence imaging was performed on a Nikon A1R confocal microscope using a 60× objective (1.4 NA). Embryos were injected with 50–200 pg of mRNA (centrin4-RFP, centrin4-GFP, GFP-CCDC11, or CLAMP-GFP) and/or 10–20 ng of morpholino oligo (MO). A CCDC11-specific translation (ATG)–blocking MO (GACGGCTTCTTTGACTATACAGCAT) and a standard control MO (Gene-Tools, Philomath, OR) were used to generate morphant embryos (Supplemental Figure S5A). Cilia imaging was performed using mouse anti–acetylated tubulin to mark cilia and phalloidin-647 (Life Sciences) to mark cell outlines. Fluid flow was recorded using a Leica M165FC microscope to visualize fluorescent microbeads as previously described (Werner et al., 2011; Werner and Mitchell, 2013).
The reversible deciliation of embryos was performed according to previously reported methods (Werner and Mitchell, 2013). Briefly, stage 28 embryos were placed in the deciliation buffer (0.1× Marc’s modified Ringer’s buffer [MMR] supplied with 75 mM of CaCl2 and 0.02% NP-40) for ∼3 min until the surface flow ceased. The embryos were then immediately transferred into 0.1× MMR solution and quickly washed twice. Washed embryos were fixed in PFA at different time points after deciliation. Embryos were stained with anti–acetylated tubulin antibody to observe ciliary regrowth after deciliation. Analysis of ciliary orientation was scored as described previously (Mitchell et al., 2007, 2009; Park et al., 2008). Specifically, rootlet orientation (GFP-CLAMP relative to centrin4-RFP) was scored manually by measuring the angle of orientation of the rootlets relative to the anterior–posterior axis of the embryo, using Nikon Elements Software. Oriana 2.0 Software was used for graphical representation of the data sets and to calculate mean vector length and circular SD.
Analysis of Ccdc11 in zebrafish
Wild-type (AB*) Danio rerio and mutant strains carrying mutations in CCDC11 (CCDC11−/−) were raised and maintained as described previously (Solnica-Krezel et al., 1996). Embryos were gathered from natural breeding events, grown at 28°C, and staged according to their morphology (Kimmel et al., 1995). TALEN gene-targeting constructs were designed against the BsmAI cleavage site within exon 2 of CCDC11 and engineered by the Hope Center Transgenic Vectors Core at Washington University. From 50 to 100 pg of TALEN mRNA pairs was injected into one-cell-stage WT embryos, which were then raised to adulthood. Sequencing identified a number of independent strains carrying deletions in the targeted site. Two specific strains used in this study carried either a 5– or a 2–base pair deletion, resulting in a frameshift and premature stop codon. Strains carrying these mutant alleles were outcrossed to WT and heterozygous progeny raised and maintained under standard conditions. Genotyping was performed by PCR amplification of a 576–base pair fragment using the forward primer 5′-CAGGCCTACTATATCATAACAGACAGGC-3′ and reverse primer 5′-AATGCCCATCTGGTACTGCTCTAGTGCATC-3′. PCR products were digested with BsmAI to identify a cut 388–base pair (WT) fragment and uncut 576–base pair mutant allele. Homozygous CCDC11−/− zebrafish were obtained by crossing CCDC11−/+ heterozygous animals and used for the various experiments. For RT-PCR, total RNA was isolated from wild-type and CCDC11−/− embryos at the 24 h postfertilization (hpf) stage using TRIZOL according to standard protocols. cDNA was reverse transcribed using the iScript cDNA Synthesis Kit according to the manufacturer’s instructions (Bio-Rad). The following primers were used for PCR analysis: CCDC11 exons 3 and 4, 5′-GTGAACTGCTGGAGTCAGAGG-3′ and 3′- CACAGATGGACATGGCTGAG-5′ (422–base pair amplicon) and CCDC11 exons 4 and 5, 5′-ATAAAGCGCAGACAGGATCAG-3′ and 3′-CACAGGAGAAAGATGAGAAGCA-5′ (473 base pairs). As a normalization standard, we used β-actin, 5′-CGAGCTGTCTTCCCATCCA-3′ and 3′-TCACCAACGTAGCTGTCTTTCTG-5′ (115 base pairs). As an additional control against genomic DNA contamination, all primer pairs were designed to span introns.
Wild-type, CCDC11−/+, and CCDC11−/− embryos were processed for whole-mount in situ hybridization as previously described (Thisse and Thisse, 2008). Briefly, embryos were fixed at the appropriate developmental stage in 4% PFA overnight at 4°C. Embryos were manually dechorionated the next day, dehydrated in methanol, and then incubated for 16 h at 65°C with the following digoxigenin (DIG)-labeled probes: cmlc2 (48 hpf, ZFIN ID, ZDB-GENE-991019-3) and southpaw (19 hpf, ZDB-GENE-030219-1). Embryos were washed and incubated in 1% Roche Blocking Reagent (Roche) and then with anti-DIG alkaline phosphatase diluted to 1:2000 overnight at 4°C. Embryos were equilibrated in Tris buffer, stained with BM Purple AP substrate (Roche), and postfixed in 4% PFA once reaction was completed. Imaging and scoring of probe localization were performed using a Nikon SMZ1500 dissecting microscope.
For immunofluorescence staining and analysis of ciliary length, embryos were fixed in 4% PFA in PBS at the eight-somite stage (Kupffer’s vesicle) and 24 hpf (pronephros). Embryos were permeabilized in 0.1% Triton in PBS for 30–60 min at room temperature, incubated for 2 h in blocking buffer (2 mg/ml BSA, 1% dimethyl sulfoxide, 2% normal goat serum, and 0.1% Tween in PBS), and then incubated overnight with anti–acetylated α-tubulin (clone 6-11b-1; 1:4000). Images were captured using a Nikon Eclipse Ti-E confocal microscope equipped with Plan Apo 20× objective. Quantification of cilia abundance and length was performed using NIS-Elements AR 4.20 software (Nikon).
Statistical analysis
Data from fluorescence intensity measurements, centriolar satellite dispersal, and ciliogenesis assays were examined by one-way analysis of variance (ANOVA) for significance (p < 0.05). Before ANOVA, the data were subjected to tests for normality (Kolmogorov–Smirnov test) and homogeneity of variance (Levene’s test). When significant differences were detected, treatment means were further analyzed by Tukey’s honest significant difference. Statistical analyses were performed using Statistica software (StatSoft, Tulsa, OK). Data are reported as mean ± SEM.
Supplementary Material
Acknowledgments
We thank S. Brody (Washington University, St. Louis, MO) for providing human tracheobronchial epithelial progenitor cells and S. Dutcher (Washington University) for careful reading of the manuscript. We also thank L. Solnica-Krezel and K. Monk (Washington University) for help with zebrafish experiments and J. Wang (Washington University) for help with some molecular biology experiments. We give a special thanks to T. Stearns (Stanford University, Stanford, CA) for helpful discussions regarding this project. This study was supported by funding from the Washington University Renal Division (93368) and the National Heart, Lung and Blood Institute (R01HL126239) to M.R.M.; from the National Institute of General Medical Sciences (R01GM0899) to B.J.M.; and from the National Center for Research Resources (5 P4 1RR011823) and National Institute of General Medical Sciences (8 P41 GM103533) to J.R.Y., III.
Abbreviations used:
- CCDC
coiled-coil domain containing
- Cep
centrosomal protein
- HTEC
human tracheal epithelial cell
- KV
Kupffer’s vesicle
- PCD
primary cilia dyskinesia
- PCM
pericentriolar material.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0474) on November 4, 2015.
REFERENCES
- Anderson RG, Brenner RM. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am J Hum Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu D, Roy S. Left-right asymmetry: cilia stir up new surprises in the node. Open Biol. 2013;3:130052. doi: 10.1098/rsob.130052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner M. Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation. 2007;115:2793–2795. doi: 10.1161/CIRCULATIONAHA.107.699256. [DOI] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Shah JV. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen ER. Centriole and basal body formation during ciliogenesis revisited. Biol Cell. 1991;72:31–38. doi: 10.1016/0248-4900(91)90075-x. [DOI] [PubMed] [Google Scholar]
- Duldulao NA, Li J, Sun Z. Cilia in cell signaling and human disorders. Protein Cell. 2010;1:726–736. doi: 10.1007/s13238-010-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Feather SA, Winyard PJ, Dodd S, Woolf AS. Oral-facial-digital syndrome type 1 is another dominant polycystic kidney disease: clinical, radiological and histopathological features of a new kindred. Nephrol Dial Transplant. 1997;12:1354–1361. doi: 10.1093/ndt/12.7.1354. [DOI] [PubMed] [Google Scholar]
- Ferkol TW, Leigh MW. Ciliopathies: the central role of cilia in a spectrum of pediatric disorders. J Pediatrics. 2012;160:366–371. doi: 10.1016/j.jpeds.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Sante J, Elliott S, Stearns T. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. J Cell Sci. 2014;127:4128–4133. doi: 10.1242/jcs.157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. Acute versus chronic loss of mammalian Azi1/Cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9:e1003928. doi: 10.1371/journal.pgen.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JM, Kim SK, Abitua PB, Park TJ, Herrington ER, Kitayama A, Grow MW, Ueno N, Wallingford JB. Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev Biol. 2007;312:115–130. doi: 10.1016/j.ydbio.2007.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PloS One. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Brody SL, Ferkol TW. Picking up speed: advances in the genetics of primary ciliary dyskinesia. Pediatric Res. 2014;75:158–164. doi: 10.1038/pr.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Nath A, Wasserman MG, Huang T, Brody SL. Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. Am J Respir Cell Mol Biol. 2013;49:341–347. doi: 10.1165/rcmb.2013-0046TE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MS, Baig SM, Neumann S, Nurnberg G, Farooq M, Ahmad I, Alef T, Hennies HC, Technau M, Altmuller J, et al. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet. 2012;90:871–878. doi: 10.1016/j.ajhg.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe KM, Thiberge SY, Bisher ME, Burdine RD. Imaging cilia in zebrafish. Methods Cell Biol. 2010;97:415–435. doi: 10.1016/S0091-679X(10)97022-2. [DOI] [PubMed] [Google Scholar]
- Katsanis N. The oligogenic properties of Bardet-Biedl syndrome. Hum Mol Genet. 2004;13:R65–R71. doi: 10.1093/hmg/ddh092. (Spec No 1) [DOI] [PubMed] [Google Scholar]
- Kennedy MP, Ostrowski LE. Primary ciliary dyskinesia and upper airway diseases. Curr Allergy Asthma Rep. 2006;6:513–517. doi: 10.1007/s11882-006-0030-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klinger M, Wang W, Kuhns S, Barenz F, Drager-Meurer S, Pereira G, Gruss OJ. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Tonthat V, Wu B, Sutterlin C. Par6 alpha interacts with the dynactin subunit p150 Glued and is a critical regulator of centrosomal protein recruitment. Mol Biol Cell. 2010;21:3376–3385. doi: 10.1091/mbc.E10-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki K, Bassi MT, Kosaki R, Lewin M, Belmont J, Schauer G, Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet. 1999a;64:712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999b;82:70–76. doi: 10.1002/(sici)1096-8628(19990101)82:1<70::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Tsukita S. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkowiak M, Zietkiewicz E, Witt M. Recent advances in primary ciliary dyskinesia genetics. J Med Genet. 2015;52:1–9. doi: 10.1136/jmedgenet-2014-102755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Lopes CA, Prosser SL, Romio L, Hirst RA, O’Callaghan C, Woolf AS, Fry AM. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol. 2010;191:331–346. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol. 2009;19:924–929. doi: 10.1016/j.cub.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, Molinari L, Niesh SR, Jefferies JL, Craigen WJ, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- Nachury MV. How do cilia organize signalling cascades. Philos Trans R Soc Lond B Biol Sci. 2014:369. doi: 10.1098/rstb.2013.0465. 20130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012;125:2232–2242. doi: 10.1161/CIRCULATIONAHA.111.079780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V, Hjeij R, Vij S, Loges NT, Wallmeier J, Koerner-Rettberg C, Werner C, Thamilselvam SK, Boey A, Choksi SP, et al. Mutations in CCDC11, which encodes a coiled-coil containing ciliary protein, causes situs inversus due to dysmotility of monocilia in the left-right organizer. Hum Mutat. 2015;36:307–318. doi: 10.1002/humu.22738. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Norris DP. Cilia, calcium and the basis of left-right asymmetry. BMC Biol. 2012;10:102. doi: 10.1186/1741-7007-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich H, Haffner K, Kispert A, Volkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- Papon JF, Perrault I, Coste A, Louis B, Gerard X, Hanein S, Fares-Taie L, Gerber S, Defoort-Dhellemmes S, Vojtek AM, et al. Abnormal respiratory cilia in non-syndromic Leber congenital amaurosis with CEP290 mutations. J Med Genet. 2010;47:829–834. doi: 10.1136/jmg.2010.077883. [DOI] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perles Z, Cinnamon Y, Ta-Shma A, Shaag A, Einbinder T, Rein AJ, Elpeleg O. A human laterality disorder associated with recessive CCDC11 mutation. J Med Genet. 2012;49:386–390. doi: 10.1136/jmedgenet-2011-100457. [DOI] [PubMed] [Google Scholar]
- Powles-Glover N. Cilia and ciliopathies: classic examples linking phenotype and genotype-an overview. Reprod Toxicol. 2014;48:98–105. doi: 10.1016/j.reprotox.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Ross AJ, Dailey LA, Brighton LE, Devlin RB. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol. 2007;37:169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- Saal S, Faivre L, Aral B, Gigot N, Toutain A, Van Maldergem L, Destree A, Maystadt I, Cosyns JP, Jouk PS, et al. Renal insufficiency, a frequent complication with age in oral-facial-digital syndrome type I. Clin Genet. 2010;77:258–265. doi: 10.1111/j.1399-0004.2009.01290.x. [DOI] [PubMed] [Google Scholar]
- Serluca FC, Xu B, Okabe N, Baker K, Lin SY, Sullivan-Brown J, Konieczkowski DJ, Jaffe KM, Bradner JM, Fishman MC, Burdine RD. Mutations in zebrafish leucine-rich repeat-containing six-like affect cilia motility and result in pronephric cysts, but have variable effects on left-right patterning. Development. 2009;136:1621–1631. doi: 10.1242/dev.020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. The Early Development of Xenopus laevis: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory; 1998. [Google Scholar]
- Solnica-Krezel L, Stemple DL, Mountcastle-Shah E, Rangini Z, Neuhauss SC, Malicki J, Schier AF, Stainier DY, Zwartkruis F, Abdelilah S, Driever W. Mutations affecting cell fates and cellular rearrangements during gastrulation in zebrafish. Development. 1996;123:67–80. doi: 10.1242/dev.123.1.67. [DOI] [PubMed] [Google Scholar]
- Staples CJ, Myers KN, Beveridge RD, Patil AA, Lee AJ, Swanton C, Howell M, Boulton SJ, Collis SJ. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012;125:4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- Steinman RM. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am J Anat. 1968;122:19–55. doi: 10.1002/aja.1001220103. [DOI] [PubMed] [Google Scholar]
- Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Wert SE, Costa RH, Kimura S, Whitsett JA. HNF-3/forkhead homologue-4 (HFH-4) is expressed in ciliated epithelial cells in the developing mouse lung. J Histochem Cytochem. 1999;47:823–832. doi: 10.1177/002215549904700612. [DOI] [PubMed] [Google Scholar]
- Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia dysfunction in lung disease. Annu Rev Physiol. 2015;77:379–406. doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere MA, Mailand N, Bekker-Jensen S. Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci. 2015;72:11–23. doi: 10.1007/s00018-014-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Rosti RO, Gibbs E, Gleeson JG. Primary cilia in neurodevelopmental disorders. Nat Rev Neurol. 2014;10:27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, Towbin J, Belmont JW. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74:93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner C, Onnebrink JG, Omran H. Diagnosis and management of primary ciliary dyskinesia. Cilia. 2015;4:2. doi: 10.1186/s13630-014-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J Cell Biol. 2011;195:19–26. doi: 10.1083/jcb.201106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Using Xenopus skin to study cilia development and function. Methods Enzymol. 2013;525:191–217. doi: 10.1016/B978-0-12-397944-5.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway G, Parry DA, Johnson CA. The role of primary cilia in the development and disease of the retina. Organogenesis. 2014;10:69–85. doi: 10.4161/org.26710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014;30:10–17. doi: 10.1016/j.tig.2013.09.001. [DOI] [PubMed] [Google Scholar]
- You Y, Brody SL. Culture and differentiation of mouse tracheal epithelial cells. Methods Mol Biol. 2013;945:123–143. doi: 10.1007/978-1-62703-125-7_9. [DOI] [PubMed] [Google Scholar]
- You Y, Huang T, Richer EJ, Schmidt JE, Zabner J, Borok Z, Brody SL. Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2004;286:L650–L657. doi: 10.1152/ajplung.00170.2003. [DOI] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.