Rab13 is necessary for insulin-regulated GLUT4-vesicle exocytosis in muscle. Biochemical and imaging analyses provide evidence that activated Rab13 engages a scaffold protein MICAL-L2 to form a complex with Rab13 and α-actinin-4. Through GLUT4 interaction with α-actinin-4, GLUT4 vesicles are recruited to the muscle plasma membrane.
Abstract
Insulin promotes glucose uptake into skeletal muscle through recruitment of glucose transporter 4 (GLUT4) to the plasma membrane. Rab GTPases are molecular switches mobilizing intracellular vesicles, and Rab13 is necessary for insulin-regulated GLUT4–vesicle exocytic translocation in muscle cells. We show that Rab13 engages the scaffold protein MICAL-L2 in this process. RNA interference–mediated knockdown of MICAL-L2 or truncated MICAL-L2 (MICAL-L2-CT) impaired insulin-stimulated GLUT4 translocation. Insulin increased Rab13 binding to MICAL-L2, assessed by pull down and colocalization under confocal fluorescence and structured illumination microscopies. Association was also visualized at the cell periphery using TIRF microscopy. Insulin further increased binding of MICAL-L2 to α-actinin-4 (ACTN4), a protein involved in GLUT4 translocation. Rab13, MICAL-L2, and ACTN4 formed an insulin-dependent complex assessed by pull down and confocal fluorescence imaging. Of note, GLUT4 associated with the complex in response to insulin, requiring the ACTN4-binding domain in MICAL-L2. This was demonstrated by pull down with distinct fragments of MICAL-L2 and confocal and structured illumination microscopies. Finally, expression of MICAL-L2-CT abrogated the insulin-dependent colocalization of Rab13 with ACTN4 or Rab13 with GLUT4. Our findings suggest that MICAL-L2 is an effector of insulin-activated Rab13, which links to GLUT4 through ACTN4, localizing GLUT4 vesicles at the muscle cell periphery to enable their fusion with the membrane.
INTRODUCTION
Skeletal muscle is the primary tissue responsible for dietary glucose uptake. In muscle and fat cells, insulin promotes the exocytic traffic of intracellular membranes containing GLUT4 glucose transporters to elicit a rapid increase in glucose uptake. Given that the insulin receptor is located at the cell membrane, whereas the majority of GLUT4 is found in perinuclear and cytosolic endomembranes, insulin-derived signals must affect structural and dynamic elements participating in GLUT4 vesicle mobilization, docking, and fusion. Accordingly, each of these steps is regulated in response to the hormone (Hou and Pessin, 2007; Chiu et al., 2011; Foley et al., 2011; Rowland et al., 2011; Stöckli et al., 2011; Bogan, 2012; Leto and Saltiel, 2012; Klip et al., 2014). The pivotal discovery that insulin signaling downstream of Akt (protein kinase B) leads to phosphorylation and thereby inactivation of the Rab GTPase–activating protein (Rab-GAP) Akt substrate of 160 kDa/TBC1 domain family member 4 (AS160; Sano et al., 2003; Karlsson et al., 2005; Thong et al., 2007) suggested that insulin signal transduction converges on Rab-GTPases. This is an important realization, given that Rab GTPases are considered quintessential molecular switches conferring directionality to vesicle movement within cells (Martinez and Goud, 1998; Zerial and McBride, 2001; Lindsay et al., 2015). Rab GTPases cycle between “on” and “off” states determined by binding GTP or GDP, respectively. In muscle cells, Rab8A and Rab13 become activated upon insulin stimulation (Sun et al., 2010), and we hypothesize that each of these GTPases recruits effectors for downstream regulation of vesicle traffic. Indeed, both are required for translocation of GLUT4-containing vesicles from intracellular compartments to the cell surface (Ishikura and Klip, 2008; Sun et al., 2010), whereas the same function in adipocytes is ascribed to Rab10 (Sano et al., 2007; Chen et al., 2012; Sadacca et al., 2013).
Identifying the effectors of these Rab GTPases is of paramount importance to understanding the transmission of information by signaling molecules to mechanical elements effectively mobilizing GLUT4 vesicles. In this context, we recently found that the processive, dimeric motor myosin Va (MyoVa) participates in GLUT4 vesicle traffic toward the cortex of L6 muscle cells (Klip et al., 2014; Sun et al., 2014). Of note, engagement of MyoVa was downstream of Rab8A activation in muscle cells and downstream of Rab10 in adipocytes (Chen et al., 2012; Sun et al., 2014).
On the other hand, the effect(s) of Rab13 on insulin-dependent GLUT4 vesicle traffic remain unknown. We previously reported that Rab13 localizes to endomembranes as well as to the muscle cell periphery, in contrast to the more perinuclear/endosomal localization of Rab8A. A similar segregation of cellular domains populated by Rabs 8A and 13 occurs in polarized and other nonpolarized cells (Zahraoui et al., 1994). This differential subcellular distribution, along with the slightly earlier activation of Rab8A compared with Rab13 in response to insulin (peaking at 2 and 5 min, respectively; Sun et al., 2010), raises the possibility that Rab8A functions to mobilize GLUT4 vesicles toward the periphery, where Rab13 might have a preferential action.
The main known interactor of Rab13-GTP is junctional Rab13-binding protein (JRAB), a protein of 110 kDa also known as molecule interacting with CasL–like 2 (MICAL-L2; Terai et al., 2006). Through its middle domain, MICAL-L2 can bind the actin-linker protein α-actinin-4 (ACTN4; Nakatsuji et al., 2008). Of note, this peripherally located actin-binding protein participates in GLUT4 vesicle association to cortical actin (Talior-Volodarsky et al., 2008). Given the participation of Rab13 in insulin-dependent GLUT4 traffic and the peripheral localization of this GTPase in muscle cells, along with its known interaction with MICAL-L2 in tight junction assembly (Marzesco et al., 2002) and neurite outgrowth (Sakane et al., 2010), we hypothesized that MICAL-L2 might be the Rab13 effector required for GLUT4 translocation.
Here we show that MICAL-L2 and Rab13 colocalize and interact in muscle cells and that their interaction is promoted by insulin stimulation. Silencing the expression of MICAL-L2 abrogated GLUT4 insertion into the membrane, as did overexpression of a truncated fragment of MICAL-L2 containing the Rab-binding domain but lacking its binding sites to actin and ACTN4. We further show that ACTN4 forms a complex with Rab13 and MICAL-L2 that is regulated by insulin and that ACTN4 binding to MICAL-L2 is required for association of GLUT4 with this complex. At the submembrane region, GLUT4-containing vesicles colocalize with Rab13 and MICAL-L2. We propose that insulin-induced activation of Rab13 downstream of AS160 phosphorylation leads to GLUT4 vesicle association with Rab13, MICAL-L2, and ACTN4 in preparation for GLUT4 vesicle interaction with the plasma membrane.
RESULTS
MICAL-L2 pulls down Rab13-GTP, and this response is stimulated by insulin
JRAB/MICAL-L2 belongs to the MICAL family of proteins, which comprises five members (MICAL-1, -2, and -3 and MICAL-like proteins MICAL-L1 and L2; Rahajeng et al., 2010). All MICAL family members contain a CH domain (amino acids [aa] 3–102 in MICAL-L2) and a LIM domain (aa 187–241 in MICAL-L2) that link to the actin cytoskeleton. Whereas MICAL-1, -2, and -3 contain a FAD domain that endows them with flavoprotein monooxygenase catalytic activity, MICAL-L1 and MICAL-L2 lack this domain. The latter two proteins exhibit only 30% amino acid identity. MICAL family proteins have a coiled-coil domain at the C-terminus that is required for interaction with Rab GTPases (Terai et al., 2006; Rahajeng et al., 2010). In MICAL-L2, the Rab-binding domain is within aa 806–1009 (Nakatsuji et al., 2008).
To test whether MICAL-L2 is an effector of Rab13, we first performed pull-down assays using glutathione S-transferase (GST)–tagged C-terminal fragment of MICAL-L2 (GST-MICAL-L2-CT) encoding the Rab-binding domain (aa 806–1009) and lysates from insulin-responsive and easily transfectable CHO cells stably expressing the human insulin receptor (CHO-IR), which were transfected with green fluorescent protein (GFP) chimeras of either the active GTP-locked form or its inactive GDP-locked form. GST-MICAL-L2-CT strongly pulled down the GTP-locked version of Rab13 but not the GDP-locked form (Supplemental Figure S1A). This result indicates that engaging MICAL-L2 is an effector function of activated Rab13. Next, because Rab13 and Rab8A are activated in response to insulin and, along with Rab10, participate in GLUT4 traffic, we tested whether insulin stimulation would promote binding of these other Rab GTPases to MICAL-L2. To this end, we expressed GFP-Rab13, GFP-Rab8A, or GFP-Rab10 in CHO-IR cells, stimulated the cells with insulin, and used GST-MICAL-L2-CT in pull-down assays with the corresponding cell lysates. GST-MICAL-L2-CT bound each of these Rab GTPases from lysates of unstimulated cells (basal state). However, insulin increased selectively the binding of GFP-Rab13 to GST-MICAL-L2-CT, whereas no insulin-dependent gain was observed for GFP-Rab8A or GFP-Rab10 (Supplemental Figure S1B). These results indicate that upon insulin stimulation, Rab13 is activated and capable of engaging with MICAL-L2. The lack of increase in Rab8A binding to MICAL-L2-CT was somewhat surprising, since in muscle cells, insulin activates both Rab13 and Rab8A. Hence MICAL-L2 may not be a major effector of Rab8A, and instead it may act through other effectors, such as MyoVa, as it does in insulin-stimulated muscle cells (Sun et al., 2014). With regard to Rab10, the lack of effect of insulin stimulation on its interaction with MICAL-L2 is consistent with the absence of detectable activation of this GTPase by insulin in muscle cells. Here we focus on the interaction between Rab13 and MICAL-L2.
Rab13 colocalizes with MICAL-L2 in muscle cells
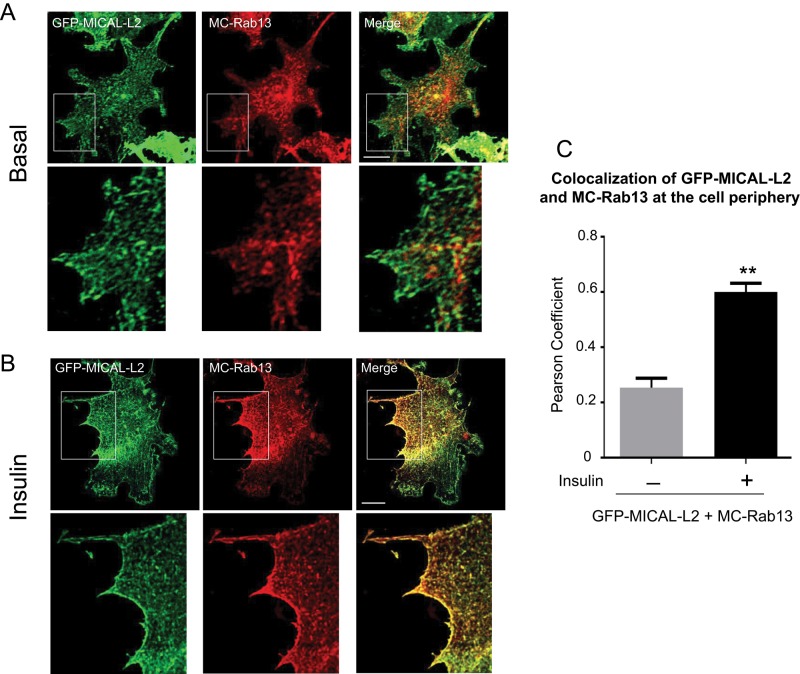
To examine the effect of insulin on the relationship of Rab13 with MICAL-L2 in muscle cells, we transfected mCherry-Rab13 (MC-Rab13) and GFP-MICAL-L2 into L6 myoblasts and determined their degree of colocalization using spinning-disk confocal fluorescence microscopy. As shown in Figure 1A, GFP-MICAL-L2 distributed along cytoplasmic filament-like structures, whereas MC-Rab13 showed a more punctate cytoplasmic distribution and also concentrated at a perinuclear pole. Of note, there was very limited colocalization of these two proteins in these unstimulated (basal) cells. Insulin stimulation caused a sharp redistribution of GFP-MICAL-L2 and MC-Rab13 that intensely outlined zones of the cell periphery (Figure 1B) and also caused colocalization of the two proteins in the cytosolic punctate structures and at the periphery. A significant Pearson coefficient of colocalization for GFP-MICAL-L2 with MC-Rab13 was obtained only in the insulin-treated cells (Figure 1C).
FIGURE 1:
Insulin increases colocalization of Rab13 with MICAL-L2 in L6 myoblasts. L6 cells were cotransfected with GFP-MICAL-L2 and MC-Rab13 for 24 h, serum starved, stimulated with insulin or not, and then processed for spinning-disk confocal microscopy. Representative collapsed optical z-stack images of GFP-MICAL-L2 (green) and MC-Rab13 (red) (>25 cells/condition per experiment). Excerpts are magnifications of the outlined region of interest. (A) Basal, (B) insulin stimulated. (C) Pearson coefficient of colocalization of MC-Rab13 to GFP-MICAL-L2 (mean ± SE, **p < 0.01).
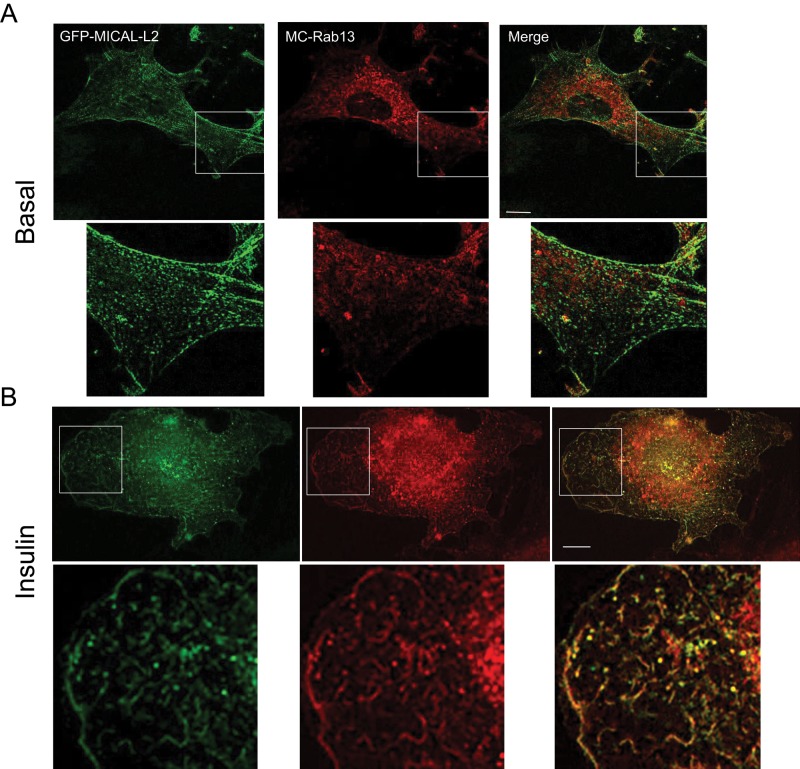
To enhance the resolution of the colocalization of GFP-MICAL-L2 with MC-Rab13, in a small number of experiments, we used structured illumination microscopy (SIM) to obtain high-resolution images of muscle cells expressing the two proteins. By this approach, the filamentous distribution of GFP-MICAL-L2 and the punctate distribution of MC-Rab13 were clearly apparent (Figure 2). The two proteins showed little colocalization in the basal state (Figure 2A), but insulin promoted evident colocalization especially toward the cell periphery (Figure 2B). This distribution is consistent with the recently reported binding of activated Rab13 to MICAL-L2 at the perimeter of HeLa cells, which contributes to the dynamic remodeling of the leading edge (Ioannou et al., 2015).
FIGURE 2:
SIM reveals detailed colocalization of MICAL-L2 with Rab13 in response to insulin. L6 cells were cotransfected with GFP-MICAL-L2 and MC-Rab13 overnight and then replated onto coverslips for 24 h, serum starved, stimulated with insulin or not, and processed for SIM. Representative single confocal planes of GFP-MICAL-L2 (green) and MC-Rab13 (red) (>10 cells/condition). (A) Basal, (B) insulin stimulated. Magnified excerpts are also shown. Scale bars, 10 μm.
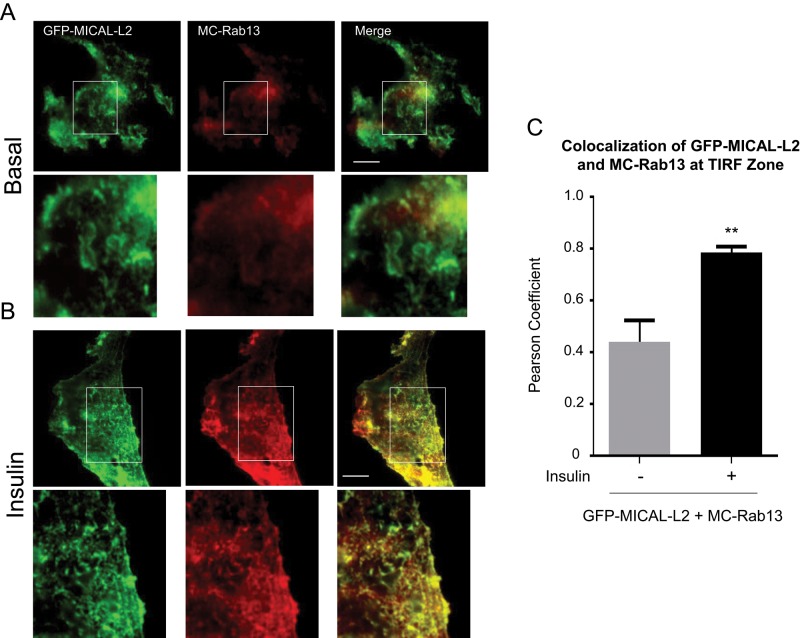
To examine in greater detail the relationship between GFP-MICAL-L2 and MC-Rab13 in the vicinity of the plasma membrane, we imaged the two proteins in the zone illuminated by total internal reflection fluorescence (TIRF) microscopy (Figure 3A). On illumination of 100 nm beneath the membrane of unstimulated cells, GFP-MICAL-L2 was clearly detected and showed a diffuse distribution, whereas MC-Rab13 was virtually absent from this region, and there was no colocalization of the two proteins. Insulin stimulation increased the organization of GFP-MICAL-L2 along distinct filamentous structures and also augmented the abundance of MC-Rab13 in the TIRF-imaged zone (Figure 3B). Moreover, GFP-MICAL-L2 and MC-Rab13 showed sharp colocalization within the TIRF-imaged zone of insulin-stimulated cells. These results corroborate that MC-Rab13 redistributes toward the cell periphery in response to insulin, where it colocalizes and possibly interacts with GFP-MICAL-L2. There was a significant Pearson coefficient of colocalization of GFP-MICAL-L2 with MC-Rab13 in the TIRF zone (Figure 3C).
FIGURE 3:
Insulin promotes colocalization of MICAL-L2 and Rab13 at the TIRF zone. L6 cells cotransfected with GFP-MICAL-L2 and MC-Rab13 were replated onto coverslips for 24 h, serum deprived, stimulated with insulin or not, and examined with an Olympus 1XR1 TIRF microscope. TIRF images of GFP-MICAL-L2 (green) and MC-Rab13 (red; >25 cells/condition). Scale bars, 10 μm. Excerpts show magnified regions. (A) Basal, (B) insulin stimulated. (C) Pearson coefficient of colocalization of MC-Rab13 to GFP-MICAL-L2 at the TIRF zone (mean ± SE, **p < 0.01).
MICAL-L2 is involved in GLUT4 translocation
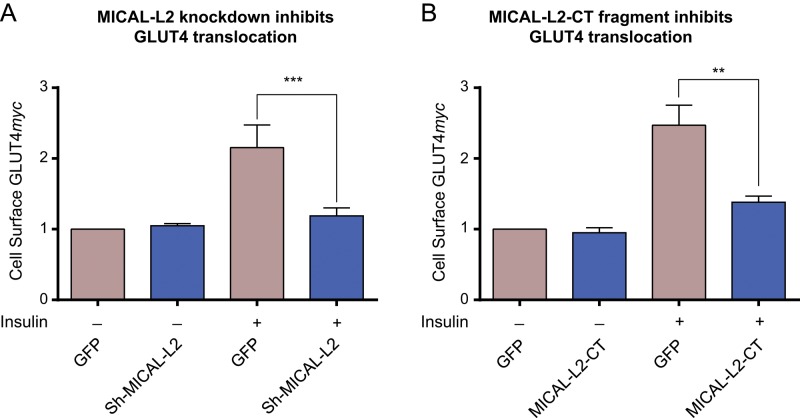
We hypothesized that MICAL-L2 is required for insulin-induced GLUT4 translocation in muscle cells. We first verified that MICAL-L2 is endogenously expressed in L6 and C2C12 muscle cell lines by using reverse transcription (RT)-PCR and to substantial levels in L6-GLUT4myc cells by quantitative PCR (Supplemental Figure S2). We then examined whether MICAL-L2 is required for GLUT4 translocation in L6-GLUT4myc myoblasts. Expression of MICAL-L2 was silenced with short hairpin RNA (shRNA) interference to MICAL-L2 (sh-MICAL-L2). This construct in pGIPZ-GFP (also encoding a GFP cDNA) was transiently transfected into L6-GLUT4myc myoblasts, and an unrelated shRNA sequence in pGIPZ-GFP was used as control. Transfected cells were identified by their GFP fluorescence, and surface GLUT4 was detected via its myc epitope using confocal fluorescence microscopy. The assay involves detection in nonpermeabilized cells, in which the exofacially facing myc epitope would only be exposed to the antibody in the medium. As shown in Figure 4A, insulin elicited a gain in cell-surface GLUT4myc levels, and this response was markedly diminished in cells expressing sh-MICAL-L2 compared with controls. In contrast, sh-MICAL-L2 expression did not affect the basal level of surface GLUT4myc.
FIGURE 4:
Insulin-dependent GLUT4 translocation is prevented by silencing MICAL-L2 or expressing MICAL-L2-CT. (A) L6-GLUT4myc cells were transfected with GFP-coexpressing vectors containing shRNA interference to rat MICAL-L2 (sh-MICAL-L2) or unrelated shRNA. Cells were replated on coverslips for 48 h, serum starved, and stimulated with insulin (15 min) or not. Surface GLUT4myc was detected with anti-myc and Alexa 555–secondary antibodies (red), imaged by confocal microscopy, and quantified as in Materials and Methods. Fold changes relative to shRNA control from three independent experiments, >25 cells/experiment (mean ± SE, ***p < 0.001 for n = 3). (B) L6-GLUT4myc cells transfected with GFP-MICAL-L2-CT or GFP were stimulated with insulin, and surface GLUT4myc was detected as in A. Results from four experiments, >25 cells/experiment (mean ± SE, **p < 0.01).
As a second strategy to test the participation of MICAL-L2 in GLUT4 translocation, we transfected into L6-GLUT4myc myoblasts the truncated fragment MICAL-L2-CT (aa 806–1009). This is the same fragment sequence linked to GST shown in Supplemental Figure S1 to pull down activated Rab13 but now subcloned in a mammalian expression vector to create a chimera with GFP. GFP-MICAL-L2-CT lacks the CH and LIM domains that link MICAL-L2 to actin filaments, and so it is expected to bind endogenous Rab13 but not allow interaction with either actin or ACTN4. Hence the transfected GFP-MICAL-L2-CT would act essentially as a Rab13 scavenger. Figure 4B shows that expression of GFP-MICAL-L2-CT markedly reduced insulin-induced GLUT4myc translocation, whereas the basal surface-GLUT4myc levels remained unchanged. Taken together, these findings indicate that MICAL-L2 participates in insulin-stimulated GLUT4 translocation in muscle cells.
ACTN4 forms a complex with Rab13 and MICAL-L2 in response to insulin
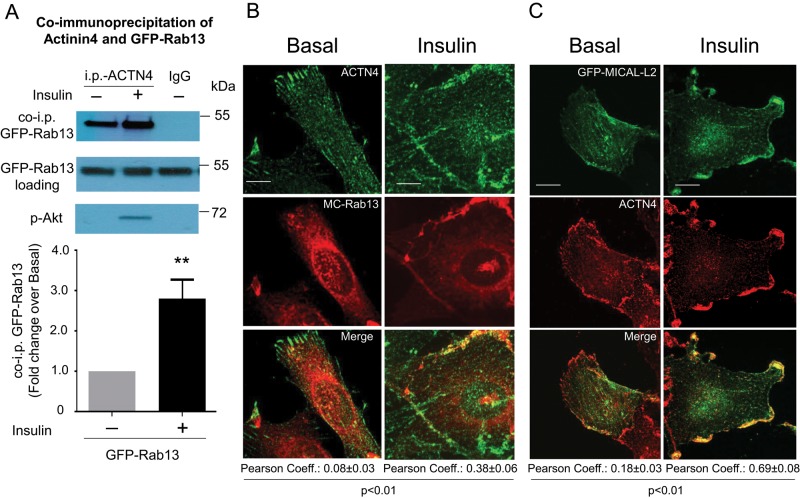
MICAL-L2 assumes a closed conformation held by interaction of its N-terminal CH and LIM domains with the coiled-coil region near the C-terminus, hindering access of residues 552–912, which recognize ACTN4. Binding of Rab13-GTP to the C-terminal region of MICAL-L2 relieves the N- and C-terminal domain interaction, exposing the ACTN4-binding site (Nakatsuji et al., 2008). We therefore hypothesized that, by activating Rab13, insulin would increase the interaction between MICAL-L2 and ACTN4 and therefore trigger the formation of a tripartite complex with MICAL-L2, essentially acting as a scaffold linking active Rab13 and ACTN4. This tripartite interaction was first examined by analyzing for the presence of GFP-Rab13 in immunoprecipitates of endogenous ACTN4 from basal and insulin-stimulated L6-GLUT4myc muscle cells. As seen in Figure 5A, insulin stimulation clearly promoted coprecipitation of GFP-Rab13 with ACTN4. The insulin dependence of this association is consistent with the activation of Rab13 in response to the hormone.
FIGURE 5:
Insulin promotes association of Rab13 with ACTN4. (A) L6 GLUT4myc-IR myoblasts were transfected with GFP-Rab13 for 48 h, cross-linked with DSP (1.5 mM, 30 min), serum deprived, and stimulated with insulin (15 min) or not. Lysates were immunoprecipitated with anti-ACTN4 or immunoglobulin G and immunoblotted with mouse anti-GFP, along with lysate aliquots. Phospho-Akt S473 confirmed insulin effectiveness. Graphs show the results of four independent experiments (mean ± SE, **p < 0.01). (B) L6 myoblasts transfected with GFP-Rab13 and stimulated with insulin or not were fixed and permeabilized, and endogenous ACTN4 was labeled with anti-ACTN4 and Alexa 546–secondary antibodies. Collapsed optical z-stack images of basal and insulin-stimulated cells, representative of three experiments. (C) L6 myoblasts grown on coverslips were transfected with GFP-MICAL-L2 and processed as in B. Representative collapsed optical z-stack images of GFP-MICAL-L2 (green) and ACTN4 (red) in basal and insulin-stimulated cells from three experiments. Scale bars, 10 μm. The Pearson correlation coefficients for the colocalization of Rab13 with ACTN4 at the cell periphery are indicated below the images of basal and insulin conditions.
The relationship between ACTN4 and Rab13 shown by coprecipitation was then examined using confocal fluorescence microscopy. In the basal state, immunodetected endogenous ACTN4 was shown to distribute upon fibers running longitudinally along the cell cytoplasm, as expected for an actin-binding protein, and delineating the cell periphery. In contrast, transfected MC-Rab13 showed the characteristic punctate and perinuclear distribution (Figure 5B). There was virtually no detectable colocalization of the two proteins in this unstimulated state. On insulin stimulation, however, the cells mount robust ruffling supported by cortical actin filaments (Chiu et al., 2010), and ACTN4 concentrated along such ruffles. Of note, a portion of MC-Rab13 also redistributed to those structures, resulting in colocalization of its fluorescence with that of immunodetected ACTN4 (Figure 5B).
The insulin-dependent coprecipitation and fluorescence colocalization of ACTN4 with Rab13 shown in Figure 5, A and B, is consistent with the ability of these two proteins to link to MICAL-L2. To support this possibility, we next examined whether ACTN4 and MICAL-L2 would also colocalize with each other and whether this occurs particularly in response to insulin. To this end, we again used confocal fluorescence microscopy in L6-GLUT4myc muscle cells. The endogenous, immunodetected ACTN4 was again observed along parallel fibers and thick cortical ribbons coincident with the cell periphery, whereas transfected, full-length GFP-MICAL-L2 showed a thinner, filamentous distribution that was not always in parallel, yet spanned the entire cell (Figure 5C). This distribution in the basal state was notably changed upon insulin stimulation, when the two proteins showed abundant colocalization at the cell periphery in thick cortical ribbons (Figure 5C). Given that insulin causes cortical actin remodeling forming membrane ruffles, the actin-binding capacity of both MICAL-L2 and ACTN4 is likely responsible for their colocalization in the region of insulin-induced actin remodeling.
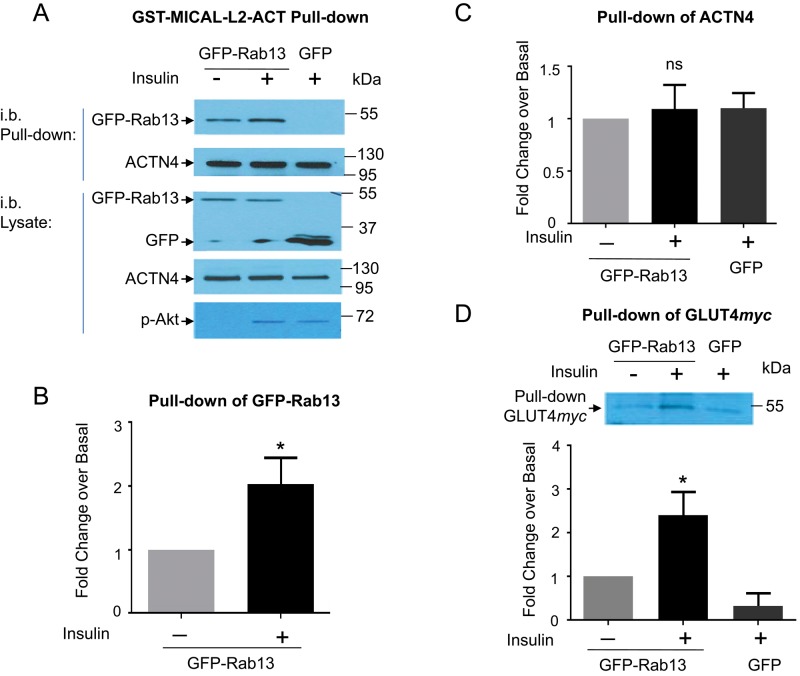
The results in Figure 5 strongly support the formation of an insulin-regulated, tripartite complex containing MICAL-L2, Rab13, and ACTN4. This scenario was tested directly by biochemical and imaging approaches in L6-GLUT4myc muscle cells. To this end, we generated a GST-linked fragment of MICAL-L2 encoding the region that can bind ACTN4 and the rest of the C-terminal tail that binds Rab13 (fragment called GST-MICAL-L2-ACT). Lysates from basal and insulin-stimulated muscle cells transiently expressing GFP-Rab13 were used for pull-down studies using GST-MICAL-L2-ACT. The fragment effectively brought down GFP-Rab13, and this association was about twofold higher in lysates from insulin-stimulated muscle cells (Figure 6, A and B). This result is consistent with the pull down of GFP-Rab13 by a smaller fragment, GST-MICAL-L2-CT (lacking the ACTN4-binding domain) from lysates prepared from insulin-stimulated CHO-IR cells shown in Supplemental Figure S1. However, as predicted from its sequence, GST-MICAL-L2-ACT also brought down ACTN4 (Figure 6A). Because GST-MICAL-L2-ACT lacks the MICAL-L2 N-terminal region, the domain recognized by ACTN4 is unobstructed and would be expected to be independent of regulation by active Rab13. Accordingly, GST-MICAL-L2-ACT pulled down ACTN4 from lysates of cells containing or lacking excess GFP-Rab13 and with or without prior insulin stimulation (Figure 6, A and C). Figure 6, A and D, shows in addition the presence of GLUT4myc in the complex (discussed later). In contrast to GST-MICAL-L2-ACT, the smaller fragment GST-MICAL-L2-CT (lacking the ACTN4-binding site) failed to pull down ACTN4 or GLUT4myc (Supplemental Figure S3).
FIGURE 6:
GST-MICAL-L2-ACT binds Rab13, ACTN4, and GLUT4. (A) Lysates from L6 GLUT4myc cells expressing GFP-Rab13 or GFP, stimulated with insulin or not, were pulled down with GST-MICAL-L2-ACT (containing Rab- and ACTN4-binding domains) and immunoblotted with anti-GFP and anti-ACTN4. Phospho-Akt illustrates insulin effectiveness. Pull down by GST-MICAL-L2-ACT of (B) GFP-Rab13, (C) ACTN4, and (D) GLUT4myc (n = 6, mean ± SE, *p < 0.05; ns, not significant).
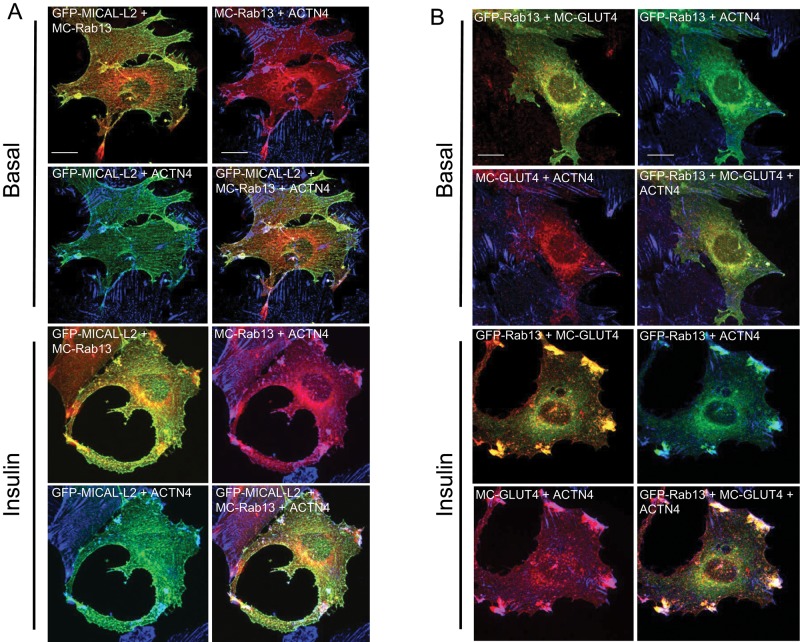
The foregoing results buttress the concept that MICAL-L2 bridges the association of Rab13 and ACTN4 and that this tripartite complex forms upon Rab13 activation in response to insulin. The three proteins then would be expected to colocalize in insulin-stimulated muscle cells. To explore this prediction, we examined their distribution by confocal fluorescence microscopy. As shown in Figure 7A, in insulin-stimulated muscle cells, GFP-MICAL-L2, MC-Rab13, and endogenous ACTN4 colocalized at the membrane ruffles. The images illustrate separately the colocalization of GFP-MCAL-L2 with MC-Rab13, of GFP-MICAL-L2 with ACTN4, and of MC-Rab13 with ACTN4, as well as all three proteins together within the same cell. These results strongly suggest that the three proteins coincide at the zone of cortical actin remodeling in insulin-stimulated muscle cells.
FIGURE 7:
Insulin-elicited three-way colocalization of MICAL-L2, ACTN4, Rab13, and GLUT4 at cellular ruffles. (A) L6 cells cotransfected with GFP-MICAL-L2 and MC-Rab13 were stimulated with insulin or not and analyzed by spinning-disk confocal fluorescence microscopy. Collapsed optical z-stack images of GFP-MICAL-L2 (green), MC-Rab13 (red), and ACTN4 (blue). (B) L6 cells cotransfected with GFP-Rab13 and MC-GLUT4 and labeled for ACTN4. Collapsed optical z-stack images of GFP-Rab13 (green), MC-GLUT4myc (red), and ACTN4 (blue). All results are representative of three experiments (>25 cells/condition). Scale bars, 10 μm.
GLUT4 is brought to the complex of Rab13, MICAL-L2, and ACTN4
We previously reported that ACTN4 is required for GLUT4 colocalization with the insulin-induced cortical actin filaments and that ACTN4 is accordingly necessary for insulin-dependent GLUT4 translocation (Talior-Volodarsky et al., 2008). Of note, ACTN4 can bind directly to the cytosolic loop of GLUT4 in vitro and ACTN4 and full-length GLUT4 coprecipitated from muscle cells, an interaction promoted by insulin stimulation (Foster et al., 2006) and confirmed here in Supplemental Figure S4. We therefore hypothesized that GLUT4 in intracellular vesicles would join the insulin-induced complex of ACTN4 with MICAL-L2 and Rab13. This was indeed borne out by the pull-down results shown in Figure 6, A and D, with GLUT4myc being pulled down from muscle cell lysates by GST-MICAL-L2-ACT but not by GST-MICAL-L2-CT (Supplemental Figure S3).
Next we analyzed the spatial relationship of GLUT4, ACTN4, and Rab13 by confocal fluorescence microscopy in L6 muscle cells. As shown in Figure 7B, in the basal state, transiently transfected mCherry-GLUT4myc (MC-GLUT4myc) showed a perinuclear, punctate distribution, consistent with the pattern observed for endogenous and stably transfected GLUT4 in muscle and adipose cells (Martin et al., 2000; Foster et al., 2001; Dugani and Klip, 2005; Ishiki and Klip, 2005; Williams and Pessin, 2008). At the perinuclear region, MC-GLUT4myc showed substantial colocalization with GFP-Rab13, confirming our previous observations in muscle cells stably transfected with GLUT4myc (Sun et al., 2010). In contrast to GLUT4 or Rab13, ACTN4 localized to longitudinal fibers. On insulin stimulation, however, all three proteins showed intense colocalization at the cell periphery, especially in ruffle-rich areas (Figure 7B). The figure also illustrates separately the exquisite colocalization of MC-GLUT4myc with GFP-Rab13,and of MC-GLUT4myc with ACTN4 and confirms the colocalization of GFP-Rab13 with ACTN4.
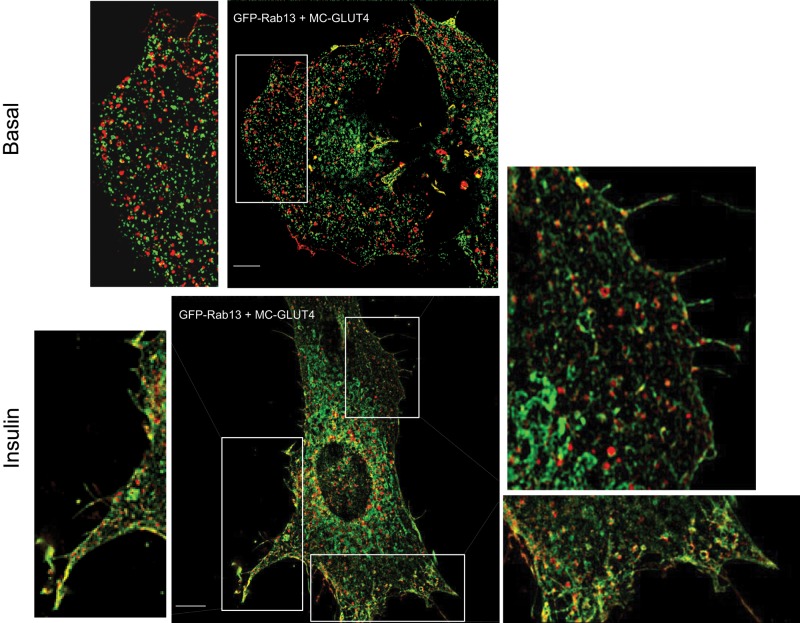
To obtain finer detail of this interaction and better discern the spatial relationship between the transporter and the GTPase, we used SIM in a few experiments to examine at high resolution the distribution of MC-GLUT4myc and GFP-Rab13. As shown in Figure 8, SIM visualized MC-GLUT4myc in vesicle-like structures throughout the cytosol. The punctate distribution of GFP-Rab13 was clearly differentiated from that of MC-GLUT4myc, and there was little if any colocalization between the two proteins in the basal state. Strikingly, however, in insulin-stimulated cells, many of the vesicular entities containing MC-GLUT4myc showed a distinctive GFP-Rab13 signal, strongly suggesting that the GTPase is mobilized to GLUT4 vesicles in response to the hormone (Figure 8).
FIGURE 8:
SIM reveals the insulin-induced colocalization of Rab13 with GLUT4 at the cell periphery. L6 cells cotransfected with GFP-Rab13 and MC-GLUT4myc were stimulated with insulin or not, fixed, and processed for SIM. Images of single focal planes of GFP-Rab13 (green) and MC-GLUT4myc (red). Scale bars, 10 μm. Excerpts show magnified regions of interest. In those peripheral regions, the Pearson coefficients of colocalization in the experiment shown were 0.05 (basal) and 0.39 (insulin).
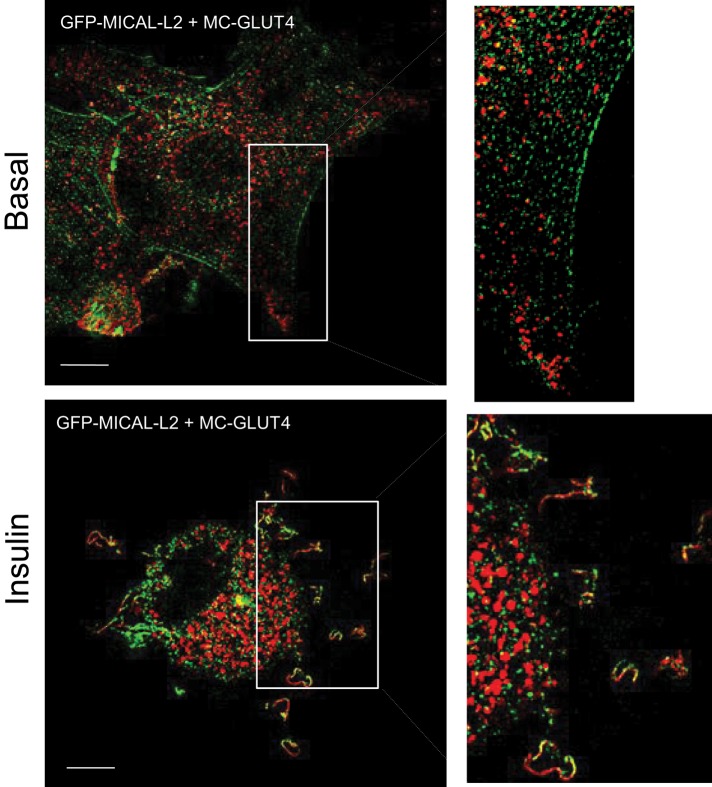
SIM also revealed the relationship of GLUT4 to MICAL-L2 (Figure 9). As expected based on the SIM images of MC-GLUT4myc (Figure 8) and MICAL-L2 (Figure 2), the transporter is distributed in vesicles around the nucleus and throughout the cytoplasm, whereas the adaptor protein aligns along fibers crossing the cytosol. There was no colocalization of the two proteins in the basal state. However, upon insulin stimulation, both proteins colocalized in selective regions specifically outlining the ruffled cell surface (Figure 9).
FIGURE 9:
SIM reveals the insulin-induced colocalization of MICAL-L2 with GLUT4 at the cell periphery. L6 cells cotransfected with GFP-MICAL-L2 and MC-GLUT4myc were stimulated or not with insulin as indicated, fixed, and processed for SIM. Images of single focal planes of GFP-MICAL-L2 (green) and MC-GLUT4myc (red). Scale bar, 10 μm. Excerpts show magnified regions of interest. The Pearson coefficients of MICAL-L2 and GLUT4 colocalization at the cell periphery in the experiment shown were 0.10 (basal) and 0.29 (insulin).
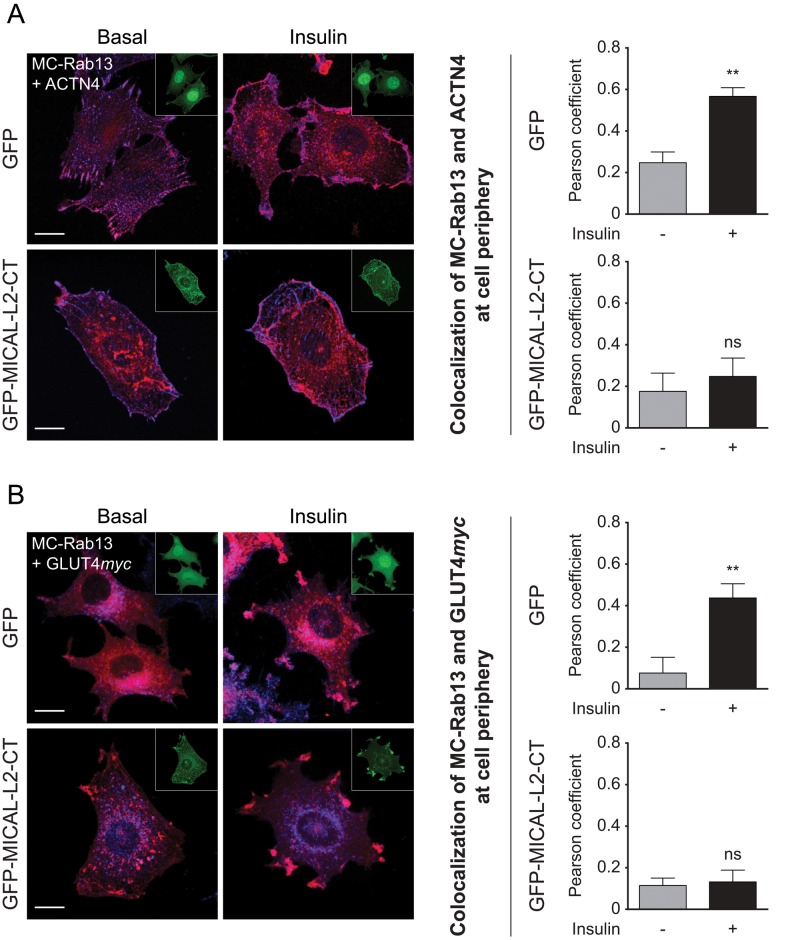
Finally, we examined whether MICAL-L2 acts as a scaffold connecting Rab13, ACTN4, and GLUT4. L6-GLUT4myc cells were transfected with the MICAL-L2-CT fragment that lacks ACTN4 binding. Under these conditions, there was minimal colocalization of Rab13 with ACTN4 or Rab13 with GLUT4myc, and insulin stimulation did not improve this colocalization at the cell periphery (Figure 10). This finding buttresses the central role of MICAL-L2 as a protein relaying the information of Rab13 activation to bring GLUT4myc to the membrane. Collectively Figures 8–10 reveal the exquisite, insulin-induced regulation of formation of this scaffold.
FIGURE 10:
The insulin-stimulated colocalization of MC-Rab13 with both ACTN4 and GLUT4myc at cell periphery is reduced in cells expressing MICAL-L2-CT. (A) L6-GLUT4myc cells cotransfected with either GFP vector and MC-Rab13 or GFP-MICAL-L2-CT and MC-Rab13 were stimulated with insulin or not, fixed, labeled for ACTN4, and analyzed by spinning-disk confocal fluorescence microscopy. Top, collapsed optical z-stack images of GFP (green, inset), MC-Rab13 (red), and ACTN4 (blue). Bottom, images of GFP-MICAL-L2-CT (green, inset), MC-Rab13 (red), and ACTN4 (blue). Graphs show Pearson coefficients of colocalization of MC-Rab13 and ACTN4 at the cell periphery (mean ± SE, **p < 0.01). (B) L6-GLUT4myc cells treated as described were labeled for GLUT4myc and otherwise analyzed as before. Top, images of GFP (green, inset), MC-Rab13 (red), and GLUT4myc (blue). Bottom, images of GFP-MICAL-L2-CT (green, inset), MC-Rab13 (red), and GLUT4myc (blue). Graphs show Pearson coefficient of colocalization of MC-Rab13 and GLUT4myc at the cell periphery (mean ± SE, **p < 0.01, n = 3). Results are representative of three independent experiments (>25 cells/condition per experiment). Scale bars, 10 μm.
DISCUSSION
Insulin induces a rapid mobilization of GLUT4-containing vesicles from intracellular depots to the muscle cell surface to promote glucose uptake. Of note, the gain in GLUT4 at the muscle membrane is reduced in human and animal models of type 2 diabetes (Gibbs et al., 1995; Zierath et al., 1996; Maianu et al., 2001; Koistinen et al., 2003; Bogan, 2012). Hence unraveling the mechanism of insulin-dependent GLUT4 translocation is of paramount importance to identifying steps that fail in insulin-resistant conditions.
Intersection between signal transduction and vesicle traffic
Signals initiated by the insulin receptor at the membrane must reach GLUT4 vesicles in a spatiotemporally regulated manner. These signals involve activation of class I phosphatidyl inositol-3-kinase and Akt. Lienhard and colleagues identified the Rab-GAP AS160 as an Akt target in adipose cells (Sano et al., 2003), and we confirmed the involvement of AS160 in GLUT4 traffic in muscle cells (Thong et al., 2007). Downstream of AS160, insulin activates Rab8A and Rab13 (Ishikura et al., 2007; Sun et al., 2010), and Rab8A and Rab13 are necessary for GLUT4 translocation in muscle cells; in contrast, Rab10 assumes a more dominant role in adipose cells (Chen et al., 2012; Sadacca et al., 2013).
Because Rab GTPases direct vesicle traffic, it became imperative to identify their effectors mediating GLUT4 traffic. We recently found that MyoVa is a Rab8A effector mobilizing GLUT4 vesicles toward the periphery. Here we show that activated Rab13 engages the molecular linker MICAL-L2. This insulin-dependent interaction allows exposure of the ACTN4-binding site within MICAL-L2, leading to formation of a cortically located scaffold that draws GLUT4. Finally, we demonstrate that MICAL-L2 is required for GLUT4 translocation.
Rab13 engages MICAL-L2 for insulin-dependent GLUT4 translocation
We here show that insulin stimulation of L6 muscle cells leads to the association of Rab13 to the MICAL-L2-ACT fragment encoding the Rab-binding site (Supplemental Figure S3), and similarly in CHO-IR cells, insulin leads to association of Rab13 with a MICAL-L2-CT fragment (also encoding the Rab-binding site; Supplemental Figure S1). In muscle cells, Rab13 and full-length MICAL-L2 colocalized in cortical regions in an insulin-dependent manner, revealed by spinning-disk confocal fluorescence microscopy, SIM, and TIRF (Figures 1–3). This is consistent with the activation of Rab13 by the hormone (Sun et al., 2010) and the selective recognition of the active form of the GTPase by MICAL-L2.
The morphological distribution of MICAL-L2 in muscle cells was filamentous and prominent in cortical areas of insulin-stimulated cells (Figures 1–3). This is consistent with the presence of an actin-binding domain at the N-terminal domain of MICAL-L2 (Terai et al., 2006) and the insulin-induced remodeling of filamentous actin at the cell cortex (Chiu et al., 2010). given that actin remodeling and Rab13 are required for insulin-dependent GLUT4 translocation, we hypothesized that MICAL-L2 would be an effector of the GTPase in insulin signal transmission. Indeed, we demonstrate a requirement for MICAL-L2 in the insulin-dependent gain in surface GLUT4, because silencing expression of the endogenous linker protein precluded GLUT4 translocation. Moreover, expression of the MICAL-L2-CT fragment that binds Rab13 but lacks ability to interact with actin also markedly reduced insulin-triggered GLUT4 translocation (Figure 4). To our knowledge, this is the first demonstrated function of MICAL-L2 in muscle cells and highlights its action at the junction of signal transduction and vesicle traffic.
As in muscle cells, the majority of MICAL-L2 selectively localizes along actin filaments in epithelial cells, where, upon binding Rab13, it mediates tight junction assembly (Terai et al., 2006; Yamamura et al., 2008; Sakane et al., 2012). In contrast, interaction between Rab13-GTP and MICAL-L2 is not required for transferrin receptor recycling, reflecting its participation in specialized rather than constitutive intracellular traffic. Consistent with a role in selective organelle movements, the interaction of Rab13 with MICAL-L2 contributes to vesicle-dependent neurite outgrowth in PC12 cells (Sakane et al., 2010). Of note, although Rab8A also interacts with MICAL-L2 in epithelial cells, this occurs in perinuclear regions. These findings underscore that distinct Rab GTPases bound to MICAL-L2 mediate distinctive cellular functions. Akin to this differential localization and function, in muscle cells, Rab8A is mostly perinuclear and associates with MyoVa, contrasting with the more peripheral distribution of Rab13 (Sun et al., 2010, 2014), where it engages MICAL-L2.
Interaction of ACTN4 with MICAL-L2 and formation of a tripartite complex with Rab13
As a consequence of Rab13–GTP interaction at the C-terminal end of MICAL-L2, the latter unfolds to expose its ACTN4-binding site within residues 552–912 (Nakatsuji et al., 2008). Accordingly, MICAL-L2-ACT, a fragment that permanently exposes its ACTN4-binding domain, pulled down ACTN4 from muscle cell lysates. As expected, MICAL-L2-ACT also pulled down Rab13 (through its C-terminal binding domain). Of importance, this interaction was higher in insulin-stimulated muscle cells (Figure 6), as observed in CHO-IR cell lysates probed with the shorter MICAL-L2-CT (Supplemental Figure S1). Moreover, ACTN4 immunoprecipitated from muscle cells brought along Rab13 but only upon insulin stimulation (Figure 5). These findings suggest that, by promoting Rab13 binding to MICAL-L2, insulin allows the complex to interact with ACTN4. Indeed, the hormone promoted colocalization of ACTN4 and Rab13, as well as MICAL-L2 and ACTN4 (Figure 5). Moreover, MICAL-L2-CT prevented the insulin-dependent colocalization of ACTN4 with Rab13 (Figure 10). Hence MICAL-L2 acts as the linker bringing together activated Rab13 and ACTN4, reminiscent of the complex that establishes tight junctions in epithelial cells (Nakatsuji et al., 2008). In muscle cells, this complex is formed in response to insulin, through the activation of Rab13.
The Rab13-induced complex of MICAL-L2 and ACTN4 positions GLUT4 at the cell cortex
We found the interaction of ACTN4 with the Rab13 and MICAL-L2 complex compelling, given that ACTN4 can interact directly with cytosolic regions of GLUT4 and this interaction is promoted by insulin (Talior-Volodarsky et al., 2008). Hence we hypothesized that insulin would induce: 1) association of GLUT4 with the complex containing ACTN4, and 2) enrichment of GLUT4 at regions of colocalization of ACTN4, MICAL-L2, and Rab13. Confirming these predictions, insulin stimulation before cell lysis promoted the association of GLUT4myc in a complex pulled down by GST-MICAL-L2-ACT, which also contained Rab13 and ACTN4 (Figure 6). The higher amount of GLUT4 pulled down by MICAL-L2-ACT in lysates from insulin-stimulated compared with basal cells is in agreement with a higher availability of GLUT4 at the cell periphery. The second prediction is borne out by the insulin-dependent colocalization of GLUT4 with ACTN4 and Rab13 (Figure 7). SIM reveals that GLUT4 is found in discrete vesicular structures that acquire Rab13 in response to insulin (Figure 8).
In summary, our results suggest the formation of a tripartite complex of active Rab13, MICAL-L2, and ACTN4 arising in response to insulin. This complex is positioned at cortical regions likely supported by insulin-dependent actin reorganization. In the basal state, GLUT4 is located in a perinuclear depot, inactive Rab8A and Rab13 are cytosolic, MICAL-L2 aligns along actin filaments throughout the cell, and ACTN4 associates with cortical actin filaments. On insulin stimulation, cortical actin remodels (Chiu et al., 2010), collecting MICAL-L2 at the cortex. Rab13, activated through insulin-dependent inhibition of AS160 (Sun et al., 2010), links to MICAL-L2, thereby codistributing along cortical actin structures and allowing association of ACTN4 with the complex. GLUT4 vesicles, propelled to the cell periphery through insulin-activated Rab8A and processive MyoVa (Sun et al., 2014), link to ACTN4 through GLUT4 itself (Talior-Volodarsky et al., 2008). Potentially, Rab13 in this scaffold associates with GLUT4 vesicles replacing Rab8A, since Rab13 and GLUT4 colocalize at the TIRF imaging zone, where Rab8A was not detected (Sun et al., 2014).
Hence insulin induces dual input on MICAL-L2 to recruit GLUT4 vesicles to the complex: one through ACTN4, the other through Rab13. Our findings further demonstrate that MICAL-L2 is required for insulin-dependent GLUT4 exocytosis, building on our previous findings that Rab13 (Sun et al., 2010) and ACTN4 (Talior-Volodarsky et al., 2008) are required for this process. We speculate that the tripartite complex of Rab13, MICAL-L2, and ACTN4 positions the incoming GLUT4 vesicles for fusion with the membrane.
MATERIALS AND METHODS
Reagents and DNA constructs
Mouse anti-myc (9E10) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-pAkt (473) was from Cell Signaling Technology (Danvers, MA). Polyclonal anti-GFP antibody was from Abcam (Cambridge, MA). Polyclonal anti–actinin-4 antibody was from Alexis Biochemicals (Farmingdale, NY). Horseradish peroxidase (HRP)–conjugated goat anti-rabbit and goat anti-mouse secondary antibodies were from Jackson ImmunoResearch (West Grove, PA). Glutathione Sepharose 4B and Protein G beads were from GE Healthcare Life Sciences (Piscataway, NJ). Alexa 546–conjugated goat anti-mouse antibody, isopropyl-β-d-thiogalactoside (IPTG) and dithiobis succinimidyl propionate (DSP) PI22585 cross-linker was obtained from Thermo Fisher Scientific (Waltham, MA). Cocktail proteinase inhibitor was purchased from Sigma-Aldrich (St. Louis, MO). Humulin human insulin was from Eli Lilly (Indianapolis, IN). A construct encoding the full-length GFP-MICAL-L2 was a kind gift from Steve Caplan (University of Nebraska, Lincoln, NE). The constitutively active, GTP-bound Rab13Q67L and the dominant-negative, GDP-bound Rab13T22N mutant cDNA constructs were kind gifts from J. Brumell (Hospital for Sick Children, Toronto, Canada). mCherry-tagged Rab13 wild type (MC-Rab13wt) was subcloned into mCherry-C1 through the SalI/BamHI restriction sites. The C-terminal mouse MICAL-L2 C-terminal (aa 806–1009) was PCR amplified from RNA of C2C12 myoblasts and subcloned into pGEX-6P-2 (Clontech, Mountain View, CA) via the BamHI/EcoRI restriction sites to generate GST-MICAL-L2-CT or into pEGFP-C2 via the XhoI/EcoRI restriction sites to generate GFP-MICAL-L2-CT. The C-terminus of MICAL-L2 (aa 261–1009) was PCR amplified from RNA of C2C12 myoblasts and was also subcloned into the pGEX-6P-2 vector via the XhoI/BamHI restriction sites to generate GST-MICAL-L2-ACT. mCherry-GLUT4myc (MC-GLUT4myc) was made by amplifying GLUT4myc cDNA by PCR from pcDNA3-GFP-GLUT4myc construct (Boguslavsky et al., 2012) and subcloned into mCherry-C1 vector via the EcoRI/BamHI restriction sites. shRNA targeting both rat and mouse MICAL-L2 sequence (CCGAAGCTGCTTCAGGTGT) in pGIPZ-GFP vector by Open Biosystems was purchased from the SPARC BioCentre (Hospital for Sick Children). A pSuper-basic vector tagged with GFP only was used as control.
RT-PCR
Total RNA was isolated from C2C12 myoblasts and L6 myoblasts using the RNeasy Mini Kit (Qiagen, Toronto, Canada). cDNA was prepared using Superscript III Reverse Transcriptase (Thermo Fisher Scientific). Specific primer pairs were designed to amplify MICAL-L2-CT, MICAL-L2-ACT, or GLUT4myc for subcloning into pGEX-6P-2, GFP-C2 vector, or mCherry-C1 vector.
Cell culture and transfection
Rat L6 muscle cells stably expressing GLUT4 with an exofacial myc epitope (L6-GLUT4myc) or L6-GLUT4myc cells also stably overexpressing the human insulin receptor (L6-GLUT4myc-IR) were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS) and antibiotics as described previously (Wang et al., 1998). CHO-IR cells were cultured in α-MEM supplemented with 10% FBS and antibiotics. Transfection of cDNA constructs was performed with FuGENE HD (Promega, Madison, WI) according to the manufacturer’s protocol. Briefly, cDNA/reagent mixture was applied to the cells in α-MEM with 10% FBS for 4–6 h without antibiotics, medium was changed, and cells were grown for 24–36 h.
Insulin stimulation, cell lysis, immunoprecipitation, and immunoblotting
Cells were incubated for 3 h in serum-free α-MEM containing 0.3% bovine serum albumin (BSA) and then not treated (basal) or treated with 100 nM insulin for 10 min unless otherwise indicated. For lysis, cells were washed twice with cold phosphate-buffered saline (PBS) and incubated in lysis buffer (1% Triton, 50 mM Tris, 140 mM NaCl, cocktail proteinase inhibitor [Sigma-Aldrich], 20 mM NaF, 2 mM Na3VO4, 5 mM MgCl2, pH 7.4). For coimmunoprecipitation, L6 rat muscle cells stably expressing GLUT4 with an exofacial myc epitope in the first extracellular loop (L6 GLUT4myc) were grown in 10-cm dishes and transfected for 48 h, followed by serum deprivation for 3 h in serum-free α-MEM. During the last 30 min, cells were treated with 1.5 mM DPS PI22585, a cell-permeant, thiol-cleavable, and amine-reactive cross-linking agent, followed by treatment with insulin or not. Lysates were passed through a 19-gauge syringe needle 10 times, and supernatants were collected by centrifugation at 11,000 × g at 4°C for 10 min. Supernatants were incubated with protein G beads conjugated to 1 μg of antibody at 4°C for 4 h, followed by three washes with washing buffer (0.4% Triton, 10 mM Tris-HCl, 140 mM NaCl, pH 7.4). Coimmunoprecipitated complexes were eluted with 2× Laemmli buffer and then separated by SDS–SPAGE. Immunoblotting was conducted on polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Richmond, CA) and incubation with respective primary antibodies. Proteins captured by GST-pull down were visualized by HRP-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare Life Sciences).
Expression of GST constructs and pull-down experiments
GST-MICAL-L2-CT or GST-MICAL-L2-ACT constructs were transformed in DH5α or BL21 bacteria, respectively, and a single clone was selected to grow overnight in 50 ml of Luria–Bertani (LB) medium containing ampicillin. The culture was expanded to 300 ml, followed by induction with 0.8 mM IPTG at 37°C for 4 h with agitation at 225 rpm. For pull-down assays, bacteria expressing GST-MICAL-L2-CT/-ACT pellets were lysed and sonicated in 1% Triton-100 and TBS buffer (20 mM Tris, 140 mM NaCl, pH 7.4), followed by mixing with 35 μl of glutathione Sepharose beads (GSHS; 60% slurry) at 4°C for 1 h. Lysates expressing different GFP-Rabs or GFP from L6-GLUT4myc cells were incubated with similar amounts of GST-MICAL-L2-CT or -ACT bound to GSHS beads at 4°C for 4 h, and after washes, bound complexes were eluted with 2× Laemmli buffer. Eluates were separated by SDS–SPAGE and transferred to PVDF for immunoblotting with antibodies to GFP (GFP-Rabs), ACTN4, and myc-epitope (GLUT4myc).
Cell-surface GLUT4myc detection
Surface GLUT4myc was detected by immunofluorescence in L6GLUT4myc myoblasts plated on glass coverslips transiently transfected with cDNA constructs, as previously described (Sun et al., 2014). Briefly, cells were serum starved for 2 h and then treated or not with 100 nM insulin for 10 min, rinsed with PBS, fixed with 3% paraformaldehyde (PFA), quenched with 50 mM NH4Cl, and blocked with 2% (vol/vol) BSA in PBS. Surface GLUT4myc was detected by anti-myc (1:100) for 1 h. After three washes, cells were incubated with Alexa 555–conjugated anti-mouse secondary antibody (1:1000) in the dark for 1 h, rinsed with PBS, and mounted on slides. Fluorescence images were acquired with a Zeiss LSM 510 laser-scanning confocal microscope (Carl Zeiss, Jena, Germany). Cells were scanned along the z-axis, and single collapsed images (collapsed xy projection) were assembled from optical stacks taken at 1-μm intervals. The pixel intensity in each cell (≥25 cells/condition) was quantified by ImageJ software (National Institutes of Health, Bethesda, MD).
Spinning-disk confocal fluorescence microscopy
L6 myoblasts grown on glass coverslips were transfected with GFP-MICAL-L2-FL or GFP or cotransfected with GFP-GLUT4myc and MC-Rab13, or GFP-MICAL-L2-FL and MC-Rab13, or GFP-MICAL-L2-FL and RFP-GLUT4myc/MC-GLUT4, for 24 h, followed by serum deprivation for 2 h. Cells were then treated or not with insulin for 10 min, fixed with 3% PFA in PBS, quenched with 50 mM NH4Cl, rinsed with PBS, and then mounted. Fluorescence images were acquired with an Olympus spinning-disk confocal microscope (IX81 using a 60×/numerical aperture [NA] 1.35 objective) equipped with Volocity 6.1.2 software. Cells were scanned along the z-axis at 0.3-μm intervals. Collapsed image stacks were exported into Photoshop CS2 (Adobe, San Jose, CA) for processing. Colocalization of GFP-MICAL-L2-FL (green) with MC-Rab13 (red) was analyzed in collapsed images using Volocity 6.1.2 software. Peripheral or cytoplasmic areas were selected and quantified for colocalization (Pearson’s coefficient). Values were analyzed by GraphPad Prism software (GraphPad, La Jolla, CA) and are presented as mean ± SE.
TIRF microscopy
L6 myoblasts grown on glass coverslips were transfected for 24 h with GFP-MICAL-L2 and MC-Rab13. Cells were serum deprived for 2 h in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) containing α-MEM and then treated or not with insulin (100 nM) for 10 min. Cells were fixed with 3% PFA on ice and then examined by TIRF microscopy using an Olympus IX81 microscope equipped with a dual laser (488 and 561 nm using a 150×/1.45 NA objective). Volocity 4.0 software was used to visualize GFP-MICAL-L2 and MC-Rab13 within 100 nm of the plasma membrane. Colocalization of GFP-MICAL-L2 with MC-Rab13 from TIRF images was quantified using Volocity 6.1.2 software, and colocalization was determined with Pearson’s coefficient. Values were analyzed by GraphPad Prism software and are presented as mean ± SE.
Structured illumination microscopy
L6 myoblasts were cotransfected with GFP-MICAL-L2 and MC-Rab13 or GFP-Rab13 with MC-GLUT4myc in six-well plates and then passaged and plated on coverslips for 18 h. Cells were then serum deprived in α-MEM supplemented with 0.3% BSA for 3 h, followed by treatment or not with insulin for 10 min. Cells were fixed with 3% PFA on ice and examined by SIM with a Zeiss Elyra PS equipped with Zen black operating software. Images were exported into Volocity 6.2.1 software and Photoshop CS2 for processing.
Supplementary Material
Acknowledgments
We thank Steve Caplan and John Brumell for GFP-MICAL-L2 and GFP-Rab13 constructs, and Paul Paroutis and Michael Woodside for guidance in TIRF and SIM analysis. This work was supported by Grant FND-143203 to A.K. from the Canadian Institutes of Health Research. Y.S. was supported by a postdoctoral fellowship from the Canadian Diabetes Association.
Abbreviations used:
- ACTN4
α-actinin-4
- AS160
Akt substrate of 160 kDa/TBC1 domain family member 4
- CHO-IR cells
CHO cells stably expressing the human insulin receptor
- L6 GLUT4myc cells
L6 rat muscle cells stably expressing GLUT4 with an exofacial myc epitope in the first extracellular loop
- MC-GLUT4
mCherry-GLUT4myc
- MC-Rab13
mCherry-Rab13
- MICAL-L2
molecule interacting with CasL-like 2
- MyoV
myosin V
- Rab-GAP
Rab GTPase-activating protein
- shRNA
short hairpin RNA
- SIM
structured illumination microscopy
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0319) on November 4, 2015.
REFERENCES
- Bogan JS. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem. 2012;81:507–532. doi: 10.1146/annurev-biochem-060109-094246. [DOI] [PubMed] [Google Scholar]
- Boguslavsky S, Chiu T, Foley KP, Osorio-Fuentealba C, Antonescu CN, Bayer KU, Bilan PJ, Klip A. Myo1c binding to submembrane actin mediates insulin-induced tethering of GLUT4 vesicles. Mol Biol Cell. 2012;23:4065–4078. doi: 10.1091/mbc.E12-04-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Zhang J, Deng Y, Jiang L, Song E, Wu XS, Hammer JA, Xu T, Lippincott-Schwartz J. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J Cell Biol. 2012;198:545–560. doi: 10.1083/jcb.201111091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu TT, Jensen TE, Sylow L, Richter EA, Klip A. Rac1 signalling towards GLUT4/glucose uptake in skeletal muscle. Cell Signal. 2011;23:1546–1554. doi: 10.1016/j.cellsig.2011.05.022. [DOI] [PubMed] [Google Scholar]
- Chiu TT, Patel N, Shaw AE, Bamburg JR, Klip A. Arp2/3- and cofilin-coordinated actin dynamics is required for insulin-mediated GLUT4 translocation to the surface of muscle cells. Mol Biol Cell. 2010;21:3529–3539. doi: 10.1091/mbc.E10-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugani CB, Klip A. Glucose transporter 4: cycling, compartments and controversies. EMBO Rep. 2005;6:1137–1142. doi: 10.1038/sj.embor.7400584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K, Boguslavsky S, Klip A. Endocytosis, recycling, and regulated exocytosis of glucose transporter 4. Biochemistry. 2011;50:3048–3061. doi: 10.1021/bi2000356. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Li D, Randhawa VK, Klip A. Insulin accelerates inter-endosomal GLUT4 traffic via phosphatidylinositol 3-kinase and protein kinase B. J Biol Chem. 2001;276:44212–44221. doi: 10.1074/jbc.M102964200. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2006;5:64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- Gibbs EM, Stock JL, McCoid SC, Stukenbrok HA, Pessin JE, Stevenson RW, Milici AJ, McNeish JD. Glycemic improvement in diabetic db/db mice by overexpression of the human insulin-regulatable glucose transporter (GLUT4) J Clin Invest. 1995;95:1512–1518. doi: 10.1172/JCI117823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr Opin Cell Biol. 2007;19:466–473. doi: 10.1016/j.ceb.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou MS, Bell ES, Girard M, Chaineau M, Hamlin JNR, Daubaras M, Monast A, Park M, Hodgson L, McPherson PS. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol. 2015;208:629–648. doi: 10.1083/jcb.201407068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiki M, Klip A. Minireview: recent developments in the regulation of glucose transporter-4 traffic: new signals, locations, and partners. Endocrinology. 2005;146:5071–5078. doi: 10.1210/en.2005-0850. [DOI] [PubMed] [Google Scholar]
- Ishikura S, Bilan PJ, Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem Biophys Res Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- Ishikura S, Klip A. Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation. Am J Physiol Cell Physiol. 2008;295:C1016–C1025. doi: 10.1152/ajpcell.00277.2008. [DOI] [PubMed] [Google Scholar]
- Karlsson HKR, Zierath JR, Kane S, Krook A, Lienhard GE, Wallberg-Henriksson H. Insulin-stimulated phosphorylation of the Akt substrate AS160 is impaired in skeletal muscle of type 2 diabetic subjects. Diabetes. 2005;54:1692–1697. doi: 10.2337/diabetes.54.6.1692. [DOI] [PubMed] [Google Scholar]
- Klip A, Sun Y, Chiu TT, Foley KP. Signal transduction meets vesicle traffic: the software and hardware of GLUT4 translocation. Am J Physiol Cell Physiol. 2014;306:C879–C886. doi: 10.1152/ajpcell.00069.2014. [DOI] [PubMed] [Google Scholar]
- Koistinen HA, Galuska D, Chibalin AV, Yang J, Zierath JR, Holman GD, Wallberg-Henriksson H. 5-Amino-imidazole carboxamide riboside increases glucose transport and cell-surface GLUT4 content in skeletal muscle from subjects with type 2 diabetes. Diabetes. 2003;52:1066–1072. doi: 10.2337/diabetes.52.5.1066. [DOI] [PubMed] [Google Scholar]
- Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- Lindsay AJ, Miserey-Lenkei S, Goud B. Analysis of the interactions between Rab GTPases and class V myosins. Methods Mol Biol. 2015;1298:73–83. doi: 10.1007/978-1-4939-2569-8_6. [DOI] [PubMed] [Google Scholar]
- Maianu L, Keller SR, Garvey WT. Adipocytes exhibit abnormal subcellular distribution and translocation of vesicles containing glucose transporter 4 and insulin-regulated aminopeptidase in type 2 diabetes mellitus: implications regarding defects in vesicle trafficking. J Clin Endocrinol Metab. 2001;86:5450–5456. doi: 10.1210/jcem.86.11.8053. [DOI] [PubMed] [Google Scholar]
- Martin S, Millar CA, Lyttle CT, Meerloo T, Marsh BJ, Gould GW, James DE. Effects of insulin on intracellular GLUT4 vesicles in adipocytes: evidence for a secretory mode of regulation. J Cell Sci. 2000;113:3427–3438. doi: 10.1242/jcs.113.19.3427. [DOI] [PubMed] [Google Scholar]
- Martinez O, Goud B. Rab proteins. Biochim Biophys Acta. 1998;1404:101–112. doi: 10.1016/s0167-4889(98)00050-0. [DOI] [PubMed] [Google Scholar]
- Marzesco A-M, Dunia I, Pandjaitan R, Recouvreur M, Dauzonne D, Benedetti EL, Louvard D, Zahraoui A. The small GTPase Rab13 regulates assembly of functional tight junctions in epithelial cells. Mol Biol Cell. 2002;13:1819–1831. doi: 10.1091/mbc.02-02-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji H, Nishimura N, Yamamura R, Kanayama H-O, Sasaki T. Involvement of actinin-4 in the recruitment of JRAB/MICAL-L2 to cell-cell junctions and the formation of functional tight junctions. Mol Cell Biol. 2008;28:3324–3335. doi: 10.1128/MCB.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahajeng J, Giridharan SSP, Cai B, Naslavsky N, Caplan S. Important relationships between Rab and MICAL proteins in endocytic trafficking. World J Biol Chem. 2010;1:254–264. doi: 10.4331/wjbc.v1.i8.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AF, Fazakerley DJ, James DE. Mapping insulin/GLUT4 circuitry. Traffic. 2011;12:672–681. doi: 10.1111/j.1600-0854.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- Sadacca LA, Bruno J, Wen J, Xiong W, McGraw TE. Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol Biol Cell. 2013;24:2544–2557. doi: 10.1091/mbc.E13-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A, Abdallah AAM, Nakano K, Honda K, Ikeda W, Nishikawa Y, Matsumoto M, Matsushita N, Kitamura T, Sasaki T. Rab13 small G protein and junctional Rab13-binding protein (JRAB) orchestrate actin cytoskeletal organization during epithelial junctional development. J Biol Chem. 2012;287:42455–42468. doi: 10.1074/jbc.M112.383653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A, Honda K, Sasaki T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol. 2010;30:1077–1087. doi: 10.1128/MCB.01067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Eguez L, Teruel MN, Fukuda M, Chuang TD, Chavez JA, Lienhard GE, McGraw TE. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sano H, Kane S, Sano E, Mîinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- Stöckli J, Fazakerley DJ, James DE. GLUT4 exocytosis. J Cell Sci. 2011;124:4147–4159. doi: 10.1242/jcs.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bilan PJ, Liu Z, Klip A. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci USA. 2010;107:19909–19914. doi: 10.1073/pnas.1009523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chiu TT, Foley KP, Bilan PJ, Klip A. Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells. Mol Biol Cell. 2014;25:1159–1170. doi: 10.1091/mbc.E13-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talior-Volodarsky I, Randhawa VK, Zaid H, Klip A. Alpha-actinin-4 is selectively required for insulin-induced GLUT4 translocation. J Biol Chem. 2008;283:25115–25123. doi: 10.1074/jbc.M801750200. [DOI] [PubMed] [Google Scholar]
- Terai T, Nishimura N, Kanda I, Yasui N, Sasaki T. JRAB/MICAL-L2 is a junctional Rab13-binding protein mediating the endocytic recycling of occludin. Mol Biol Cell. 2006;17:2465–2475. doi: 10.1091/mbc.E05-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong FSL, Bilan PJ, Klip A. The Rab GTPase-activating protein AS160 integrates Akt, protein kinase C, and AMP-activated protein kinase signals regulating GLUT4 traffic. Diabetes. 2007;56:414–423. doi: 10.2337/db06-0900. [DOI] [PubMed] [Google Scholar]
- Wang Q, Khayat Z, Kishi K, Ebina Y, Klip A. GLUT4 translocation by insulin in intact muscle cells: detection by a fast and quantitative assay. FEBS Lett. 1998;427:193–197. doi: 10.1016/s0014-5793(98)00423-2. [DOI] [PubMed] [Google Scholar]
- Williams D, Pessin JE. Mapping of R-SNARE function at distinct intracellular GLUT4 trafficking steps in adipocytes. J Cell Biol. 2008;180:375–387. doi: 10.1083/jcb.200709108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19:971–983. doi: 10.1091/mbc.E07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahraoui A, Joberty G, Arpin M, Fontaine JJ, Hellio R, Tavitian A, Louvard D. A small rab GTPase is distributed in cytoplasmic vesicles in non polarized cells but colocalizes with the tight junction marker ZO-1 in polarized epithelial cells. J Cell Biol. 1994;124:101–115. doi: 10.1083/jcb.124.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zierath JR, He L, Gumà A, Odegoard Wahlström E, Klip A, Wallberg-Henriksson H. Insulin action on glucose transport and plasma membrane GLUT4 content in skeletal muscle from patients with NIDDM. Diabetologia. 1996;39:1180–1189. doi: 10.1007/BF02658504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.