Abstract
Telomerase reverse transcriptase gene (TERT) promoter mutations are identified in many malignancies but not in hematological malignancies. Here we analyzed TERT and protection of telomeres 1 gene (POT1) mutations, and four different TERT SNVs in 226 acute myeloid leukemia (AML) patients and 806 healthy individuals in a case referent design, where also overall survival was assessed. A significant association for increased risk of AML was found for TERT SNVs, rs2853669 (OR = 2.45, p = 0.00015) and rs2736100 (OR = 1.5, p = 0.03). The overall survival for patients with CC genotype of rs2853669 was significantly shorter compared to those with TT or TC genotypes (p = 0.036 and 0.029 respectively). The influence of TERT rs2853669 CC on survival was confirmed in multivariable Cox regression analysis as an independent risk biomarker in addition to high risk group, higher age and treatment. No hot spot TERT promoter mutations at −228C > T or −250C > T or POT1 mutations could be identified in this AML cohort. We show that rs2853669 CC may be a risk factor for the development of AML that may also be used as a prognostic marker to identify high risk normal karyotype -AML (NK-AML) patients, for treatment guidance.
Keywords: TERT, SNV, AML, prognostic markers
INTRODUCTION
It is estimated that more than 90% of all cancers have increased telomerase activity [1] which correlates with immortalization, resistance to senescence and apoptosis, and telomere elongation. TERT is the transcriptional catalytic subunit for telomerase activity [2] and is considered to have a critical role in tumor formation. Therefore, genetic variants and somatic alterations in the TERT gene may affect telomerase function and contribute to the development of cancer as well as the outcome of chemotherapy. Up regulation of TERT expression is abundantly reported in somatic cells and two hot spot mutations in the TERT promoter, −228C > T and −250C > T, were recently reported in several different solid tumors e.g melanoma and gliomas [3–6]. These mutations have strong clinical implications with worse prognosis and poor survival and may represent a novel therapeutic target [6]. Hematological malignancies are not reported to be subject for somatic promoter mutations in the TERT gene but display enhanced telomerase activity and shortened telomeres [7]. An alternative or additional mechanism to altered telomere length regulation, in the absence of TERT mutations, may be the presence of single nucleotide variants (SNVs) in the TERT gene influencing activity and/or expression. Meta-analysis of 85 studies of SNV's and cancer types, but not hematological malignancies revealed associations for several TERT SNV's [8]. Telomere integrity is also regulated by the protection of telomeres 1 (POT1) gene and the POT protein is responsible for recruiting telomerase to the single-stranded 3′ telomere repeats and consecutively limiting telomere elongation by telomerase [9]. Andrew et al. identified twelve (3.5%) POT1 somatic mutations in chronic lymphocytic leukemia (CLL), the majority affecting the two oligonucleotide/oligosaccharide-binding (OB) folds suggested to have an essential role in binding to the telomeric repeats and for protein function [10]. POT1 loss-of-function mutations are prevalent in familial melanoma [11].
TERT also possess telomere independent functions in tumor formation and other human diseases, regulating Wnt-dependent transcription [12], mitochondrial function and apoptosis [13] and DNA damage response [14]. Recently, it was shown that TERT interact with NFκB and co-activate the expression of several genes, including cytokines, such as IL-6 and TNFα, that are critical for inflammatory reactions and cancer progression [15, 16].
Genetic alterations resulting in enhanced telomerase activity have recently been implicated in a variety of bone marrow failure syndromes such as acute myeloid leukemia [7, 17–18] inducing an expansion of undifferentiated myeloid hematopoietic stem cell progenitors, but so far no somatic mutations in either TERT or POT1 genes have been described in acute myeloid leukemia (AML).
AML is a genetically heterogeneous disease with various cytogenetic abnormalities affecting clinical outcome. AML with cytogenetically normal karyotype (NK-AML) have varying outcomes and there is a lack of risk markers to identify patients with worse prognosis. Mutations in the nucleophosmin 1 (NPM1), fms-related tyrosine kinase 3 (FLT3) and CCAAT/enhancer binding protein alpha (CEBPA) genes, are clinically important prognostic markers of outcome and survival particularly for NK-AML [19, 20].
Mutations in NPM1 indicates a favorable factor for achieving complete remission (CR) [21]. In contrast, FLT3 internal tandem duplication (ITD) is present in 30% of NK-AML patients, and associated with increased relapse rates, and decreased overall survival among NK-AML patients [22–25]. The CEBPA is the founding member of a family of related leucine zipper transcription factors that play important roles in myeloid differentiation [26]. Mutations in CEBPAdouble-mut are seen in 5% to 14% of AML and have been associated with a favorable clinical outcome. Only double mut is considred. Still a large proportion of NK-AML patients with intermediate risk are lacking prognostic markers to guide in treatment decisions.
Several studies investigating the impact of individual SNVs on prognosis and survival in AML have been suggested, but so far not reached the clinic e.g XPA and XPD variants, ABCB1, WT1 and IDH variants [27–31]. Most of them are based on candidate pathway approaches suggesting some significant SNV(s) to reflect the prognosis of the patients. We selected four TERT SNVs on the basis of previous reports of their association with the risk for several other forms and hallmarks of cancer and its potential as a biomarker of clinical outcome in AML.
RESULTS
TERT mutation and polymorphisms genotyping analysis
Mutation analysis of the TERT promoter and selected POT1 exons disclosed no mutations in our AML patient cohort. TERT 1062A> T (rs35719940) that has been identified as a susceptibility mutation in an Egyptian AML population, was found in 10/249 patients (4%) and in 22 of 806 healthy control (2.7%), (OR = 1.47, 95% CI 0.68–3.14, p = 0.21). Nevertheless rs35719940 showed no association with an increased risk for AML or effect on survival.
Genotyping of the four SNVs in the TERT gene in this study, revealed an increased risk for AML of the rs2853669 CC genotype (OR = 2.45, 95% CI 1.54–3.88, p = 0.00015) (Table 1). Homozygosity for the minor allele of rs2736100 (CC) disclosed a modest but still significantly increased risk for AML (OR = 1.5, 95% CI 0.98–2.29, p = 0.03) (Table 1). None of the other analyzed TERT SNV's showed any overall association with risk for AML. rs2853669 (C > T) is located in the TERT promoter at −246 upstream the ATG start codon, in proximity to the somatic mutation hot spots identified in several solid cancers. rs2736100 (C > A), rs10069690 (C > T) and rs4246742 (C > T) are located in introns 2, 4 and 8 respectively of the TERT gene (Figure 1A) and linkage disequilibrium (LD) analysis for the different SNV's showed a modest linkage between rs2853669 and rs2736100 (Figure 1B).
Table 1. Genotype distribution of the different polymorphisms in AML patients and normal control population investigated in the study and their association with AML susceptibility.
| Polymorphisms | AML n (%) | Controls n (%) | OR (95% CI) | p-Value |
|---|---|---|---|---|
| rs2853669 | ||||
| TT | 89 (39.38) | 373 (47.88) | ||
| TC | 99 (43.8) | 341 (43.77) | 1.21 (0.88 – 1.67) | 0.13 |
| CC | 38 (16.81) | 65(8.34) | 2.45 (1.54 – 3.88) | 0.00015 |
| TC + CC | 137 | 406 | 1.41 (1.04 – 1.91) | 0.01 |
| Total | 226 | 779 | ||
| rs10069690 | ||||
| CC | 118 (52.21) | 409 (53.32) | ||
| CT | 93 (41.15) | 319 (41.60) | 1.01 (0.74 – 1.37) | 0.5 |
| TT | 15 (6.63) | 39 (4.68) | 1.33 (0.71 – 2.5) | 0.22 |
| CT + TT | 108 | 358 | 1.04 (0.77 – 1.4) | 0.41 |
| Total | 226 | 767 | ||
| rs2736100 | ||||
| AA | 48 (21.23) | 201 (25.5) | ||
| AC | 113 (50) | 406 (51.53) | 1.16 (0.79 – 1.7) | 0.24 |
| CC | 65 (28.76) | 181 (22.97) | 1.5 (0.98 – 2.29) | 0.03 |
| AC + CC | 178 | 587 | 1.26 (0.88 – 1.81) | 0.1 |
| Total | 226 | 788 | ||
| rs4246742 | ||||
| TT | 156 (69.02) | 520 (66.41) | ||
| TA | 65 (28.76) | 240 (30.66) | 0.9 (0.65 – 1.25) | 0.29 |
| AA | 05 (2.21) | 23 (2.93) | 0.72 (0.27 – 1.93) | 0.35 |
| TA + AA | 70 | 263 | 0.88 (0.64 – 1.22) | 0.25 |
| Total | 226 | 783 |
Figure 1. TERT gene, SNVs localization and disequilibrium.
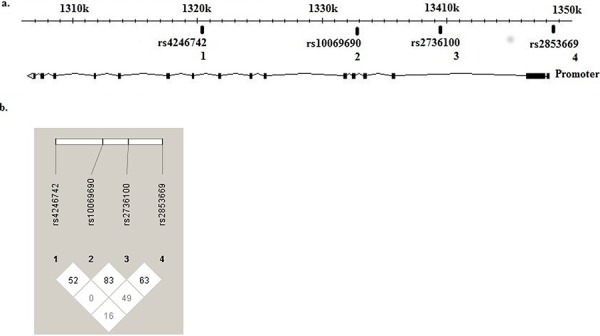
Localization of TERT SNVs, SNV1 (rs4246742), SNV2 (rs10069690), SNV3 (rs2736100) located at introns 8, 4 and 2 respectively and SNV 4 (rs2853669) located at the promoter region. (HapMap Data Rel 28/phase II+III, October 2010, on NCBI B36 assembly, dbSNP b126) A. Linkage disequilibrium showed a modest linkage between rs2853669 and rs2736100. (HaploView version 4.2) B.
Impact of TERT rs2853669 on overall survival
The rs2853669 CC genotype is firmly associated with an increased risk for AML (p = 0.00015) and Kaplan Meier survival analysis revealed a decrease in overall survival (OS). A significant difference in OS was evident for all AML patients as well as NK-AML patients, being homozygous CC for rs2853669 compared to TT or TC heterozygotes (p = 0.036 and 0.029, respectively) for all AML and p = 0.05 and 0.03, respectively, for NK-AML patients, (Figure 2A and 2B). Stratification of NK-AML patients according to FLT3 or NPM1 status, revealed a significantly reduced OS for NK-AML rs2853669 CC genotypes among FLT3-ITD or NPM1 non-mutated patients (4.6 months compared to 11.8 or 12.6 months ( p < 0.001) for the TC or TT genotypes, respectively (Figure 2C). A similar pattern was observed when NK-AML patients were stratified according NPM1 status. NPM1-mutated patients with a CC genotype showed a significantly shorter OS compared to TC or TT (5.4 vs 13.84 and 18.4 months, p < 0.001) (Figure 2D). However rs2736100 that showed a modest risk for AML, had no effect on survival in our AML cohort.
Figure 2. Differences in overall survival (OS) depending on TERT rs2853669 genotypes.
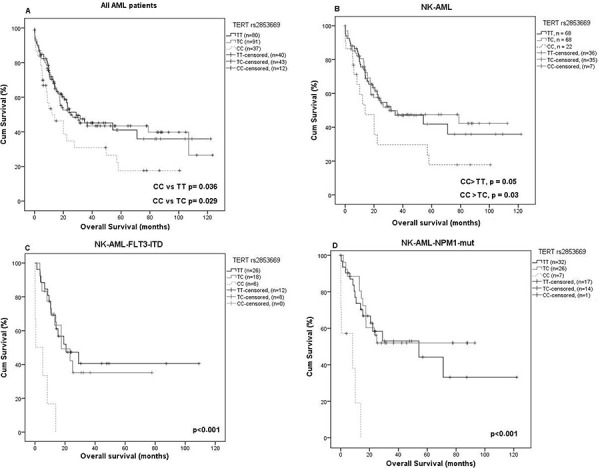
In entire group and in NK-AML group homozygous CC genotype was significantly associated with a shorter OS A. and B. In FLT3-ITD-positive patients, homozygous CC genotype was significantly associated with a shorter OS C. the mean OS was 4.6 vs. 11.8 and 12.6 months for CC vs. TT and TC genotype, respectively, p < 0.001, D. The same result was found for NPM1 mutated patients, the mean OS was 5.4 vs. 18.4 and 13.8 months CC vs. TT and TC respectively p < 0.001.
To evaluate whether TERT rs2853669 is an independent prognostic marker for survival in the entire AML group (Table 2) and when stratified according to karyotype (Table 3 and 4), multivariable Cox-regression analysis was performed with covariates including age, risk group and treatment (chemo only or chemo + allogeneic stem cell transplantation) (Table 2). The rs2853669 CC genotype was identified as an independent predictor for patient survival, in entire cohort and in NK-AML patients ( p = 0.024 and 0.022, respectively), in addition to older age ( p = 0.001 and 0.003, respectively) and classification in a high risk group ( p = 0.002 and 0.061, respectively). The Cox-regression analysis also showed that transplantation is an independent predictor for increased survival (p = 0.002) (Table 2).
Table 2. Cox regression of overall survival in entire AML cohort.
| Covariates | HR | 95% CI | p-Value |
|---|---|---|---|
| Age | 1.03 | 1.01 – 1.05 | 0.001 |
| Risk group | |||
| • *Intermediate riska | 1.76 | 0.92 – 3.37 | 0.038 |
| • *High riska | 3.23 | 1.51 – 6.91 | 0.002 |
| TERT rs2853669 C/Tb | 1.35 | 0.87 – 2.10 | 0.110 |
| TERT rs2853669 C/Cb | 1.74 | 1.03 – 2.94 | 0.024 |
| Treatmentc | 0.5 | 0.29 – 0.84 | 0.002 |
Compared to low risk,
Compared to T/T
Chemo + allo-SCT compared to chemo only
*Intermediate risk: FLT3-ITD positive/NPM1 mutation positive or FLT3-ITD negative/NPM1 mutation negative or FLT3-ITD negative/monoallelic and or CEBPA wild type patients, *High risk: FLT3-ITD positive/NPM1 mutation negative NK-AML patients, *Low risk: FLT3-ITD negative/NPM1 mutation positive or FLT3-ITD negative/biallelic CEBPA mutation patients.
Table 3. Cox regression of overall survival in NK-AML patients.
| Covariates | HR | 95% CI | p-Value |
|---|---|---|---|
| Age | 1.03 | 1.01 – 1.06 | 0.003 |
| Risk group | |||
| • *NK-intermediate riska | 1.49 | 0.77 – 2.87 | 0.22 |
| • *NK-high riska | 2.32 | 0.94 – 5.67 | 0.061 |
| TERT rs2853669 C/Tb | 1.22 | 0.75 – 2 | 0.40 |
| TERT rs2853669 C/Cb | 2.08 | 1.11 – 3.88 | 0.022 |
| Treatmentc | 0.56 | 0.3 – 1.02 | 0.061 |
Compared to low risk
Compared to T/T
Chemo + allo-SCT compared to chemo only
Table 4. Cox regression of overall survival in aberrant AML karyotype.
| Covariates | HR | 95% CI | p-Value |
|---|---|---|---|
| Age | 1.04 | 1.001 – 1.086 | 0.046 |
| Risk group | |||
| • *Aberrant K-intermediate riska | 1.49 | 0.77 – 2.87 | 0.235 |
| • *Aberrant K-high riska | 2.46 | 0.92 – 6.58 | 0.073 |
| TERT rs2853669 C/Tb | 1.22 | 0.75 – 2 | 0.409 |
| TERT rs2853669 C/Cb | 0.8 | 0.22 – 2.89 | 0.736 |
| Treatmentc | 0.34 | 0.1 – 1.16 | 0.086 |
Compared to low risk
Compared to T/T
Chemo + allo-SCT compared to chemo only
Correlation between TERT expression and other pro-inflammatory cytokines
Since the TERT protein is proposed to co-activate NFκB to enhance gene-expression of e.g IL-6, TNFα and other pro-inflammatory cytokines, contributing to tumor progression, we analyzed the relative mRNA levels on thirty-six AML patients with different rs2853669 genotypes (Supplementary Figure S1A–S1C). The CC genotype increased the TERT mRNA levels 2.17 fold, a non-significant increase. On the other hand, both IL-6 and TNFα were significantly increased, 6.25 and 3.58 fold respectively, for the CC genotypes compared to CT and TT genotypes ( p < 0.04 and 0.05, respectively, Supplementary Figure S1B and S1C).
DISCUSSION
In the promoter region, close to the melanoma and glioblastoma hot spot TERT mutations, there is a SNV rs2853669 (−245 T > C), which minor allele destroy a binding site for the Ets2 transcription factor, with an allele frequency in Europeans of 29% (1000 Genome project, http://www.Ensembl.org) and in the Swedish population of 28.6% (Table 1). We analyzed this and three additional intragenic SNVs for association to the risk of developing AML, in relation to clinical prognostic and predictive biomarkers and to overall survival in AML (n = 226). We found that the CC genotype of the TERT promoter SNV (rs2853669) and also the intronic SNV rs2736100 were associated with increased risk of AML. Previous studies have recognized this SNV as having an important role in other cancers, like lung and breast cancers [32, 33] and in non-small cell lung cancer (NSCLC) tissues [16].
The rs2736100 is located in intron 2 of TERT within a putative regulatory region [8, 34] and is the most studied SNV of the TERT gene. It has been described in 46 studies enrolling in total 74785 case subjects with 11 solid tumor types and 115726 control subjects [8]. However, no blood tumors were included in this meta-analysis and our study now add AML to the list of tumors with a moderate influence on AML risk associated to the rs2736100 SNV. Sheng and colleagues showed that rs2736100 CC genotype is associated with lower telomerase activity and longer telomere length, compared with the wild type allele, but not affecting TERT mRNA expression [35]. Otherwise, rs2736100 is close to mutations known to alter telomerase activity [8]. Rachakonda and Hsu showed that rs2853669 CC genotype influence telomerase activity and telomere length maintenance in bladder and non-small cell lung cancer [36, 37]. None of the other analyzed TERT SNVs in our study showed any association with the risk of having AML. Recently, TERT 1062A> T SNV (rs35719940) was identified as a mutation with prognostic value and shorter overall survival and it was suggested as an independent negative prognostic factor in AML patients [38]. In our cohort of AML patients, the SNV was identified in 10/226 (4.4%) but it was also found in a similar frequency among Swedish population healthy controls 22/806 (2.7%). This mutations did not significantly alter the risk, nor affecting the OS in our AML cohort, contrary to the study by Salah et al where they identified this mutation in 18 of 153 AML patients and only in one in 197 control group subjects [38]. In the present study we also show that the TERT promoter polymorphism rs2853669 influence OS with a significantly shorter survival in AML patients with the CC genotype. In the clinical setting, NPM1, CEBPA, and FLT3-ITD mutation status is used for risk categorization and prognostic assessment and to guide treatment. Some data suggest that NPM1 mutant/FLT3-ITD wild-type and CEBPA double mutation/FLT3-ITD wild-type AML patients may not benefit from early allogeneic stem cell transplantation, while stem cell transplantations reduce risk of relapse and death for AML patients with FLT3-ITD in the absence of NPM1 or CEBPA double mutation and, thus, represents a preferred treatment option in this high risk group [39]. However, there are still patients without FLT3-ITD/NPM1 or CEBPA double mutations in which additional prognostic biomarkers would be of high value to further assess the prognosis of the intermediate risk NK-AML patient group. Our results indicate that the TERT SNV rs2853669 CC may present an independent biomarker associated with poor prognosis in NK-AML patients.
In this study, we also screened our AML cohort for the two frequent promoter mutations, −228C> T and −250C > T, found in several other types of cancer. Both these mutations were absent in our cohort, confirming the findings by Killela et al and Yan et al on a smaller collection of 15 and 72 AML respectively [5, 18]. However, Yan et al. describe three novel mutations in the N-terminal of TERT (c.896 G > A, Glu280Lys; c.1079 C > G, Leu341Val and c.1451 G > C, Val465Leu) in 72 AML which may lead to shortened telomeres and telomerase dysfunction. AML patients with shorter overhang length had a higher frequency of unfavorable karyotype abnormalities. Therefore, Yan et al deduced that shorter telomere overhang length may indicate poor prognosis for AML patients [18]. Claudia et al showed that modulation of the telomerase complex is a highly effective strategy for targeting leukemia stem cells (LSCc) in AML. They proved that treatment with telomerase inhibitor (imetelstat) prevented the in vivo expansion of AML cells, and this was sustained during treatment. Telomerase deficient LSC population, depletes LSCs via the DNA damage, differentiation and apoptosis cascade, suggesting that clinical trials should address the efficacy of telomerase inhibitors as a strategy for preventing relapse or potentially together with chemotherapy, to improve outcomes in AML patients [40].
In addition to TERT, POT 1 is known as a negative regulator of telomere length by directly inhibiting telomerase activity, or by controlling telomeric DNA access to telomerase in human cells [9]. Andrew and collaborators found twelve somatic mutations in POT1 in 5% of CLL cases, nine among them were detected in N-terminal OB domains for POT1 and three of the twelve mutations lead to a truncated protein [10]. These somatic mutations were absent in our AML patients. The absence of TERT and POT1 mutations in our cohort indicate that in AML patients, SNVs in TERT may be of a relatively high importance for gene expression and/or activity.
Further, TERT has been implicated to possess several other cellular functions, such as cell differentiation and apoptosis and most recently as a co-activator to the nuclear transcription factor NFκB, to stimulate the transcription of pro-inflammatory and angiogenic factors [15]. In the present study, we found on a subset of samples where RNA were available, that the CC variant of rs2853669 is associated with an increased expression of IL-6 and TNFα, cytokines considered as markers for inflammation and cancer progression [15]. In the same context Fuxia et al showed that TERT rs2736100 CC enhance the expression of IL-6 in non-small cell lung cancer (NSCLC) in a NFkB dependent way [16]. Inflammation is a beneficial acute response activated to restore tissue injury and as a response to exposure for pathogenic agents. However, if unregulated, it may become chronic, inducing malignant cell transformation in the surrounding tissue [41]. The inflammatory response shares and influences various molecular targets and signaling pathways in carcinogenesis such as apoptosis, increased proliferation rate, and angiogenesis and represent an enabling characteristic that facilitates tumor cell survival [42]. Furthermore, the use of nonsteroidal anti-inflammatory drugs (NSAIDs) has been shown to decrease the amount on several pro-inflammatory mediators as well as incidence and mortality of several cancers [43]. Wang et al observed increased cell apoptosis and demonstrated a strong inhibition of tumor growth, after inhibiting the expression of FLT3 and NFkB p65 simultaneously in THP-1 cell line, and that a combined treatment strategy may be effective for AML in humans [44]. Although, we were not able to demonstrate an increased mRNA expression of TERT, in relation to SNV genotypes, an increased IL-6 expression was clearly observed (Supplementary Figure S1) and demonstrates that TERT orchestrates several activities in addition to increased telomere length and cell survival, which contributes to tumor progression.
MATERIALS AND METHODS
Study subjects
Two hundred twenty six (n = 226) patients mean age 57.7 (range 18–81) with AML from three different Swedish centers with clinical characteristics are shown in Table 5. Blood and bone marrow samples collected at diagnosis before treatment initiation was used for genotyping. Patients diagnosed before 2005 received treatment according to regional guidelines, which most commonly comprised of Cytarabine (AraC) doses of 200 mg/m2 as 24 h i.v infusions for 7 days, together with either Daunorubicin or Idarubicin for three days (24). From 2005, patients were treated according to national guidelines (http://www.sfhem.se/Filarkiv/Nationella-riktlinjer, accessed 2013–08-30), where the large majority of cases received induction treatment regimens including Daunorubicin 60 mg/m2/day for three days combined with AraC as 1000 mg/m2 twice a day in 2 h i.v infusions for 5 days. Other drugs used in combination with AraC and Daunorubicin or Idarubicin included mitoxantrone, 6-Thioguanine, etoposide and cladribine. Survival times were calculated as the time from diagnosis until an event (progression or death) or the latest follow up date. Patients were divided into high, intermediate or low risk group according to standard cytogenetic analysis, or, for NK-AML patients, according to FLT3-ITD/NPM1/CEBPA mutation status. Thus, FLT3-ITD positive/NPM1 mutation negative NK-AML were considered as high risk, FLT3-ITD negative/NPM1 mutation positive or FLT3-ITD negative/biallelic CEBPA mutation patients were considered as low risk, and FLT3-ITD positive/NPM1 mutation positive or FLT3-ITD negative/NPM1 mutation negative or FLT3-ITD negative/monoallelic and or CEBPA wild type patients were considered as intermediate risk. The control population comprised of 806 healthy individuals (50% women, 50% men) with mean age of 54 years (range 20–80 years), randomly collected from the population register from the same geographic region, south-eastern Sweden, as the patients. The study was approved by the local ethical committee and conducted in compliance with the Helsinki declaration.
Table 5. AML patient characteristics.
| AML patient characteristics | Total N = 226 |
|---|---|
| Gender | |
| Male | 124 (54.86%) |
| Female | 102 (45.13%) |
| Age at diagnosis, mean (range) | 57.7 (18–81) |
| FLT3 status | |
| FLT3 wild type | 160 (70.79%) |
| FLT3 mutated | 61 (26.99%) |
| Not determined | 5 (2.21%) |
| NPM1 status | |
| NPM1 wild type | 147 (65.04%) |
| NPM1 mutated | 72 (31.85%) |
| Not determined | 7 (12.04%) |
| CEBPA status | |
| Monoallelic | 29 (12.83%) |
| Biallelic | 2 (0.88%) |
| Karyotype | |
| Normal Karyotype | 169 (74.77%) |
| NK-low risk | 31 (13.71%) |
| NK-intermediate risk | 121 (53.53%) |
| NK-high risk | 17 (7.52%) |
| Aberrant Karyotype: | 43 (19.02%) |
| Aberrant -low risk | 5 (2.21%) |
| Aberrant -intermediate risk | 21 (9.29%) |
| Aberrant -high risk | 17 (7.52%) |
| Not determined | 14 (6.19%) |
| Treatmenta | |
| Dnr + AraC | 163 (72.12%) |
| Ida + AraC | 20 (8.85%) |
| Ida + AraC + CdA | 15 (6.63%) |
| AraC | 10 (4.42%) |
| Dnr + AraC + 6-TG | 4 (1.76%) |
| Dnr + mylotarg | 2 (0.88%) |
| Dnr + AraC + Mitox | 2 (0.88%) |
| Others | 10 (4.42%) |
| Treatment responseb | |
| CR | 167 (73.89%) |
| Non-CR | 32 (11.16%) |
| Not evaluated | 27 (11.94%) |
Dnr = Daunorubicine; AraC = Cytarabine; 6-TG = 6-thioguanine; Ida = Idarubicine; Cda = Cladribine; Mitox = Mitoxantrone; mylotarg = Gemtuzumab ozogamicin
CR = complete remission
DNA and RNA extraction
Genomic DNA was isolated from blood samples with the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) according to the supplier's recommendations. Total RNA was isolated from pelleted cells and vital frozen cells stored at −70°C using RNeasy Mini Kit (Qiagen, Hilden, Germany).
Mutation analysis
The TERT core promoter and 1062A> T polymorphism were amplified using MyTaqTM DNA polymerase (Bioline, USA) and published primers [5, 6], Supplementary Table S1. PCR products were purified with ExoSap-IT (GE Healthcare, USA), and standard Sanger sequencing was performed according to BigDye 3.1 protocol (Applied Biosystems, USA) and capillary electrophoresis on ABI 3500 Genetic Analyzer (Applied Biosystems, USA). PCR and mutation analysis of POT1 were performed according to the same protocol as for TERT, for the exons previously shown to harbor mutations (exons 5–10 and 18) including the oligonucleotide-/oligosaccharide-binding (OB) regions. Primers are listed in Supplementary Table S1.
Insertion mutations in exon 12 of the NPM1 gene and ITDs in the FLT3 gene were identified by PCR as previously described [45, 46].
Four overlapping primer pairs were used to amplify the entire CEBPA coding region, with subsequent DNA sequencing according to the Big Dye 3.1 protocol (Supplementary Table S1).
TERT SNVs and genotyping
For this study we selected 4 tag TERT SNVs described to be associated with risk in other tumors [8, 47–50]. The rs2853669, rs2736100 and rs4246742 in TERT, were genotyped using TaqMan® SNV Genotyping assays (C_8773290_10, C_1844009_10, C_11772271_20, respectively). All analysis were performed in ABI Prism 7500 or 7900 Sequence Detection System (Applied Biosystems), using the SDS 1.3 and 2.4 software for allelic discrimination, respectively.
The rs10069690 and TERT 1062A > T (rs35719940) were genotyped in AML patients and healthy controls by pyrosequencing. Each PCR reaction consisted of 5X My Taq Reaction Buffer, 1 U My Taq DNA polymerase (Bioline, USA), 1 μM of each primer (Supplementary Table S1), and 20 ng of template DNA in a final volume of 20 μl.
cDNA synthesis and quantitative real-time PCR
Thirty six samples were available for RNA isolation and investigated for TERT, IL-6, IL-1β and TNFα gene expression in relation to the rs2853669 genotype. Twelve samples were (TT), eleven (TC) and thirteen (CC). Total RNA from each sample was reversely transcribed into cDNA with MaximaR First Strand cDNA synthesis Kit for RT-qPCR (Fermentas, St Leon-Rot, Germany) according to supplier's instructions. The relative mRNA expression of TERT, IL-6, IL-1β and TNFα was determined by real-time PCR [7900HT Fast Real-Time PCR System (Applied Biosystems)], and normalized to the expression of two control genes β-glucuronidase (GUSB) (4333767F, amplicon length 81 bp) and Hypoxanthin-guanine phosphoribosyltransferase 1 (HPRT1) (4333768F, amplicon length 100 bp) (Applied Biosystems). All samples were run as duplicates. The calculation of the normalized gene expression was based on established methods, using the 2−ΔCT formula to calculate final relative expression in relation to HPRT1 and GUSB mRNA expression as house-keeping genes [51].
Statistical analysis
The genotype distribution in the control population was tested for Hardy-Weinberg equilibrium using a χ2 test and all SNV's displayed an expected Hardy-Weinberg distribution. The association between each SNVs and risk of AML are presented as odds ratios (OR) with 95% confidence intervals (CI). For survival analysis Kaplan-Meier, curves were generated and tested for significance by the log-rank test. Overall survival was the time elapsed from diagnosis of cancer to the date of death or date of the latest follow up. These tests were performed with both the entire material and in the NK-AML subset. Significant findings were also further investigated by multivariable Cox regression analysis using the forced entry method. Statistical analysis was performed with IBM SPSS Statistics software version 20 (IBM Corporation, Somers, NY) and Epi info™7. A p-value of < 0.05 was considered as significant.
CONCLUSIONS
In conclusion, our results show an association of the TERT SNV rs2853669 with the risk of having AML, a possible co-regulation of cytokine expression, as well as an association between TERT SNV genotype and OS. Our results indicate that the SNV rs2853669 has potential as a prognostic marker of survival in AML, in addition to the clinically used biomarkers FLT3-ITD, NPM1 and CEBPA mutations, which may further aid in treatment decisions such as allocation of patients to early stem cell transplantation.
SUPPLEMENTARY METHODS FIGURE AND TABLE
Acknowledgments
We would like to thank Åsa Schippert and Annette Molbaek for technical assistance.
Footnotes
FUNDING
This work was supported by the Swedish Research Council, the Swedish Cancer Society, County Council of Östergötland, AFA Insurance, and FORSS.
CONFLICTS OF INTEREST
The authors declare no potential conflict of interests.
REFERENCES
- 1.Chen CH, Chen RJ. Prevalence of telomerase activity in human cancer. J Formos Med Assoc. 2011;110:275–289. doi: 10.1016/S0929-6646(11)60043-0. [DOI] [PubMed] [Google Scholar]
- 2.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 3.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 5.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Wu G, Shan Y, Hartmann C, von Deimling A, Xing M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle. 2013;12:1637–1638. doi: 10.4161/cc.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aalbers AM, Calado RT, Young NS, Zwaan CM, Wu C, Kajigaya S. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia. 2013;27:1786–9. doi: 10.1038/leu.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, Baird DM, Prescott J, De Vivo I, Nitti D. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104:840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrew JR, Víctor Q, Miguel F, Conde L, Martínez-Trillos A, Villamor N, Rodríguez D, Kwarciak A, Garabaya C, Gallardo M, López-Guerra M, López-Guillermo A, Puente XS, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–532. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 11.Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, Pooley KA, Pritchard AL, Tiffen JC, Petljak M, Palmer JM, Symmons J, Johansson P, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46:478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, McLaughlin M, Veenstra TD, Nusse R, et al. Telomerase modulates Wnt signaling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Indran IR, Hande MP, Pervaiz S. hTERT overexpression alleviates intracellular ROS production, imnproves mitochondrial function and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 2011;71:266–276. doi: 10.1158/0008-5472.CAN-10-1588. [DOI] [PubMed] [Google Scholar]
- 14.Nitta E, Yamashita M, Hosokawa K, Xian M, Takubo K, Arai F, Nakada S, Suda T. Telomerase reverse transcriptase protects ATM deficient hematopoietic stem cells from ROS induced apoptosis through a telomere independent mechanism. Blood. 2011;117:4169–4180. doi: 10.1182/blood-2010-08-297390. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, Zhou J, Chng WJ, Li S, et al. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol. 2012;14:1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, Fu P, Pang Y, Liu C, Shao Z, Zhu J, Li J, Wang T, Zhang X, Liu J. TERT rs2736100T/G polymorphism upregulates interleukin 6 expression in non-small cell lung cancer especially in adenocarcinoma. Tumour Biol. 2014;35:4667–4672. doi: 10.1007/s13277-014-1611-z. [DOI] [PubMed] [Google Scholar]
- 17.Calado RT, Regal JA, Kajigaya S, Young NS. Erosion of telomeric single-stranded overhang in patients with aplastic anemia carrying telomerase complex mutations. Eur J Clin Invest. 2009;39:1025–1032. doi: 10.1111/j.1365-2362.2009.02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S, Han B, Wu Y, Zhou D, Zhao Y. Telomerase gene mutation screening and telomere overhang detection in Chinese patients with acute myeloid leukemia. Leuk Lymphoma. 2013;54:1437–1441. doi: 10.3109/10428194.2012.729834. [DOI] [PubMed] [Google Scholar]
- 19.Felicetto F, Charles A S. Acute myeloid leukemia in adults. The Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 20.Iriyama N, Asou N, Miyazaki Y, Yamaguchi S, Sato S, Sakura T, Maeda T, Handa H, Takahashi M, Ohtake S, Hatta Y, Sakamaki H, Honda S, et al. Normal karyotype acute myeloid leukemia with the CD7+ CD15+ CD34+ HLA-DR + immunophenotype is a clinically distinct entity with a favorable outcome. Ann Hematol. 2014;93:957–63. doi: 10.1007/s00277-014-2013-4. [DOI] [PubMed] [Google Scholar]
- 21.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, Akiyama H, Saito K, Nishimura M, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 23.Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, Löffler H, Sauerland CM, Serve H, Büchner T, Haferlach T, Hiddemann W. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 24.Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, Döhner H, Döhner K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 25.Wahlin A, Billstrom R, Bjor O, Ahlgren T, Hedenus M, Höglund M, Lindmark A, Markevärn B, Nilsson B, Sallerfors B, Brune M. Results of risk-adapted therapy in acute myeloid leukaemia. A long-term population-based follow-up study. European Journal of Haematology. 2009:99–107. doi: 10.1111/j.1600-0609.2009.01256.x. [DOI] [PubMed] [Google Scholar]
- 26.Xiang-Mei W, Jiang L, Jing Y, Yao DM, Deng ZQ, Tang CY, Xiao GF, Yang L, Ma JC, Hu JB, Qian W, Qian J. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7:6832–6840. [PMC free article] [PubMed] [Google Scholar]
- 27.Monzo M, Brunet S, Urbano-Ispizua A, Navarro A, Perea G, Esteve J, Artells R, Granell M, Berlanga J, Ribera JM, Bueno J, Llorente A, Guardia R, et al. Genomic polymorphisms provide prognostic information in intermediate-risk acute myeloblastic leukemia. Blood. 2006;107:4871–4879. doi: 10.1182/blood-2005-08-3272. [DOI] [PubMed] [Google Scholar]
- 28.Willander K, Falk IJ, Chaireti R, Paul E, Hermansson M, Gréen H, Lotfi K, Söderkvist P. Mutations in the isocitrate dehydrogenase 2 gene and IDH1 SNP 105C> T have a prognostic value in acute myeloid leukemia. Biomark Res. 2014;2:18. doi: 10.1186/2050-7771-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho PA, Kopecky KJ, Alonzo TA, Gerbing RB, Miller KL, Kuhn J, Zeng R, Ries RE, Raimondi SC, Hirsch BA, Oehler V, Hurwitz CA, Franklin JL, et al. Prognostic implications of the IDH1 synonymous SNP rs11554137 in pediatricand adult AML: a report from the Children's Oncology Group and SWOG. Blood. 2011;118:4561–4566. doi: 10.1182/blood-2011-04-348888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner K, Damm F, Gohring G, Görlich K, Heuser M, Schäfer I, Ottmann O, Lübbert M, Heit W, Kanz L, Schlimok G, Raghavachar AA, Fiedler W, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. Journal of Clinical Oncology. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 31.Damm F, Heuser M, Morgan M, Yun H, Grosshennig A, Göhring G, Schlegelberger B, Döhner K, Ottmann O, Lübbert M, Heit W, Kanz L, Schlimok G, et al. Single nucleotide polymorphism in the mutational hotspot of WT1 predicts a favorable outcome in patients with cytogenetically normal acute myeloid leukemia. Journal of Clinical Oncology. 2010;28:578–585. doi: 10.1200/JCO.2009.23.0342. [DOI] [PubMed] [Google Scholar]
- 32.Zhong R, Liu L, Zou L, Zhu Y, Chen W, Zhu B, Shen N, Rui R, Long L, Ke J, Lu X, Zhang T, Zhang Y, et al. Genetic variations in TERT-CLPTM1L locus are associated with risk of lung cancer in Chinese population. Mol Carcinog. 2013;52:118–126. doi: 10.1002/mc.22043. [DOI] [PubMed] [Google Scholar]
- 33.Shen J, Gammon MD, Wu HC, Terry MB, Wang Q, Bradshaw PT, Teitelbaum SL, Neugut AI, Santella RM. Multiple genetic variants in telomere pathway genes and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2010;19:219–228. doi: 10.1158/1055-9965.EPI-09-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng X, Tong N, Tao G, Luo D, Wang M, Fang Y, Li J, Xu M, Zhang Z, Wu D. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children. Carcinogenesis. 2013;34:228–235. doi: 10.1093/carcin/bgs325. [DOI] [PubMed] [Google Scholar]
- 36.Rachakonda PS, Hosen I, de Verdier PJ, Fallah M, Heidenreich B, Ryk C, Wiklund NP, Steineck G, Schadendorf D, Hemminki K, Kumar R. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. PNAS. 2013;110:17426–17431. doi: 10.1073/pnas.1310522110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu CP, Hsu NY, Lee LW, Ko JL. Ets2 binding site single nucleotide polymorphism at the hTERT gene promoter—effect on telomerase expression and telomere length maintenance in non-small cell lung cancer. Eur J Cancer. 2006;42:1466–1474. doi: 10.1016/j.ejca.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Aref S, El-Ghonemy MS, Abouzeid TE, El-Sabbagh AM, El-Baiomy MA. Telomerase reverse transcriptase (TERT) A1062T mutation as a prognostic factor in Egyptian patients with acute myeloid leukemia (AML) Med Oncol. 2014;31:158. doi: 10.1007/s12032-014-0158-6. [DOI] [PubMed] [Google Scholar]
- 39.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;21(115):453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 40.Bruedigam C, Bagger FO, Heidel FH, Paine Kuhn C, Guignes S, Song A, Austin R, Vu T, Lee E, Riyat S, Moore AS, Lock RB, Bullinger L, et al. Telomerase Inhibition Effectively Targets Mouse and Human AML Stem Cells and Delays Relapse following Chemotherapy. Cell Stem Cell. 2014;15:775–90. doi: 10.1016/j.stem.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic Inflammation and Cytokines in the Tumor Microenvironment. J Immunol Res. 2014 doi: 10.1155/2014/149185. ID 149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Li YG, Mark A, Mark D, Marian TN. Therapeutic potential of cytokine and chemokine antagonists in cancer therapy. European Journal of Cancer. 2006;42:793–802. doi: 10.1016/j.ejca.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Lu J, Wang Y, Bai S, Wang Y, Wang L, Sheng G. Combined effects of FLT3 and NF-κB selective inhibitors on acute myeloid leukemia in vivo. J Biochem Mol Toxicol. 2012;26:35–43. doi: 10.1002/jbt.20411. [DOI] [PubMed] [Google Scholar]
- 45.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 46.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A, Ehninger G, Illmer T. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 47.Simon M, Hosen I, Gousias K, Rachakonda S, Heidenreich B, Gessi M, Schramm J, Hemminki K, Waha A, Kumar R. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-Oncology. 2014;0:1–8. doi: 10.1093/neuonc/nou158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao L, Thakur A, Liang Y, Zhang S, Wang T, Chen T, Meng J, Wang L, Wu F, Jin T, Li X, Liu JJ, Chen C, Chen M. Eur J Cancer Prev. Polymorphisms in the TERT gene are associated with lung cancer risk in the Chinese Han population. Eur J Cancer Prev. 2014;23:497–501. doi: 10.1097/CEJ.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Jiao S. Increased lung cancer risk associated with the TERT rs2736100 polymorphism: an updated meta-analysis. Tumour Biol. 2014;35:5763–9. doi: 10.1007/s13277-014-1765-8. [DOI] [PubMed] [Google Scholar]
- 50.Terry KL, Tworoger SS, Vitonis AF, Wong J, Titus-Ernstoff L, De Vivo I, Cramer DW. Telomere length and genetic variation in telomere maintenance genes in relation to ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21:504–12. doi: 10.1158/1055-9965.EPI-11-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


