Abstract
Fast and accurate diagnostic systems are needed for further implementation of precision therapy of BRAF-mutant and other cancers. The novel IdyllaTM BRAF Mutation Test has high sensitivity and shorter turnaround times compared to other methods. We used Idylla to detect BRAF V600 mutations in archived formalin-fixed paraffin-embedded (FFPE) tumor samples and compared these results with those obtained using the cobas 4800 BRAF V600 Mutation Test or MiSeq deep sequencing system and with those obtained by a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory employing polymerase chain reaction–based sequencing, mass spectrometric detection, or next-generation sequencing. In one set of 60 FFPE tumor samples (15 with BRAF mutations per Idylla), the Idylla and cobas results had an agreement of 97%. Idylla detected BRAF V600 mutations in two additional samples. The Idylla and MiSeq results had 100% concordance. In a separate set of 100 FFPE tumor samples (64 with BRAF mutation per Idylla), the Idylla and CLIA-certified laboratory results demonstrated an agreement of 96% even though the tests were not performed simultaneously and different FFPE blocks had to be used for 9 cases. The IdyllaTM BRAF Mutation Test produced results quickly (sample to results time was about 90 minutes with about 2 minutes of hands on time) and the closed nature of the cartridge eliminates the risk of PCR contamination. In conclusion, our observations demonstrate that the Idylla test is rapid and has high concordance with other routinely used but more complex BRAF mutation–detecting tests.
Keywords: BRAF, rapid, integrated, qPCR
INTRODUCTION
With the identification of druggable molecular aberrations in cancer, we have increased our understanding of cancer biology and identified novel molecular targets for cancer therapy. [1–6] In particular, the identification of the BRAF V600 mutation hotspot in melanoma and other malignancies has led to the development of small-molecule kinase inhibitors targeting the BRAF oncogene. These BRAF inhibitors have revolutionized therapy for patients with BRAF V600–mutant advanced melanoma and demonstrated promising results in patients with other BRAF mutation–harboring diseases, including histiocytosis, hairy cell leukemia, non–small cell lung cancer, and biliary cancer. [4, 7–10]
BRAF inhibitors are contraindicated in the absence of a BRAF mutation. To determine whether a disease harbors a BRAF mutation, and thus whether treatment with BRAF inhibitors is appropriate, various methods to detect BRAF mutations in archived formalin-fixed, paraffin-embedded (FFPE) tissue samples are used. However, the methods that are currently routinely used for this purpose often require several hours to perform owing to time-consuming steps that include incubation, pipetting, and other processes. [11] In addition, for these molecular testing methods to be cost-effective, they are often performed on batches of samples, rather than single samples, thereby further increasing the turnaround time from sample to result. Consequently, results from these tests may not be available for at least several days or even weeks. [12] Such delays can hinder the delivery of effective treatment and thus have negative implications for care, especially in patients with a rapidly progressing disease such as advanced melanoma. [13]
A faster and simplified method for detecting BRAF mutations in FFPE tumor samples is the novel, fully integrated, real-time polymerase chain reaction (PCR)-based IdyllaTM system. Unlike current routinely used technologies for BRAF mutation detection, the IdyllaTM BRAF Mutation Test does not require manual sample preprocessing steps such as deparaffinization, FFPE tissue digestion, or DNA extraction because all these steps are integrated within a single-use cartridge. A complete FFPE tissue section or macrodissected FFPE material is placed directly into the cartridge and is subsequently processed by the IdyllaTM system, which provides automated sample processing, real-time PCR–based mutation detection, and result reporting. In the present study, we compared the performance of the IdyllaTM BRAF Mutation Test with that of other routinely used diagnostic methods for detecting BRAF V600 mutations.
RESULTS
Analytical sensitivity and specificity
To determine the sensitivity of the IdyllaTM BRAF Mutation Test (hereafter referred to as Idylla) in detecting BRAF V600 mutations in FFPE material, we used the test to analyze eight different sections of commercially available FFPE cell-line blends containing wild-type BRAF only, 1% BRAF V600E or 1% BRAF V600K in a wild-type BRAF background and demonstrated 100% agreement among eight independent experiments for each respective mutation (Table 1, Figure 1A–C).
Table 1. Performance of the IdyllaTM BRAF Mutation Test on control formalin-fixed, paraffin-embedded samples containing 1% or no BRAF V600 mutation.
| Control sample | V600E mutation detected | V600K mutation detected | No mutation detected |
|---|---|---|---|
| 1% BRAF V600E | 8/8 | 0/8 | 0/8 |
| 1% BRAF V600K | 0/8 | 8/8 | 0/8 |
| BRAF V600 wild-type | 0/8 | 0/8 | 8/8 |
Figure 1. Representative examples of polymerase chain reaction curves for formalin-fixed paraffin-embedded cell line mixtures containing.
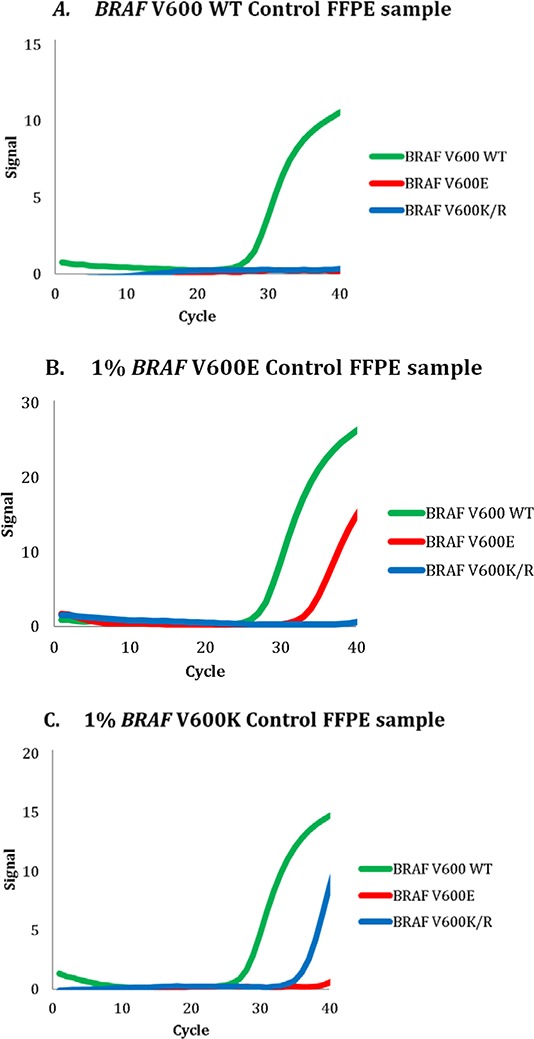
A. wild-type BRAF, B. 1% BRAF V600E, or C. 1% BRAF V600K. WT, wild type.
To assess the reproducibility of the Idylla TMBRAF V600 Mutation Test across different IdyllaTM instruments and among different operators, three operators used the test to repeatedly analyze consecutive sections from the same control 1% BRAF V600K FFPE sample 139 times on 7 different instruments using cartridges from two different production batches. BRAF V600K mutations were identified in 100% of the sections, indicating the high reproducibility of the test's results, even in cases with low abundant mutations, regardless of instrument and operator variation (Supplementary Figure S1).
To determine the specificity of the IdyllaTM BRAF Mutation Test in detecting BRAF V600 mutations, we used the test to analyze a high number of wild-type BRAF genomic DNA copies (8 × 104 per PCR reaction) from the BRAF wild-type CHL-1 cell line to identify the extent of cross-reactivity between the BRAF wild-type allele and the BRAF V600E and V600K reactions. Unlike the DNA recovered from FFPE samples, which is degraded by the formalin fixation and only partially amplifiable by PCR, the unfixed, high-quality genomic DNA recovered from the CHL-1 cell line enabled us to maximally challenge the specificity of the assay. In this experiment, the delta Ct between the specific signal of the BRAF V600 wild-type reaction and the cross-reactivity signal in the BRAF V600E and BRAF V600K reactions was greater than 20, demonstrating that the mutation detection reactions are highly specific even in the presence of a high number of wild-type alleles (Supplementary Figure S2).
Performance of Idylla versus cobas and MiSeq
We initially tested the IdyllaTM BRAF Mutation Test (hereafter referred to as Idylla) capacity to detect BRAF V600 mutations using 73 unprocessed (i.e., without macro- or micro dissection) FFPE tumor samples from patients with diverse cancers (melanoma, n = 30; colorectal cancer, n = 23; non-small cell lung cancer [NSCLC], n = 12; papillary thyroid cancer, n = 5; breast cancer, n = 3) obtained from commercial suppliers as outlined in Methods section (Table 2). No samples produced an invalid result. Idylla detected BRAF V600 mutations in 12 of 30 melanomas (40%), 1 of 23 colorectal cancers (4%), and 2 of 5 papillary thyroid cancers (40%) but did not detect BRAF V600 mutations in breast cancers or NSCLC.
Table 2. BRAF V600 mutations in different tumor types detected by Idylla.
| Tumor type (n) | BRAF V600E | BRAF V600K/R | BRAF V600 wild-type |
|---|---|---|---|
| Melanoma (30) | 9 | 3 | 18 |
| Colorectal cancer (23) | 1 | 0 | 22 |
| Lung cancer (12) | 0 | 0 | 12 |
| Thyroid cancer (5) | 2 | 0 | 3 |
| Breast cancer (3) | 0 | 0 | 3 |
From the 73 FFPE tumor samples, we randomly selected 45 samples with wild-type BRAF and all 15 samples with BRAF V600 mutations as per Idylla and subjected these 60 samples to the cobas BRAF V600 Mutation Test, a U.S. Food and Drug Administration–approved companion diagnostic, according to manufacturer's instructions. The sample size of 60 samples was expected to be adequate to demonstrate concordance of 95%–100% (kappa 0.9, 95% confidence interval [CI] +/− 0.15). To help maximize the sensitivity of the cobas test, we used macrodissection to enrich the tumor area of 42 of the 60 tumor samples (70%). Of these 60 FFPE tumor samples, 2 (1 melanoma and 1 breast cancer) could not be analyzed with cobas owing to insufficient DNA concentrations. For 56 of the remaining 58 samples (97%), the Idylla and cobas results were in overall agreement (kappa 0.91, standard error [SE], 0.07, 95% CI, 0.78–1.00). Compared with Idylla, cobas had a sensitivity of 87% (95% CI, 0.60–0.98), specificity of 100% (95% CI, 0.92–1.00), positive predictive value of 100% (95% CI, 0.75–1.00), and negative predictive value of 96% (95% CI, 0.85–0.99; Table 3). Of interest, the 2 samples in which Idylla but not cobas detected BRAF mutations (1 BRAF V600E mutation and 1 BRAF V600R mutation) contained less than 25% tumor cells. As described below, the Idylla results for both these samples were confirmed using an independent method.
Table 3. Concordance between Idylla and other methods in detecting BRAF V600 mutations in formalin-fixed paraffin-embedded (FFPE) tumor tissue samples.
| Concordance between cobas and Idylla testing of FFPE tumor tissue samples (N = 58) | ||
|---|---|---|
| BRAF mutation (cobas) | BRAF wild-type (cobas) | |
| BRAF mutation (Idylla) | 13 | 2 |
| BRAF wild-type (Idylla) | 0 | 43 |
| Observed agreements | 56 (97%); kappa, 0.91, SE, 0.07; 95% CI, 0.78–1.00 | |
| Concordance between MiSeq and Idylla testing of FFPE tumor tissue samples (N = 58) | ||
| BRAF mutation (MiSeq) | BRAF wild-type (MiSeq) | |
| BRAF mutation (Idylla) | 15 | 0 |
| BRAF wild-type (Idylla) | 0 | 43 |
| Observed agreements | 58 (100%); kappa, 1.00, SE, 0.00; 95% CI, 1.00–1.00 | |
| Concordance between laboratory and Idylla testing of FFPE tumor tissue samples (N = 100) | ||
| BRAF mutation (CLIA) | BRAF wild-type (CLIA) | |
| BRAF mutation (Idylla) | 61 | 1 |
| BRAF wild-type (Idylla) | 3 | 35 |
| Observed agreements | 96 (96%); kappa, 0.91, SE, 0.04; 95% CI, 0.83–1.00 | |
| Sensitivity | 95% (95% CI, 0.87–0.99) | |
| Specificity | 97% (95% CI, 0.85–1.00) | |
| Positive predictive value | 98% (95% CI, 0.91–1.00) | |
| Negative predictive value | 92% (95% CI, 0.79–0.98) | |
Abbreviations: SE, standard error; CI, confidence interval.
To validate the results obtained with Idylla and cobas, we subjected the 60 FFPE tumor samples (45 with wild-type BRAF and 15 with BRAF V600 mutations as per Idylla) to mutation analysis using the MiSeq deep sequencing system, in which the BRAF V600–surrounding region was amplified by PCR and sequenced at high depth (coverage >5000x) to facilitate the identification of mutations present in at least 1% of the DNA. Of these 60 samples, 2 had insufficient coverage and were excluded from further analysis. For the remaining 58 samples, the Idylla and MiSeq results demonstrated an overall agreement of 100% (kappa 1.00, SE 0.00; 95% CI, 1.00–1.00). Compared with MiSeq, Idylla had a sensitivity of 100% (95% CI, 0.78–1.00), specificity of 100% (95% CI, 0.92–1.00), positive predictive value of 100% (95% CI, 0.78–1.00), and negative predictive value of 100% (95% CI, 0.92–1.00; Table 3). The MiSeq analysis also revealed that the two low-tumor-cellularity samples in which Idylla but not cobas had identified a BRAF mutation contained 4.6% V600E and 5.7% V600R, respectively, which is in line with the overall specifications of the cobas test.
Independent clinical validation of the IdyllaTM BRAF mutation test
We then used Idylla in a retrospective study to test FFPE tumor samples from 100 patients with advanced cancers (melanoma, n = 38; colorectal cancer, n = 25; papillary thyroid carcinoma, n = 12; ovarian cancer, n = 5; cholangiocarcinoma, n = 3; head and neck cancer, n = 3; gastrointestinal stromal tumor, n = 2; NSCLC, n = 2; other, n = 10, Table 4) whose BRAF V600 mutation status had been previously determined in MD Anderson's Clinical Laboratory Improvement Amendments (CLIA)-certified Molecular Diagnostics Laboratory from routine clinical biopsies and resections. The CLIA laboratory detected BRAF V600 mutations in 66 of the 100 specimens (66%), and Idylla detected BRAF V600 mutations in 63 of the specimens (63%). The results of Idylla and the CLIA laboratory had overall agreement in 96 cases (96%; kappa, 0.91, SE, 0.04; 95% CI, 0.83–1.00). Compared with the CLIA laboratory, Idylla had a sensitivity of 95% (95% CI, 0.87–0.99), specificity of 97% (95% CI, 0.85–1.00), positive predictive value of 98% (95% CI, 0.91–1.00), and negative predictive value of 92% (95% CI, 0.79–0.98; Table 3). We always attempted to obtain the identical tissue blocks that were used by the CLIA laboratory for testing with Idylla; however, this was not possible in 9 cases. Of interest, of the 9 samples for which a different block was analyzed by the Idylla test, only 1 had discrepant results with the CLIA laboratory (BRAF V600K by CLIA, but wild-type by Idylla) resulting in an overall agreement of 89% for this subset (Supplementary Table S1). One (colorectal cancer with BRAF V600K mutation by the CLIA, but not Idylla) of four patients with discrepant BRAF V600 mutation status between the CLIA laboratory and Idylla received a MEK inhibitor with disease progression and one patient (melanoma with BRAF V600E mutation by the CLIA, but not Idylla) received a combination of BRAF inhibitor with chemotherapy and responded for 3 months. In addition, a patient with prostate carcinoma (BRAF V600E mutation by the CLIA, but not Idylla) and colorectal cancer (BRAF V600E mutation by the Idylla, but not CLIA) never received BRAF or MEK inhibitors.
Table 4. Tumor types of 100 patients with advanced cancers with known BRAF V600 status from the CLIA laboratory tested with Idylla.
| Tumor type | No. of patients | No. of BRAF V600 mutations detected (CLIA) | No. of BRAF V600 mutations detected (Idylla) |
|---|---|---|---|
| Melanoma | 38 | 34 | 33 |
| Colorectal cancer* | 25 | 9 | 9 |
| Papillary thyroid cancer | 12 | 12 | 12 |
| Ovarian cancer | 5 | 1 | 1 |
| Cholangiocarcinoma | 3 | 2 | 2 |
| Head and neck cancer | 3 | 0 | 0 |
| Sarcoma | 3 | 0 | 0 |
| Gastrointestinal stromal tumor | 2 | 1 | 1 |
| Non-small cell lung cancer | 2 | 2 | 2 |
| Esophageal cancer | 1 | 0 | 0 |
| Adrenocortical cancer | 1 | 0 | 0 |
| Prostate cancer | 1 | 1 | 0 |
| Appendiceal cancer | 1 | 1 | 1 |
| Neuroendocrine carcinoma of unknown primary | 1 | 0 | 0 |
| Glioblastoma | 1 | 1 | 1 |
| Thymoma | 1 | 0 | 0 |
One wild-type sample from CLIA showed BRAF V600 mutation on Idylla and one BRAF V600-mutant sample from CLIA demonstrated wild-type on Idylla
DISCUSSION
The present study's findings demonstrate that the IdyllaTM BRAF Mutation Test can reproducibly detect BRAF V600 mutations in FFPE samples with as little as 1% mutant DNA in a wild-type background and that the test's performance is on par with that of other routinely used but more complex methods of BRAF mutation detection.
First, we found an overall agreement of 97% between Idylla and cobas on 60 FFPE tumor samples. Idylla detected BRAF V600 mutations (4.6% V600E and 5.7% V600R according to MiSeq) in two samples that had relatively low tumor cellularity and that cobas identified as having wild-type BRAF, plausibly due to the test's higher detection limit (at least 5% of V600E mutant DNA) and because cobas is not validated for the detection of V600R. Furthermore, the Idylla results were confirmed using the MiSeq next-generation sequencing platform with 100% concordance. In an independent study, we found an overall agreement of 96% between Idylla and a CLIA-certified laboratory on 100 FFPE samples despite the fact that the testing was not performed simultaneously and that a different block had to be used for Idylla testing in 9% of cases. Although intratumoral heterogeneity for the BRAF V600 mutation might confound the concordance analysis, this effect appears to have a limited impact in our study, since the overall agreement level between both tests reached 96%. [14]
The workflow complexities and turnaround times of the different BRAF mutation–detecting methods used in the present study differ considerably. Unlike the FFPE sample processing steps in methods such as cobas, the FFPE sample processing steps in Idylla are completely integrated within one cartridge, which eliminates the need for manually performing time-consuming procedures such as deparaffinization, tissue digestion, and DNA extraction. Both the cobas test and MiSeq platform require a separate upfront DNA extraction step, which can take up to 3 hours, including about 1 hour for manual pipetting and incubation; subsequent PCR testing requires an additional 2 hours, including 30 minutes for manual procedures, when cobas is used. While these procedures are mostly performed in a sample batching mode, Idylla allows random access analysis of individual samples, leading to a turnaround time of hours instead of up to several weeks. The MiSeq workflow is even more complex and time-consuming than that of cobas owing to the different steps needed for preparing the sequencing library and performing the sequencing itself. In addition, for both cobas and MiSeq, the use of separate pre-PCR and post-PCR rooms to prevent sample contamination is strongly recommended. In contrast, the IdyllaTM BRAF Mutation Test produces results in about 90 minutes with about 2 minutes of hands on time and the closed nature of the cartridge eliminates the risk of PCR contamination. Furthermore, given its simple workflow and quick turnaround time, the IdyllaTM system can be used at nearly any facility, including those that would not be able to implement technologies with more complex workflows such as cobas or MiSeq.
Although Idylla detects BRAF V600E, V600K, V600R, V600M, and V600D mutations, it does not distinguish between BRAF mutations occurring on the same nucleotide, i.e., the V600E/D or V600K/R/M mutations. However, this is clinically acceptable, because patients carrying either one of these mutations have been reported to benefit from treatment with a BRAF inhibitor.[15] Whereas the MiSeq platform can detect all of these mutations, the cobas test only detects BRAF V600E down to at least 5% of mutant allele and BRAF V600K down to at least 30% of mutant allele. In the present study, 70% of the samples analyzed with the cobas test were macrodissected to maximize the test's sensitivity; in contrast, no samples analyzed with the Idylla test were macrodissected, although the test is compatible with micro- or macrodissected tissue. In addition, the Idylla test detected a BRAF mutation in one unprocessed FFPE sample that MiSeq analysis revealed to have less than 5% BRAF mutation (i.e., 4.6% V600E), whereas the cobas test did not detect this mutation even in tissue that had been macrodissected. These findings suggest that Idylla can detect all clinically relevant BRAF V600 mutations and that, compared with the cobas test, Idylla has a higher sensitivity for detecting BRAF V600 mutations and thus could be used to identify a greater number of patients who may benefit from treatment with a BRAF inhibitor.
In conclusion, the IdyllaTM BRAF Mutation Test, which offers an integrated and sensitive “sample-to-result” approach to detecting BRAF V600 mutations in FFPE samples, has a high concordance with routinely used methods for detecting BRAF V600 mutations in such samples.
MATERIALS AND METHODS
Tumor samples
From May 2012 until April 2014, patients with advanced cancers referred to the Department of Investigational Cancer Therapeutics at MD Anderson Cancer Center were enrolled in the study. FFPE tumor tissue samples from these patients were tested for BRAF V600 mutations in MD Anderson's CLIA-accredited Molecular Diagnostics Laboratory. Patients had to have enough archived tissue to allow the study and patient accrual was enriched to ensure that at least 60% of patients in the study had BRAF V600 mutations. Patient registration in the database, tumor pathology assessment, and tumor mutation analysis were performed at MD Anderson. The study was conducted in accordance with MD Anderson's Institutional Review Board guidelines. Additional FFPE tumor tissue samples from cancer patients who had signed an Institutional Review Board– or Ethical Committee–approved informed consent form were acquired through commercial suppliers (OriGene Technologies, Inc., Rockville, MD; Asterand Europe, Royston, Hertfordshire, United Kingdom).
BRAF V600 mutation detection with the IdyllaTM BRAF mutation test
The IdyllaTM system (Biocartis, Mechelen, Belgium) is a random-access molecular diagnostic system that provides quantitative allele-specific real-time PCR–based sample-to-result functionality using a disposable cartridge that can detect and quantify up to 30 molecular biomarker groups from a variety of solid and liquid samples, including plasma and FFPE tissue. The instrument is composed of a sample preparation module integrated with a combined PCR thermocycling and fluorescence detection module. For FFPE specimens, the sample preparation module uses high intensity focused ultrasound technology to emulsify the paraffin and simultaneously rehydrate the tissue sample in an aqueous solution, thereby liberating DNA. Nucleic acids are then transported via microfluidic channels in the cartridge into 5 separate PCR chambers that contain pre-deposited dried PCR reagents (i.e., primers, probes, and enzymes). Each PCR chamber allows for the identification of up to 6 different biomarker groups (30 biomarker groups total), each of which can be composed of multiple individual biomarkers.
The Idylla BRAF Mutation Test (Biocartis, Mechelen, Belgium) is a single-use cartridge-based test designed to detect the nucleotide G1798 > A and T1799 > A changes in the BRAF gene with a sensitivity limit of 1% BRAF mutant DNA in wild-type background. The G1798 > A change is present in patients with V600K, V600R, and V600M mutations, whereas the T1799 > A change is present in patients with V600E, V600K, V600E2, and V600D mutations. The test requires an analytic time of about 90 minutes and a hands-on time of about 2 minutes. Once the sample is inserted into the cartridge and the lid is closed, the cartridge is sealed, thereby eliminating the possibility of cross-contamination between different samples. The test does not require that FFPE samples be manually deparaffinized or preprocessed; all reagents required for sample preparation (i.e., liberation of DNA from the FFPE section) and real-time PCR detection are included in the IdyllaTM cartridge. Although the test is compatible with macrodissected FFPE material, unprocessed FFPE sections were used in this study. For the clinical concordance analyses in the present study, a single 10-μm section per FFPE sample was processed according to the manufacturer's instructions by a molecular biology-trained operator who was blinded for the results of the reference methods.
BRAF V600 mutation detection with the cobas 4800 BRAF V600 mutation test
The cobas 4800 BRAF V600 mutation test (Roche Molecular Systems, Pleasonton, CA) was performed according to the manufacturer's instructions at the College of American Pathologists– and CLIA-accredited molecular laboratory of HistoGeneX (Antwerp, Belgium). Briefly, 5-μm sections were stained with hematoxylin and eosin and examined by a certified pathologist who delineated the tumor areas and determined the tumor cell content. For optimal sensitivity, macrodissection was used to enrich the tumor area. DNA from two 5-μm sections was isolated using the cobas DNA Sample Preparation Kit (Roche Molecular Systems, Pleasonton, CA), diluted to 5 ng/μl, and tested on a cobas 4800 System v2.0 (Roche Molecular Systems) according to the manufacturer's instructions by a molecular biology-trained operator who was blinded for the IdyllaTM and MiSeq results.
BRAF V600 mutation detection with the MiSeq deep sequencing system
For BRAF V600 mutation detection with the MiSeq deep sequencing system (Illumina, San Diego, CA), DNA was first extracted from 10-μm FFPE sections using a QIAamp FFPE tissue DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and eluted in 50 μl of elution buffer. Different samples' DNA was normalized to 10 ng/μl based on measurement by Nanodrop (Thermo Fisher Scientific, USA), and an amplicon surrounding the BRAF V600 codon was generated using the forward primer 5′-CTACTGTTTTCCTTTACTTACTACACCTCAGA-3′ and the reverse primer 5′-ATCCAGACAACTGTTCAA ACTGATG-3′. The DNA samples were combined with a PCR reaction mixture consisting of 10 mM Tris, 50 mM KCl, 500 μM each PCR primer, 3 mM MgCl2, 0.2 mM dNTPs, 2 U FastStart Taq DNA polymerase (Roche Diagnostics, Rotkreuz, Switzerland), and 2 μl DNA input. The mixture was then subjected to PCR thermocycling for 10 minutes at 95°C followed by 50 cycles of denaturation at 95°C for 10 seconds, annealing at 62°C for 15 seconds, an extension step of 1 minute at 72°C, and a final extension step of 7 minutes at 72°C. The PCR products were visualized on Experion gel to confirm the presence of a single band of the correct length. Subsequent steps, including PCR product purification, sample barcoding, preparation of the MiSeq sequencing library, MiSeq sequencing, and bioinformatics data analysis, were performed by the Nucleomics Core at the Vlaams Instituut voor Biotechnologie (Gent, Belgium). At least 5000x coverage of the target region in each sample was required for MiSeq to have sufficient sensitivity. During the data analysis, the mutation threshold percentage for variant reporting was set at 1% mutant allele in a wild-type background to identify all mutations occurring in at least 1% of the sample DNA.
BRAF V600 mutation testing in MD Anderson's Molecular Diagnostics Laboratory
Archived tumor tissues obtained from enrolled patients' primary or metastatic sites using routine diagnostic and/or therapeutic procedures were subjected to mutation testing in the CLIA–certified Molecular Diagnostics Laboratory in the Division of Pathology and Laboratory Medicine at MD Anderson. All histologies were centrally reviewed at MD Anderson. DNA was extracted from microdissected paraffin-embedded tumor sections and analyzed using a PCR-based DNA sequencing method for BRAF V600 mutations utilizing primers designed by the Molecular Diagnostics Laboratory. In January 2011, the assay was changed to mass spectrometric detection (MassARRAY, Sequenom, San Diego, CA), and in March 2012, the assay was changed to next-generation sequencing (Ion Torrent, Life Technologies, Carlsbad, CA). The lower limit of detection is approximately 5–10%.
Statistical analysis
Concordance among mutation analyses was assessed using the kappa coefficient, sensitivity, specificity, and positive and negative predictive values, which were calculated using the GraphPad software program (GraphPad Software, Inc.; La Jolla; CA).
SUPPLEMENTARY FIGURES AND TABLE
Footnotes
CONFLICTS OF INTEREST
Filip Janku has research support from Novartis, Biocartis, Trovagene, and Foundation Medicine. Bart Claes, Martin Reijans, Geert Maertens, and Erwin Sablon are employees of Biocartis NV, and Benoit Devogelaere was previously employed by Biocartis NV. Mark Kockx and Isabelle Vanden Bempt are employees of HistoGeneX NV. Razelle Kurzrock has research support from EMD Serono, Genentech, Foundation Medicine and Pfizer, consultantship fees from SEquenom, and ownership interest in RScueRx.
FINANCIAL SUPPORT
This study was supported by Biocartis NV, the Elsa U. Pardee Foundation, the Sidney Kimmel Foundation for Cancer Research, the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, the National Center for Advancing Translational Sciences (grant no. UL1 TR000371), and the National Institutes of Health through MD Anderson's Cancer Center Support Grant (P30 CA016672).
REFERENCES
- 1.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the bcr-abl tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: A phase 1 dose-escalation trial. Lancet. 2012;379:1893–901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated braf in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, et al. Assessing pik3ca and pten in early-phase trials with pi3k/akt/mtor inhibitors. Cell Rep. 2014;6:377–87. doi: 10.1016/j.celrep.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters S, Michielin O, Zimmermann S. Dramatic response induced by vemurafenib in a braf v600e-mutated lung adenocarcinoma. J Clin Oncol. 2013;31:e341–4. doi: 10.1200/JCO.2012.47.6143. [DOI] [PubMed] [Google Scholar]
- 8.Munoz J, Schlette E, Kurzrock R. Rapid response to vemurafenib in a heavily pretreated patient with hairy cell leukemia and a braf mutation. J Clin Oncol. 2013;31:e351–2. doi: 10.1200/JCO.2012.45.7739. [DOI] [PubMed] [Google Scholar]
- 9.Hyman DM, Diamond EL, Vibat CR, Hassaine L, Poole JC, Patel M, et al. Prospective blinded study of brafv600e mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with braf v600e mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sam SS, Lebel KA, Bissaillon CL, Tafe LJ, Tsongalis GJ, Lefferts JA. Automation of genomic DNA isolation from formalin-fixed, paraffin-embedded tissues. Pathol Res Pract. 2012;208:705–7. doi: 10.1016/j.prp.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3003161. 111ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. Raf inhibitors prime wild-type raf to activate the mapk pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 14.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, et al. Intra- and inter-tumor heterogeneity of braf(v600e) mutations in primary and metastatic melanoma. PLoS One. 2012;7:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein O, Clements A, Menzies AM, O'Toole S, Kefford RF, Long GV. Braf inhibitor activity in v600r metastatic melanoma. Eur J Cancer. 2013;49:1073–9. doi: 10.1016/j.ejca.2012.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


