Abstract
Alternative splicing plays an important role in regulation of normal cellular function. Alternative splicing of pre-mRNA leads to the diversity of downstream protein products in the cell. The Affymetrix Exon arrays allow for a high throughput evaluation of the differences in spliced mRNA expressed in a biological system. In this study, we describe a method using this technology to study the generation of alternative mRNA transcripts in breast cancer cells that differ in the levels of a particular integrin, α3β1.
Keywords: Alternative splicing, gene regulation, expression profiling, microarray, exon splicing, integrins
1. Introduction
α3β1 integrin belongs to a family of heterodimeric cell surface receptors that mediate cell adhesion to the extracellular matrix. Integrins can mediate both inside-out and outside-in signal transduction, and they have been demonstrated to be involved in many aspects of cellular biology such as adhesion, migration and survival. Laminin-332 is the primary ligand for α3β1 which is expressed in a variety of epithelial cell types. α3β1 is overexpressed in a variety of human cancers and experiments conducted in breast cancer cells have indicated an important role for this integrin in invasion (1,2). In addition, we have shown that α3β1 in epithelial cells can induce the expression of EMT and angiogenesis promoting genes such as MMP9 and Mrp3 (3-5). α3β1-dependent induction of MMP9 gene expression was established to occur via enhanced stability of the MMP9 mRNA transcript in mouse keratinocytes, resulting in increased protein expression (3) To investigate the role of α3β1 in regulating differential gene expression as well as gene splicing events in the breast cancer cell line MDA-MB-231, we performed microarray analysis using the Affymetrix Human Exon 1.0 ST array platform (6). We have identified various candidate genes that are differentially spliced in cells that stably express an shRNA that targets the α3 integrin subunit (α3-knockdown cells), compared to cells that express a control shRNA. One of these genes was identified as POLR2I, which encodes a subunit of RNA Polymerase II. POLR2I mRNA was found to be differentially spliced at the 5’ end, where exon 1 was excluded from mRNA isolated from control breast cancer cells, but was included in mRNA from the α3-knockdown cells. This difference in exon1 processing could be attributed to differential usage of the 5’ untranslated region of the gene or variations in promoter usage.
2. Materials and Reagents
2.1. Equipment
Agilent Bioanalyzer 2100 system
Nanodrop ND-1000 UV-Vis spectrophotometer
Affymetrix Genechip® System
2.2. Materials for cell culture
MDA-MB-231 breast cancer cell lines were stably infected with lentivirus expressing a control shRNA (control cells; MISSION™ shRNA, Sigma)
MDA-MB-231 breast cancer cell lines stably infected with lentivirus expressing shRNA that targets the human α3 mRNA (α3-knockdown cells; MISSION™ shRNA, Sigma)
Phosphate buffered saline (PBS)- 137mM NaCl, 2.7mM KCl, 4.3 mM Na2HPO4.7H2O, 1.4mM KH2PO4, pH-7.4
2.3. Materials for RNA isolation
All tips, tubes and reagent bottles must be DNAse and RNase free (see Note 1)
Tri-reagent (Molecular Research Inc, cat#TR118) or TRIzol (Invitrogen cat#15586-026)
1-bromo-3-chloropropane (Molecular Research Inc, cat# BP151) or chloroform
Isopropanol
We recommend the use of nuclease-free water (Ambion cat# AM9932) to prepare all buffers and solutions
RNeasy mini RNA isolation kit (Qiagen cat# 74104)
DNase I (Ambion cat#AM2222)
RNase Zap (Ambion cat # AM9780)
2.4. Materials for RNA QC and microarray experiment
RNA 6000 Nanokit (Agilent cat#5067-1511)
GeneChip WT Sense Target Labeling and Control reagents (Affymetrix cat#900652). This catalog number includes all kits required for this protocol including cDNA synthesis, amplification, labeling, cleanup and hybridization.
GeneChip® Human Exon 1.0 ST arrays (Affymetrix cat# 900650)
RiboMinus™ Transcriptome Isolation Kit (Human/Mouse) (Invitrogen cat# K1550-02)
Magna-Sep™ Magnetic Particle Separator (invitrogen cat# K1585-01)
Betaine 5M (Sigma cat# B-0300)
3. Methods
3.1 Cell culture and harvesting of cells for RNA isolation
Indicated cell lines were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Hyclone) 100 U/ml penicillin and 100 μg/ml streptomycin and 2mM L-glutamine. Cells were maintained in 10-cm2 dishes in a 37°C incubator under 5% CO2.
Wash the cells with PBS to remove any residual media prior to harvesting.
Add 1 ml Tri-reagent or TRIzol directly to the cells in each 10-cm2 dish. DO NOT trypsinize the cells prior to treatment with tri-reagent or TRIzol. (see Note 2). Move the TRIzol around the flask and gently tap to slough off all attached cells. Pipette into a clean tube and store at −20°C till further use.
3.2. RNA isolation
The specific RNA isolation method that you choose will depend on your downstream application. Generally either method is acceptable for microarray, RT-PCR, or Northern blotting. The Qiagen spin-column cleanup offers the advantage of performing an optional DNase I digestion while purifying the RNA so further processing is avoided. However, detection of RNA molecules of 200bp or smaller will be limited if using the Qiagen cleanup procedure and hence not advised if you intend to use the RNA for miRNA analysis. While using arrays such as the Exon ST 1.0, Gene ST 1.0 or Tiling arrays, ensure that the RNA is DNAse treated since DNA contaminants will be amplified and labeled in the array protocol.
3.3. Qiagen RNEasy mini-cleanup
Add nuclease free water to the 150-200μL RNA from the RNA isolation to adjust the volume to 200μl (see Note 3)
Perform the RNEasy micro-cleanup as per the manufacturer's protocol.
3.4. Assessment of RNA Quality
Using a NanoDrop® spectrophotometer, measure the optical absorbance characteristics of the sample. (see Note 4). The A260/A280 as well as the A260/A230 ratio will ideally be close to 2.0, signifying the purification of nucleic acids away from protein and other organics, respectively. If either ratio is lower than 1.6, expect problems with downstream applications of the RNA (see Note 5).
Performance of a NanoChip assay using Agilent's BioAnalyzer allows for measurement of the molecular weight profile of the isolated RNA. In this way, you may evaluate the 28S/18S ratio measurements. A total RNA ratio between 1.8 and 2.0 is desirable however ratios 1.6-1.8 may be acceptable. A RNA Integrity number (RIN) score should be between 7 to 10 if the samples are to be used in a microarray or QPCR experiment downstream. (see Note 6).
3.5. Expression analysis of mRNA from cells
While we have used many different microarray platforms for standard gene expression analysis, we recommend the use of Affymetrix Exon 1.0 ST arrays for experiments where alternate splicing is of interest.
There are two methods recommended by Affymetrix to amplify and label the RNA for hybridization to Exon arrays starting with 100ng or 1μg of total RNA. We will demonstrate use of the 1μg protocol in this example (7). (Note 7) We have had good results with both protocols and also with the Nugen protocol which enables starting with much lower amounts of RNA as seen with LCM or flow sorted samples. Please remember that since data generated by each of these protocols are not directly cross-comparable, process all samples of a given study using the same protocol.
3.6. Synthesis of labeled cDNA and microarray hybridization
3.6.1. Ribosomal reduction of 1μg total RNA
Make serial dilutions of the GenChip PolyA controls (1:20; 1:50 and finally 1:50) using the polyA dilution buffer supplied with the kit. The final concentration of the polyA controls is 1: 50000 of the original stock
Add 1 part 5M betaine to 2 parts hybridization buffer supplied in the Invitrogen ribominus kit (162 μl/sample).
Aliquot 1 μg total RNA in RNAse free tube. The total volume of the sample should not exceed 3.2 μl. Add 2 μl of the diluted poly A controls to the sample
Prepare a master mix composing of 1 μl ribominus probe (100pmol/μl) and 30 μl of the betaine buffer per reaction. Add this to the tube from step 3. Incubate at 70 °C for 5 min and then place on ice.
Resuspend the bottle containing the magnetic beads by flicking it until no sediment is seen at the bottom. Aliquot 50μl of the resuspended bead solution per reaction to a fresh tube. Add 50 μl of RNAse-free water, briefly spin and place the tube on the magnetic stand for 1 min. Gently aspirate and discard the supernatant. Repeat this wash again with 50 μl l water and then resuspend the beads in the hybridization buffer with betaine prepared in step 2. Spin briefly and place on the magnetic stand. Aspirate and discard the supernatant. Resuspend the beads in 20 μl hybridization buffer with betaine and incubate at 37 °C for 10 min mixing once during incubation.
Transfer the cooled sample mix from step 4 to the bead suspension from step 5. Mix gently and incubate at 37 °C for 10 min, mixing once during incubation. Place on the magnetic stand and aspirate the supernatant into a clean labeled tube. Add 50 μl of hybridization buffer with betaine to the beads, mix, place on magnetic stand and aspirate the supernatant and combine into the previously labeled tube. The total volume of this rRNA reduced sample is approximately 100 μl.
Add 350 μl of cRNA binding buffer (containing ethanol) from the GenChip IVT cRNA cleanup kit, to each rRNA reduced sample. Vortex and then add 250 μl of 100% ethanol to each reaction. Mix well and apply the sample to the IVT cRNA cleanup column. Centrifuge 15 sec at 8000g, transfer column to a fresh tube, add 500 μl cRNA wash buffer and centrifuge again for 15 sec at 8000g. Discard the flow through, add 500 μl of 80% ethanol to the column and spin again for 15 sec at 8000g. Discard the flow through, open the column cap and centrifuge for 5 min at 20000g with the cap open. Transfer the column to a fresh tube and add 11 μl of RNase-free water directly to the membrane. Spin at 20000g for 1 min to elute the rRNA reduced total RNA/Poly A RNA control mix.
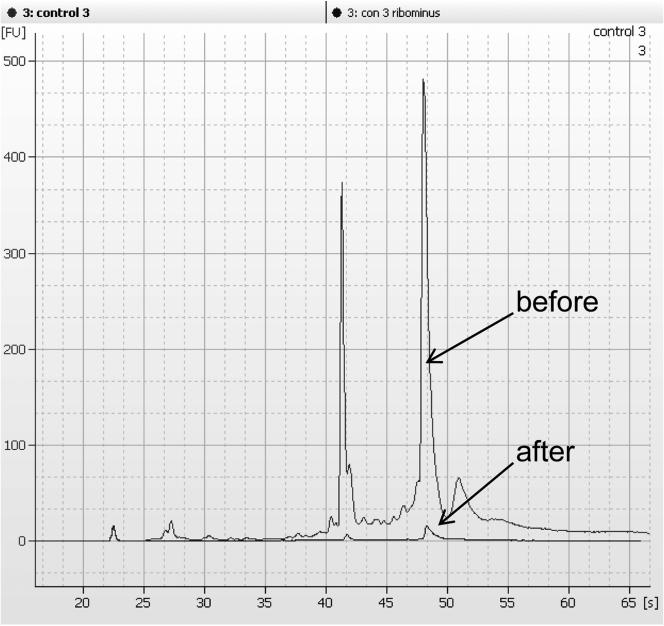
Check the sample from step 7 on a bioanalyzer to ensure that ribosomal peaks are reduced in the sample. We typically see greater than 80% reduction after this protocol (See Fig 1). Samples with less than optimal reduction may be subjected to an additional ribosomal reduction step.
Figure 1.
Electropherogram traces of total RNA before and after ribosomal reduction
3.6.2 Synthesis of labeled cDNA
Prepare a 1:5 dilution of the supplied T7-(N)6 primers and add 1 μl of the diluted solution to 4 μl of the rRNA reduced total RNA/Poly A RNA control mix from Step 3.6.1.8. Flick the tube to mix, spin down and incubate 5 min at 70 °C followed by 2 min at 4 °C. Place on ice.
Prepare the double stranded cDNA using the GeneChip WT cDNA synthesis kit as per the manufacturer's protocol (see Note 8)
This is then converted to complimentary RNA (cRNA) by in vitro transcription using the GeneChip WT cDNA amplification kit as per the manufacturer's protocol. This protocol should yield at least 15 – 30 μg cRNA
This cRNA (10 μg) from the first cycle is then reverse transcribed back to cDNA using random primers and a 10 mM nucleotide mix containing dNTP and dUTP. Typical yields of the sense DNA is in the range of 6-7.5 μg.
The uridylated single stranded cDNA (5.5 μg) is then fragmented using Uracil DNA glycolase (UDG) and human apurinic/apyrimidinic endonuclease (APE 1). This procedure fragments the cDNA in a reproducibly at locations where dUTP is incorporated in the DNA during the second-cycle first-strand reverse transcription step.
The fragmented cDNA is end labeled using terminal deoxynucleotidyl transferase (TdT) and the kit supplied DNA labeling reagent that is covalently linked to biotin.
3.6.3 Hybridization and scanning
The labeled cDNA (5.5 μg) is mixed with 20× eukaryotic hybridization controls, denatured and hybridized to Human Exon 1.0 ST arrays as recommended in the kit (Note 9).
After 18 hr hybridization, the arrays are subjected to a fluidics protocol that washes and stains the array with streptavidin phycoerithrin.
The stained arrays are then scanned in a GeneChip 3000G scanner and the data is exported as CEL files.
3.7 Analysis of Human Exon 1.0 ST array data
3.7.1 Gene level analysis
Traditional microarray analysis methods present a steep learning curve for the average user. The problem resides primarily in the normalization techniques used to distribute the signal intensities on the array. To obtain a robustly confident list of genes associated with a given condition, we use the iterPLIER algorithm as the probe intensity summarization method (6, 8). We have successfully used Agilent GeneSpring GX v10, Biotique X-ray as well as Partek Genomics software to analyze Exon array data.
We strongly recommend the use of replicates in the experiments using microarray technology for gene expression profiling. While we realize that these experiments can be cost prohibitive, confidence in that data from microarray experiments requires the use of at least 2-3 biological replicates. While generating preliminary data, one could resort to pooling of multiple samples to neutralize the biological variance however this could lead to loss of meaningful important data
After summarization, we routinely conduct a Principle Component Analysis to identify any outliers in the samples. We also evaluate the control spikes and hybridization metrics as described by Affymetrix (9)
Next we filter the data to exclude probesets that fall in the bottom 20th percentile for signal intensity and do not show good signal in all replicates of any given condition. This reduces the noise in the data and makes it manageable.
A statistical test (Students t-test or ANOVA) with a p-value <0.05 and a false discovery rate correction (Benjamini Hochberg or Bonforoni) routine is most appropriate at this step. The stringency of the statistics will determine how many differentially expressed targets are identified.
We further reduce the data by applying a filter on fold change of expression values between the two conditions. While a 2-fold cutoff seems to be used in many microarray experiments we prefer to use a 1.5 fold cut off. This enables us to have enough probe sets in our lists while performing secondary analysis such as Gene Ontology or Pathway Analysis.
3.7.2 Exon Splicing Analysis (core-level)
Exon array analysis can be done on multiple levels; core probe set (17,800 transcript clusters of RefSeq and full-length GenBank mRNAs); extended probe set (core + EST and partial mRNA-based annotation) and full probe set (262000 transcript clusters including extended + ab-initio gene predictions). The analysis describe here is based on core level probes.
For Exon level analysis, the summarized probeset values are filtered using the DABG (detection above background) algorithm and p-value (probeset) <= 0.05. For a transcript to be called as Present, a substantial number of core probe sets should be “Present” (as designated by the DABG generated p-value). The default value specifies 50% of core probe sets to be ‘Present’. The percentage of samples (within a condition) in which a gene must be present for it to be retained is set at 50% and can be increased for more stringency.
This is followed by a Splicing ANOVA with a p-value <0.05. This uses a gene-normalized intensity value i.e. ratio of probe set intensity to expression level of the gene.
A Splicing Index value is then calculated. This is similar to a fold-change filter where the gene normalized intensity values are compared between the two experimental conditions. For a given transcript, this difference is computed for each probeset; if any of the probesets has an absolute value difference greater than the specified threshold (0.5 by default) then the transcript will pass this filter.
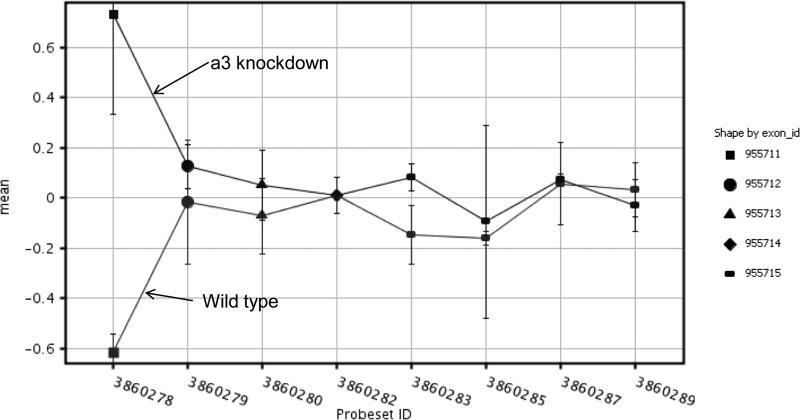
The list of probes that pass the above steps can then be visualized. See figure 2 for an example of alternative splicing in the 5'end of POLR2I.
Figure 2.
Signal intensities of exons for POLR2L from control and a3b1 knockdown samples show that exon 1 is alternatively spliced and expressed in the knockdown cells
The results of any microarray experiment should be verified using an independent technique such as quantitative PCR or sequencing. Additional functional analysis is also recommended.
Footnotes
All instruments, glassware and plastic-ware that touch cells or cell lysates should be certified DNase-free and RNase-free or should be pre-washed with RNase Zap (Ambion, cat. #9780; 9782) or RNase Away (Molecular BioProducts cat. #7001) followed by DEPC water and allowed to air dry.
The number of cells required for each microarray experiment can vary from cell type to cell type. Typically we utilize a 25 or 75 cm2 flask of confluent cells per condition. This corresponds to about 2-10×106 cells and provides enough material for both the microarray experiment as well as other validation and QC experiments.
If using this RNA for any miRNA analysis, AVOID the Qiagen cleanup step since it results in loss of small RNAs.
If limited in the amount of available sample, one can analyze the RNA via NanoDrop® and then recover material to use for BioAnalyzer runs.
Ambion and Affymetrix protocols and technical literature (and our experience) suggest that samples failing to meet either (or both) of these criteria may (or will) perform poorly in molecular techniques, which are based on reverse transcription followed by amplification. This is likely due to the interference of protein, carbohydrate, or phenolic contaminants on the reverse transcription process.
The Agilent BioAnalyzer is a preferable substitute to MOPS-formaldehyde agarose gel analysis due to the reduced sample required, increased sensitivity, and reduced exposure to toxic reagents.
The Agilent 2100 Expert software provides a RIN or RNA integrity number (8) for the RNA nano and pico assays (series II). It is recommended that this RIN number be between 7-10 if the RNA sample is to be used in a microarray experiment. We generally use the RIN number as a secondary QC criteria along with 260/280, 260/230 and 28S/18S ratios.
All the reagents for this protocol are supplied in the Affymetrix WT Sense Target Labeling and Control reagents kit (7). It is recommended that polyA RNA controls be spiked in to the starting RNA samples since this will allow to QC for any degradation occurring during the protocol. The signals from these spikes can also be used for normalization. We have also successfully used the NuGEN WT-Ovation Pico and the WT-Ovation Exon module to generate data from exon arrays.
It is recommended that hybridization controls be prepared from a master mix. The signal from the controls (bioB, bioC, bioD and Cre) can be used to qualitatively compare chips being hybridized over time.
References
- 1.Carpenter PM, Dao AV, Arain ZS, Chang MK, Nguyen HP, Arain S, Wang-Rodriguez J, Kwon SY, Wilczynski SP. Motility induction in breast carcinoma by mammary epithelial laminin 332 (laminin 5). Mol Cancer Res. 2009;7:462–75. doi: 10.1158/1541-7786.MCR-08-0148. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter PM, Wang-Rodriguez J, Chan OT, Wilczynski SP. Laminin 5 expression in metaplastic breast carcinomas. Am J Surg Pathol. 2008;32:345–53. doi: 10.1097/PAS.0b013e3181592201. [DOI] [PubMed] [Google Scholar]
- 3.Iyer V, Pumiglia K, DiPersio CM. Alpha3beta1 integrin regulates MMP-9 mRNA stability in immortalized keratinocytes: a novel mechanism of integrin-mediated MMP gene expression. J Cell Sci. 2005;118:1185–95. doi: 10.1242/jcs.01708. [DOI] [PubMed] [Google Scholar]
- 4.Lamar JM, Pumiglia KM, DiPersio CM. An immortalization-dependent switch in integrin function up-regulates MMP-9 to enhance tumor cell invasion. Cancer Res. 2008;68:7371–9. doi: 10.1158/0008-5472.CAN-08-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell K, Szekeres C, Milano V, Svenson KB, Nilsen-Hamilton M, Kreidberg JA, DiPersio CM. Alpha3beta1 integrin in epidermis promotes wound angiogenesis and keratinocyte-to-endothelial-cell crosstalk through the induction of MRP3. J Cell Sci. 2009;122:1778–87. doi: 10.1242/jcs.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8(4):R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Affymetrix GenChip Whole Transcript Sense target Labeling Assay Manual. :701880. http://www.affymetrix.com/support/downloads/manuals/wt_sensetarget_label_manual.
- 8.Xing Y, Kapur K, Wong WH. Probe selection and expression index computation of Affymetrix Exon Arrays. PloS one. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Affymetrix Alternative transcript analysis for Exon Arrays. 2005 white paper v 1.1 http://www.affymetrix.com/support/technical/whitepapers/exon_alt_transcript_analysis_whitepaper.pdf.