Abstract
Molecular mechanisms leading to high level drug resistance have been analyzed for the clinical variant of HIV-1 protease bearing 20 mutations (PR20); which has several orders of magnitude worse affinity for tested drugs. Two crystal structures of ligand-free PR20 with the D25N mutation of the catalytic aspartate (PR20D25N) revealed three dimers with different flap conformations. The diverse conformations of PR20D25N included a dimer with one flap in a unique “tucked” conformation; directed into the active site. Analysis of molecular dynamics (MD) simulations of the ligand-free PR20 and wild-type enzymes showed that the mutations in PR20 alter the correlated interactions between two monomers in the dimer. The two flaps tend to fluctuate more independently in PR20 than in the wild type enzyme. Combining the results of structural analysis by X-ray crystallography and MD simulations; unusual flap conformations and weakly correlated inter-subunit motions may contribute to the high level resistance of PR20.
Keywords: HIV-1 protease, drug resistance mutations, flap dynamics, resistance mechanisms, X-ray crystallography, molecular dynamics simulations, coordinated motions
Graphical Abstract
Introduction
Human immunodeficiency virus type 1 (HIV-1) protease (PR) is an important therapeutic target for HIV/AIDS. HIV-1 PR is a retroviral protease with an essential role in viral replication by cleaving the viral polyproteins Gag and Gag-Pol into individual enzymes and structural proteins [1]. HIV PR is a small enzyme; acting as a dimer of two 99-residue subunits; and is tractable for structural and computational analyses. Potent antiviral agents have been designed using crystal structures of PR-inhibitor complexes [2; 3] and structure-based computational techniques [4–6].
Currently; HIV/AIDS patients receive highly active antiretroviral therapy (HAART) using combinations of different drugs. PR inhibitors are more effective than reverse transcriptase inhibitors in preventing cell-free and cell to cell transmission of the virus [7]. Drug resistance; however; is a critical barrier to successful treatment. HIV rapidly evolves drug resistance because of the lack of proof reading by the viral reverse transcriptase; in addition to the fast replication of the virus [8]. Thus; compliance to HAART is critical to alleviate the emergence of resistance [9; 10]. Drug resistant viral strains are common in infected patients; and to date; 36 missense mutations have been associated with resistance to one or more of the nine FDA approved PR inhibitors [11]. The challenge of drug resistance is compounded by the existence of many possible combinations of mutations in PR. Highly resistant mutants often have 20 or more mutations and strategies for developing effective inhibitors are reviewed in [12].
We are studying a highly drug resistant variant of PR from a clinical isolate; which bears 20 mutations (PR20) [13]. Compared to wild-type enzyme; PR20 showed drastically worse affinity of greater than three orders of magnitude for current drugs [14]. Additionally; PR20 had a higher dimer dissociation constant (kd) and about 10-fold lower catalytic efficiency (kcat/Km) for cleavage of peptide substrate [14]. The stability of the core monomer structure was enhanced for PR20 relative to the wild type enzyme [15]. Crystallographic analysis on PR20 showed altered intersubunit interactions and an unusually wide separation of the two flexible flaps in the absence of inhibitors [16]. We have pioneered studies of PR20 as a highly resistant mutant for evaluation of new inhibitors. Recently; we reported two promising antiviral compounds with 10-fold better inhibition of PR20 than seen for the potent drug; darunavir (DRV) [17]; including a fluorinated compound with improved brain penetration [18].
The active site cavity in the PR dimer is capped by two flaps (residues 45–55 in each subunit); which have a key role in the proteolytic reaction. The two flaps exist in a closed conformation when inhibitor or substrate is bound; while in the apo-enzyme; the flaps can exhibit an open conformation with the flap tips further from the catalytic site. Three categories; defined as closed; semi-open; and open conformations; have been identified in different structures of the apo-enzyme [19]. The relationship between drug resistant mutations and flap dynamics is complex. Recently; for example; the effects of single mutations V32I and M46L were studied by molecular dynamics in [20]. However; the observation of widely separated flaps in the structures of drug resistant mutants such as PR20 [13] and MDR769 [21] suggests that altered flap flexibility contributes to resistance. Here; we have explored the conformational variation for the PR20 mutant with the inactivating D25N mutation (PR20D25N). The D25N mutation is commonly introduced to prevent autoproteolytic degradation when studying the protease structure. Introduction of the D25N mutation into wild-type PR in experimental systems increased the equilibrium dimer dissociation constant by more than 100-fold and decreased the binding affinity of DRV by about 106 fold; although no significant alteration was observed in the inhibitor-bound dimer structure [22].
Crystallographic analysis was complemented by molecular dynamics (MD) simulations with AMMP [23; 24] in order to understand the major effects of the mutations in PR20 on the structure and dynamics in the absence of inhibitor. The effect of the D25N mutation was evaluated in the MD simulations for comparison with the experimental studies. AMMP is the molecular mechanics and dynamics engine of VegaZZ [25] and as part of the SPEC2000 benchmark has undergone extensive professional code review [26]. It is designed to be a computational backend that is plugged into other programs. In tests on small molecule benchmarks; the current SP5 and Tuna potential sets compare well to the CHARMM and AMBER sets [27]. We have used AMMP molecular mechanics and MD simulations to investigate substrate binding to HIV protease [23; 28]; and the effects of drug resistant mutants on inhibitor binding [29; 30].
Our new crystal structures of ligand-free PR20D25N exhibited various flap conformations. In particular; an unusual “tucked” conformation with one flap penetrating into the active site cavity was observed in one crystal structure; and a similar conformation also occurred in the MD simulations of the dimers. Extreme flap conformations were found for PR20D25N relative to wild type enzyme in both the crystal structures and the MD simulations; consistent with the idea that increased flap mobility contributes to drug resistance in this highly resistant variant. Moreover; the intersubunit motions were less correlated in PR20 than in the wild type enzyme. These insights into the molecular mechanisms of resistance may assist in strategies to combat drug resistant HIV infections.
MATERIALS AND METHODS
Preparation of PR20 with D25N mutation
Plasmid DNA encoding PR (subtype B of group M) with 20 mutations Q7K; L10F; I13V; I15V; D30N; V32I; L33F; E35D; M36I; S37N; I47V; I54L; Q58E; I62V; L63P; A71V; I84V; N88D; L89T and L90M (termed PR20) [16] cloned between the Nde1 and BamH1 sites of pET11a vector (Novagen; San Diego; CA) was used to introduce the D25N mutation by the Quick-Change mutagenesis kit (Stratagene). This construct (PR20D25N) was transformed into E. coli BL-21 (DE3; Stratagene) for protein expression; purification; and folding as described [16].
Protein crystallization; X-ray data collection and structure determination
Protein crystals were grown by the hanging drop vapor diffusion method. Crystals of PR20D25N were obtained by mixing 1 µL of protein (2.1 mg/mL) and 1 µL of reservoir solution under different conditions for the two crystal forms: 0.2M magnesium chloride and 20% PEG 3350 at pH 5.9; and 0.9M sodium chloride and 0.2 M sodium acetate at pH 4.8. The crystals were cooled in a mixture of the mother liquor and 30% glycerol for X-ray data collection.
Diffraction data were collected at 100 K on beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source; Argonne National Laboratory. The data were integrated and scaled with HKL2000 [31]. The structure designated PR20D25N open was solved by molecular replacement with the wild-type HIV-1 PR in complex with p1/p6 (2AOI) as the starting model by PHASER [32]. The PR20D25Nopen structure was refined using Refmac5.5 with TLS [33; 34]; and the model building was carried out in COOT [35]. The second crystal structure of PR20D25N was solved by PHASER using 3UCB as the starting model [16; 32]; and CNS and Refmac5.5 were used for refinement including anisotopic B factors [34; 36]. This crystal structure contains two PR20D25N dimers in the asymmetric unit. The flaps in both dimers were deleted initially and rebuilt during refinement. The solvent molecules were identified from the shape of the electron density; and interatomic distances and angles consistent with hydrogen bond interactions with other molecules. Secondary-structure matching (SSM) was used for superposition of protein structures [37]. Molecular figures were prepared with PyMOL [38].
The coordinates and structure factors for the two new crystal structures have been deposited in the Protein Data Bank for release upon publication. The accession numbers are 4Z50 for PR20D25N (with two dimers in the asymmetric unit) and 4Z4X for PR20D25Nopen.
Molecular dynamics simulations of wild-type PR and PR20
The starting models for MD simulations were the DRV complexes of PR20 (3UCB) [16] and wild type PR (2IEN) [39] with resolutions of 1.4Å and 1.3Å; respectively. DRV was removed from the starting models for MD runs termed PRWTMD and PR20MD. A second set of simulations termed PRD25NMD and PR20D25NMD was prepared by substituting asparagines for the catalytic aspartic acids. Water molecules from the crystal structures were included in the MD simulations. Furthermore; each dimer was solvated with 50 sodium ions; 50 chloride ions and about nine thousand water molecules randomly generated to fill the free space within a 10Å shell of the protein. The overall system was neutral. Simulations were performed with a NVT ensemble (atom number (N); volume (V) and temperature (T) are fixed) and an amortized fast multipole algorithm for the calculation of non-bonded terms. All atoms and interatomic forces were included in the non-bonded terms without the use of a cutoff radius. The amortization parameter (the mxdq parameter in AMMP) was 0.75Å; the long range forces were calculated when any atom moved more than 0.75Å. The initial box size for the fast multipole expansion (mmbox parameter in AMMP) was 10Å; and the volume of the system was constrained to a sphere of 36Å radius (bbox parameter in AMMP) using a detailed balance Markov chain. The temperature was set to 300K and constrained with the Nose algorithm as implemented with a stiffly stable predict and correction integration in AMMP [23; 24; 30]. These constraints correspond to a mean pressure of approximately 1 atmosphere. The program; AMMP; was used for the MD simulations with the Tuna potential set [40]; a modification of the SP5 set described in [27].
At least one of the four carboxylate oxygen atoms of Asp25 and Asp25’ is expected to be protonated due to their proximity in the dimer. The correct protonation state is expected to produce the highest affinity of PR for inhibitor [41]. Calculations on PRWT (PDB 3NU3) complexed with amprenavir [42] showed the lowest internal energy (ΔU = ΔUPI+WAT; where ΔUPI+WAT is the potential energy for protease-inhibitor complex and solvent) when the Oδ2 of Asp25 and Oδ1 of Asp25’ were protonated. This protonation state agrees with the 20ns MD simulations performed by AMBER and analyzed by the Poisson-Boltzmann surface area (MM-PBSA) method [43]. This protonation state was used for the MD simulations. Initially; the system was equilibrated for the randomly generated solvent molecules prior to MD simulation; and the substrate binding site was visually checked to ensure full occupation by water molecules. A total of 1000 frames; one for every 10 ps; were saved from the 10 ns simulation. Each frame was superimposed on the starting model to remove rotational and translational motion from the conformations before calculating the RMSD values and averaged structure.
Trajectories of the MD simulations are represented by a large number of snapshots; and therefore; two statistical analyses were applied to the simulations. The major conformations of the protein were extracted using k-means clustering; and the individual variation was analyzed with a cross-correlation coefficient. The optimal number of clusters was decided by evaluating within the group sum of squares [44]; and trajectories were partitioned by applying k-means algorithm that recursively assigns data points to its nearest centroid until the cluster converges [45]. The cross-correlation coefficient obtained by calculation of displacement vector of any two Cα atoms i and j by ; where Δri is a vector representing the displacement of the atom for every frame from the mean position of the atom derived from superimposing all of the frames [46]. Thus; the Dynamic Cross-Correlation map (DCCM) shows the parts of the molecule that tend to move in the same directions (positive correlation) or the opposite directions (negative correlation) over the entire trajectory. The DCCM for an unstructured system; for example the trajectory of a gas or fluid; would be a diagonal line reflecting the unit correlation of each atom with itself and zero everywhere else. The features in the DCCM reveal the degree to which the motions of the system are non-random. With a thousand frames; the Student T-test indicates statistical significance for correlations with a magnitude larger than 0.063. The DCCMs were plotted by gnuplot 4.6 for each simulation.
RESULTS
Crystal structures of ligand-free PR20D25N
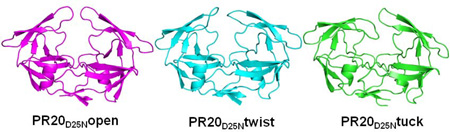
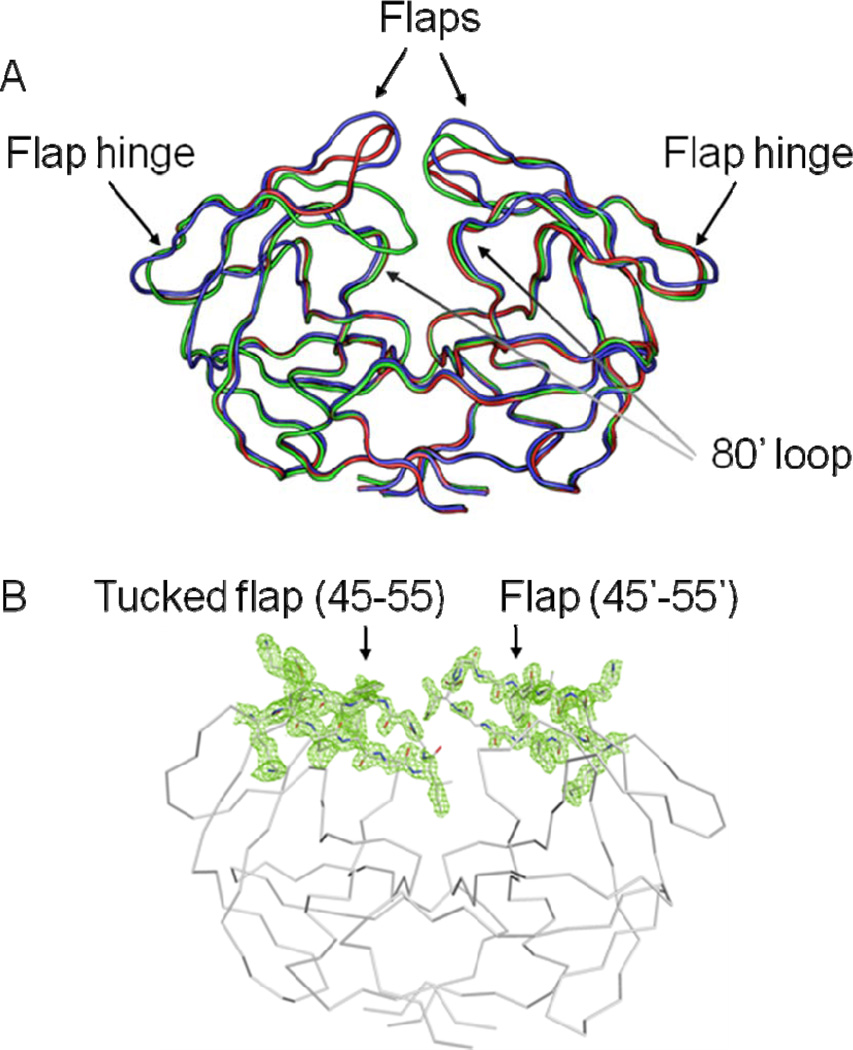
Two crystal structures were solved for ligand-free PR20D25N comprising three distinct dimers with varied flap conformations. The two crystal structures were refined to resolutions of 1.75 and 1.45Å with R-factors of 19.3% and 13.8%; respectively; as summarized in the crystallographic statistics shown in Table 1. One structure termed PR20D25Nopen was refined with one dimer (residues 1–99 and 1’–99’) in the asymmetric unit (Figure 1A). The second crystal structure contained two dimers per asymmetric unit exhibiting different flap conformations: one dimer (PR20D25Ntwist) showed twisted flap conformations relative to those of PR20open; and the other dimer (PR20D25Ntuck) had one flap tucked or inserted into the active site cavity and the opposite flap raised out of the cavity. The electron density map for the flaps of PR20D25Ntuck is shown in Figure 1B.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| PR20D25N Open | PR20D25N (2 dimers) |
|
|---|---|---|
| Space group | P41 | P 1211 |
| Unit cell dimensions: (Å) | ||
| a b c |
45.52 45.52 104.14 |
54.15 48.57 69.68 |
| α = β = γ =90 | α = γ = 90 β = 99.15 |
|
| Resolution range (Å) | 41.71-1.75 | 50-1.45 |
| Unique reflections | 21;444 | 59;088 |
| Rmerge (%) overall (final shell) | 6.5(41.4) | 4.9(31.6) |
| I/σ(I) overall (final shell) | 17.9(4.5) | 27.3(4.2) |
| Completeness (%) overall (final shell) | 97.3 (98.6) | 93.0 (65.2) |
| Data range for refinement (Å) | 41.71-1.75 | 45.9-1.45 |
| R (%) | 19.3 | 14.0 |
| Rfree (%) | 23.4 | 19.2 |
| No. of solvent atoms (total occupancies) | 110 (110) | 417 (415.5) |
| RMS deviation from ideality | ||
| Bonds (Å) | 0.027 | 0.026 |
| Angle distance (degrees) | 2.125 | 2.039 |
| Average B-factors (Å2) | ||
| Main-chain atoms | 11.7 | 14.6 |
| Side-chain atoms | 15.7 | 18.6 |
| Solvent | 17.2 | 30.9 |
Figure 1.
(A) Superposition of ligand-free dimers of PR20D25Nopen (blue); PR20D25Ntwist (red) and PR20D25Ntuck (green) showing the different flap conformations. The protein has the following mutations relative to the standard sequence for HIV-1 subtype B of group M: Q7K; L10F; I13V; I15V; D25N; D30N; V32I; L33F; E35D; M36I; S37N; I47V; I54L; Q58E; I62V; L63P; A71V; I84V; N88D; L89T and L90M. (B) 2 Fo-Fc electron density map contoured at 1 σ.level for the flap region in the crystal structure of PR20D25Ntuck.
Overall RMSD values calculated for Cα atoms are: 1.5 Å (0.9 Å) for comparison of the PR20D25Nopen and PR20D25Ntwist dimers; 1.1 Å (0.8 Å) for PR20D25Nopen and PR20D25Ntuck; and 1.4 Å (0.3 Å) for PR20D25Ntwist and PR20D25Ntuck. [Values inside parentheses were calculated without the flap residues.] The largest differences appear in the flaps of the three dimers. The structures of the PR20D25Nopen and PR20D25Ntwist dimers are in the category designated as open conformation [19]; while PR20D25Ntuck shows a novel conformation of the flap; which has not been described in previous crystal structures.
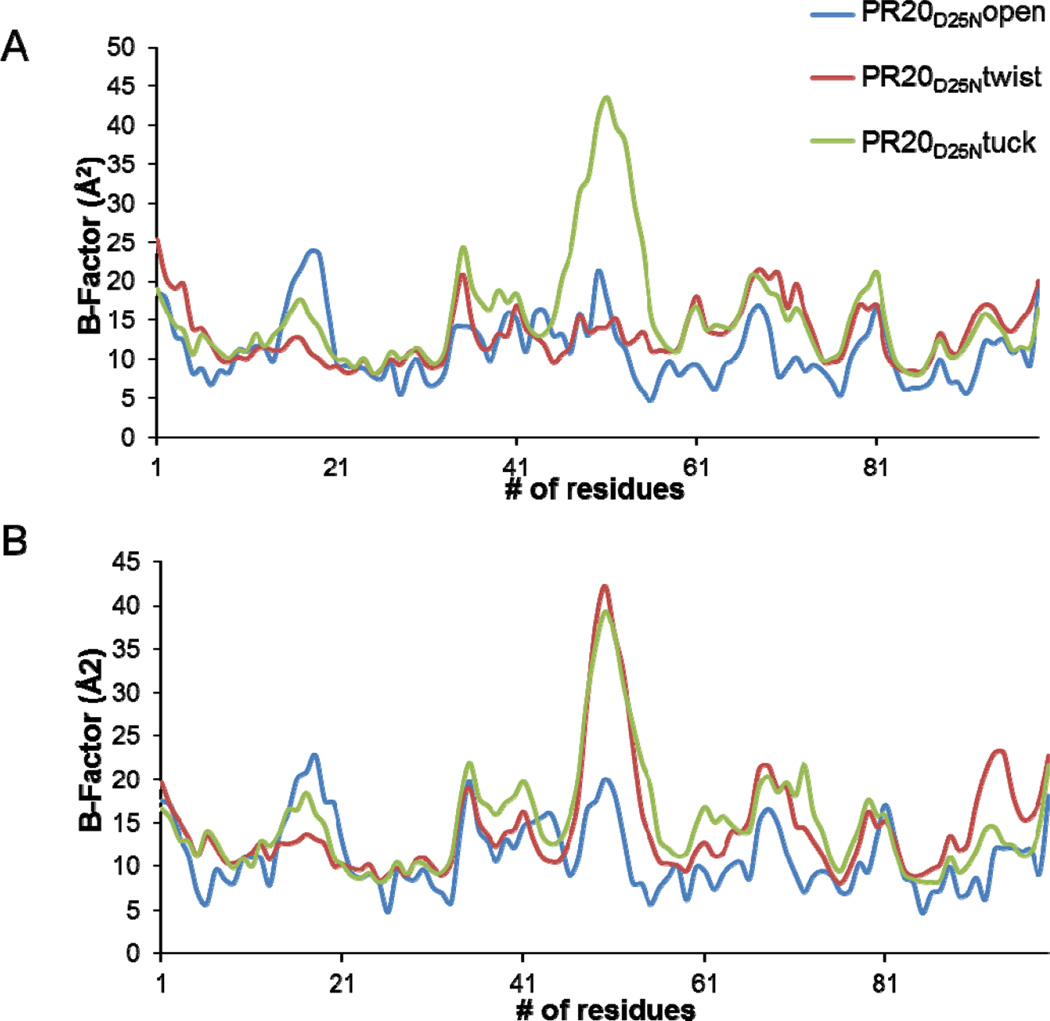
The positional disorder in the backbone of each dimer was assessed by plotting the B-factor averaged over the main chain atoms per residue (Figure 2). The highest variation among the structures was seen for the flap residues. The largest B-factors of about 40 Å2 occurred in one flap (residues 45’–55’) in the PR20D25Ntwist dimer (Figure 1B) and in both flaps of the PR20D25Ntuck structure; while the flap residues in the PR20D25Nopen dimer had the maximum B-factor of about 20 Å2. The B factors showed similar peaks in both subunits of the three structures at the N- and C-termini in the dimer and the variable surface loops of residues 33–38; 65–70; and 76–83.
Figure 2.
B values averaged over the main chain atoms of each residue for PR20D25Nopen (blue); PR20D25Ntwist (red) and PR20D25Ntuck (green). Upper panel: residues from subunit A. Lower panel: residues from subunit B.
Flaps exhibit diverse conformations
The flap conformations in PR20D25Nopen and PR20D25Ntwist were compared to the closed and open conformations for wild type PR and the PR20 mutant [19]. In the PR20D25Nopen structure; the tips of the flaps are separated widely and raised vertically compared to the flaps in the closed conformation (Figure 3A). No van der Waals contacts occur between the atoms of the two flaps. The minimum separation of 7.5 Å occurs between the carbonyl oxygen of Ile50 and the Cγ2 atom of Ile50’ at the tips of the two flaps. Besides losing contacts between the two flaps; Ile50 also lacks van der Waals interactions with Pro81’; which are generally retained in both open and closed conformation dimers [21]. An example of the van der Waals interactions of Ile50 with Pro81’ in the closed conformation of PR20 is shown in Figure 4A. In PR20D25Nopen; the shortest distances between the side chain atoms of Ile50/Ile50’ and Pro81’/Pro81 are 4.7 Å and 4.0 Å; respectively.
Figure 3.
Comparison of the dimers of PR20D25N with wild type PRopen and PR20 in open and closed conformations. (A) PR20D25Nopen (blue); PR20D25Ntwist (red) and PRopen (pink). (B) PR20D25Ntwist (red) and PR20open (cyan). The closed conformation of PR20 with bound APV PR20/APV(green) is shown in both panels as a reference.
Figure 4.
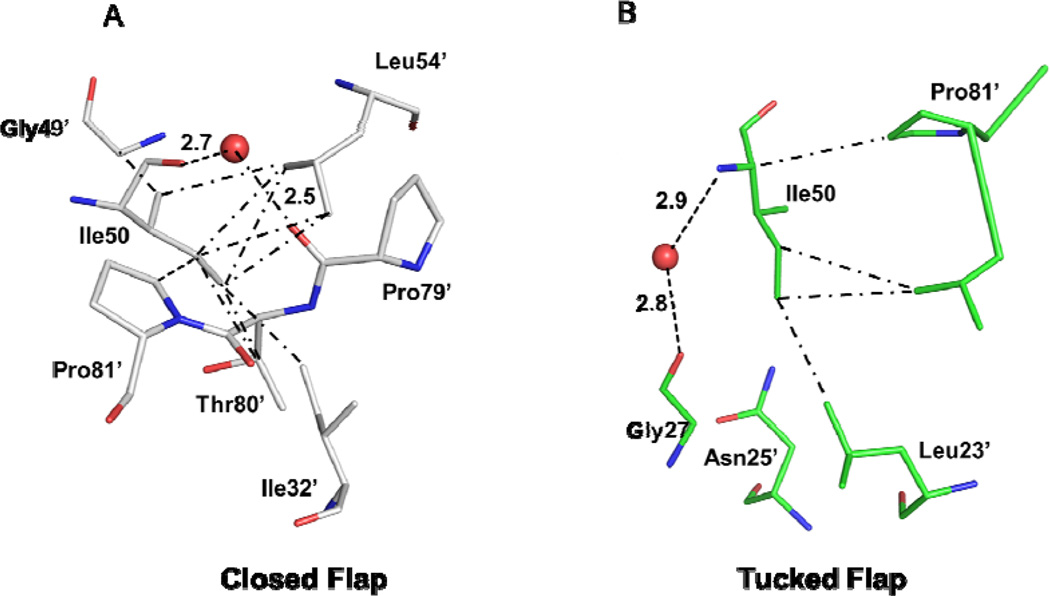
Interactions of Ile50 with neighboring residues of PR20. (A) The closed conformation observed in PR20 in complex with inhibitor GRL02031 (4J55). The protein is shown with gray carbons. (B) The tucked conformation flap of PR20D25Ntuck with green carbons. Water molecules are shown as red spheres. Hydrogen bonds are indicated by dashed lines and van der Waals contacts in dot-dash lines.
The PR20D25Ntwist dimer has the most widely separated flaps reported in any crystal structure to date (Figure 3). In PR20D25Ntwist; the flap tips lie nearly in the same plane but are further from the catalytic Asp25/25’; and almost perpendicular to their arrangement in the closed conformation. The shortest distance observed between the two flaps is 9.6 Å between the carbonyl oxygen atoms of Ile50 and Ile50’; and the intersubunit separations between Ile50 and Pro81’ and Ile50’ and Pro81 are 8.5 Å and 5.1 Å; respectively. The wide open flaps of PR20D25Nopen and PR20D25Ntwist show few direct interactions with the residues near Asp25/25’ at the base of the active site cavity.
Several water molecules occupy the active site cavity of the ligand-free dimers instead of inhibitor or substrate. In the closed conformation dimer; the water mediated hydrogen bond interactions connecting the carboxyl oxygen of Pro79’/Pro79 to the carboxyl oxygen of Ile50 and the amide nitrogen of Gly51’ have been proposed to stabilize the flaps (Figure 4A) [47]. In the structures of PR20D25Nopen and PR20D25Ntwist; the carbonyl oxygen of Pro79’/Pro79 retains a hydrogen bond interaction with water; however; the structures lack the interactions with Ile50 or Gly51’ because of the greater separation of the tips of the flaps.
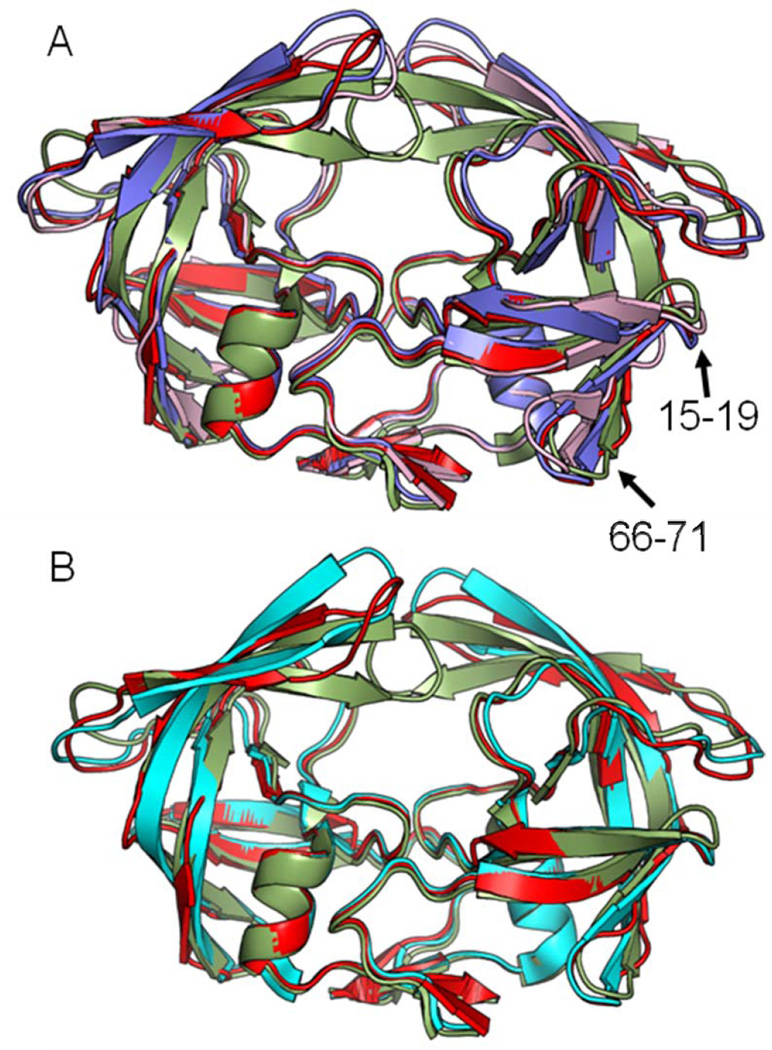
The unliganded PR20D25Nopen and PR20D25Ntwist structures can be compared to the open conformation of wild-type PRopen (2PC0) complex with a magnesium ion bound near the catalytic aspartates (Figure 3A) [48]. The dimers of PR20D25Nopen and wild type PRopen superimposed with an RMSD value of 0.9 Å for the Cα atoms. The main differences of PR20D25Nopen structure are observed at hinge loop; flap; residues 16 – 17 and 66 – 71 in both subunits. The tips of the flaps of PR20D25Nopen are 1.8 – 2.0 Å further from the catalytic residues than the tips of the flaps of PRopen. Comparison of the PR20D25Ntwist and wild type PRopen structures gives an overall RMSD value of 1.3 Å for the Cα atoms. Larger variations are observed at hinge loop; flap; 80` loop; residues 15 – 19 and 66 – 71 for both monomers. In addition; the tips of the flaps of PR20D25Ntwist are almost perpendicular to the tips of the flaps of wild type PRopen.
The structures of PR20D25Nopen and PR20D25Ntwist were compared to the wide open structure of PR20 (3UF3) with yttrium bound near the catalytic aspartates (Figure 3B) [16]. Although PR20D25Nopen and PR20open dimer structures were solved in two different space groups; no significant difference was apparent in the main chain conformation as indicated by the low RMSD value of 0.3 Å for Cα atoms. The PR20D25Ntwist dimer is more different from the PR20open dimer; as shown by the 1.2 Å RMS value. The major differences occur for residues in the surface loops: the flap hinge; the 80’s loop and the flap.
Unusual tucked flap conformation
The flap conformation of the PR20D25Ntuck structure (Figure 1) does not belong to the categories defined as closed; semi-open; and open [19]. In the closed conformation; the flap tips interact closely (Figure 3A). The main chain carbonyl oxygen of Ile50 forms a hydrogen bond interaction with the amide of Gly49’ in the adjacent flap. Instead; the two flaps of PR20D25Ntuck exhibits asymmetrical conformations with one flap in an open conformation and the other in a unique conformation. The tip of the flap tucks or inserts into the active site cavity so that Ile50 lies next to the two catalytic Asp25/25`; while the other flap is directed away from the catalytic residues (Figure 1A). In the typical closed conformation; the side chain atoms of Ile50 form van der Waals contacts with the side chains of Val32’; Val54’; Thr80’ and Pro81’ (Figure 4A). In PR20D25Ntuck; Ile50 in the tucked flap forms completely different hydrophobic interactions with the carbonyl oxygen of Gly27; the side chain atoms of Leu23’; Asn25’; and Val82’; while water mediated hydrogen bond interactions connect the amide nitrogen of Ile50 and the carbonyl oxygen of Gly27 (Figure 4B). The typical water-mediated interactions linking the 80’s loop residues and the tips of the flaps are missing in the PR20D25Ntuck structure. The lack of stabilizing interactions with the rest of the protein is consistent with the asymmetric conformations observed for the two flaps; however; the flap tips show a van der Waals contact of 4.2 Å between Cγ1 of Ile50’ and the carbonyl oxygen of Gly51.
Molecular Dynamics simulations
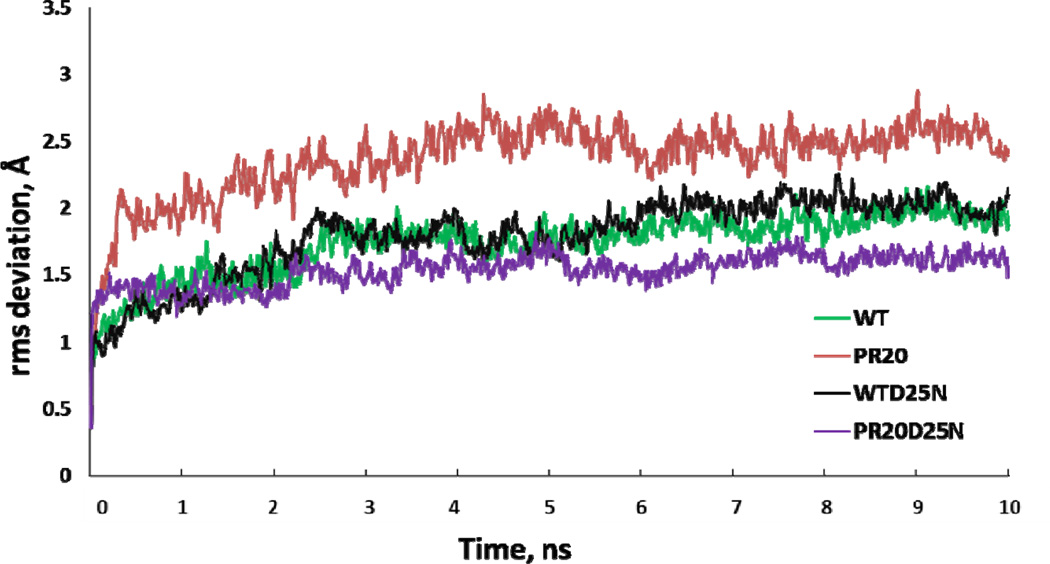
In parallel with the crystallographic analysis; MD simulations were performed to assess the conformational dynamics of PR20 and wild type PR dimers in the absence of bound inhibitor. Simulations were run on the catalytically active enzyme forms with Asp25 and 25’ (PR20MD and PRMD); and on a second set incorporating the active site mutation D25N (PR20D25NMD and PRD25NMD) for comparison with the new crystal structures reported here. In order to maintain consistent initial structures; the four simulations started from the closed conformation of the dimer after removing inhibitor; and ended after 10 ns. The trajectories of the RMS deviations from the initial model are shown in Figure 5. Simulations show rapid equilibration within the first 500 ps followed by a longer time evolution. PRMD and PRD25NMD equilibrated to similar RMS values of 1.7 ± 0.2 Å and 1.8 ± 0.3 Å; respectively. The PR20MD simulation equilibrated to the highest RMS deviation of 2.4 ± 0.3 Å; while PR20D25NMD gave the lowest RMS value of 1.5 ± 0.1 Å. The scale of the fluctuations is similar to the range described in previous simulations of HIV-1 protease and its mutants by other groups [20; 49–51].
Figure 5.
Trajectories of the MD simulations. The time course is plotted for the RMSD calculated by superposing Cα atoms of the dimer at each time point with the corresponding atoms in the starting crystal structure. The simulations are shown for PRMD (green); PR20MD (red); inactive PRD25NMD (black) and PR20D25NMD (purple).
Cluster Analysis of simulations
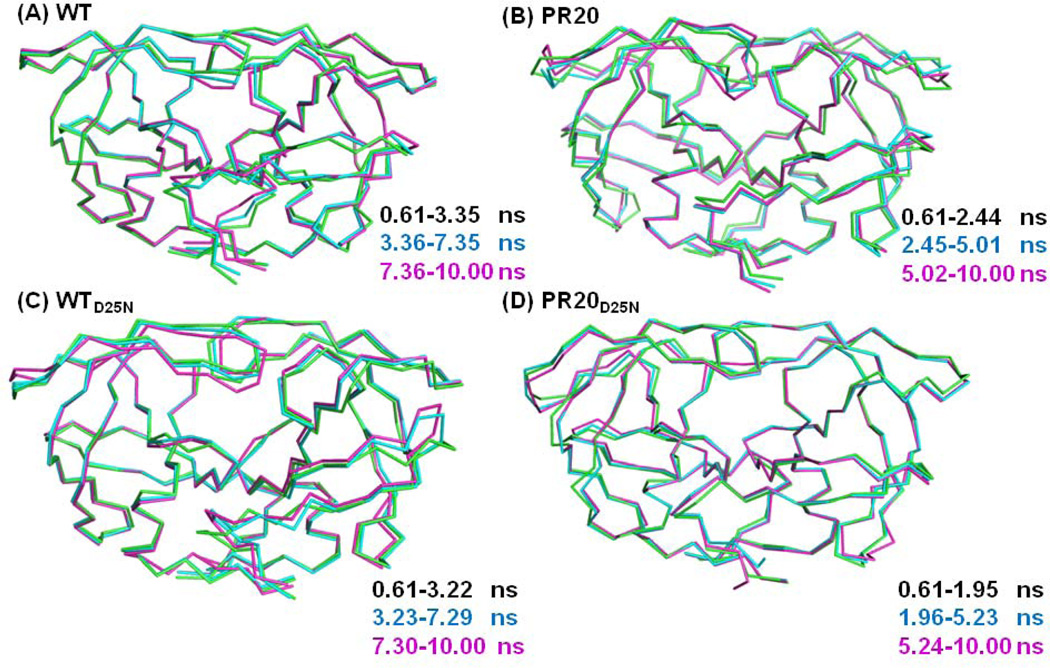
Clustering is a data-mining technique that partitions geometrically closer conformations into disjoint sets. Thus; conformational information from relatively long trajectories is simplified into a small conformational space and the comparison between clusters provides insight into protein flexibility. The major conformations of the protein were extracted using k-means clustering on the frames 61–1000 from the 10 ns simulations; after removing the first 60 frames of the run. Each trajectory was well described with only three clusters. A similar number of clusters was obtained from a visual inspection of the dendrogram drawn from hierarchical cluster analysis using the R package Bio3D [52].
The averaged structures for each cluster were superimposed and displayed in Figure 6. Generally; the main chain structures are similar to their corresponding starting models; with the exception of the flaps and other surface loops; which display a larger range of conformations. The features of the tertiary structure of the dimer are preserved over the simulations with good agreement for the elements of regular secondary structure; such as β-strands between the surface loops of residues 9–15; 33–42; and 65–72 and the alpha helix at residues 86–94. The tips of the flaps move toward the catalytic dyad in all the simulations; in spite of the presence of water in the active site cavity of the initial structure. The antiparallel β-sheet structure of the flaps and conformation of the hinge loop (residues 34−43) are preserved in all clusters of the simulations for PRMD; PRD25NMD and PR20D25NMD. In contrast; for the mutant PR20MD; the β-sheet at the tip of one flap is lost during the simulation; as shown in clusters 2 and 3 (Figure 6). The hinge loop and residues 14’–17’ of PR20MD also vary in conformation among the three clusters. Differences between the clusters occur for the residues at the N- and C- termini of PRMD; PRD25NMD and PR20D25NMD.
Figure 6.
Superposition of averaged structures of each cluster calculated for the simulations: (A) PRMD; (B) PR20MD; (C) PRD25NMD and (D) PR20D25NMD.
In summary; the wild type PR and PR20 simulations cluster into similar conformational spaces. The tips of the flaps move from the closed conformation toward the catalytic aspartates during the 10 ns simulations. In addition; the tip of the flap in one subunit of the PR20MD dimer loses the β-sheet conformation during the simulation; and this asymmetry is presumably due to the effects of the 20 mutations relative to the wild type PR.
Correlated motions extracted from MD simulations
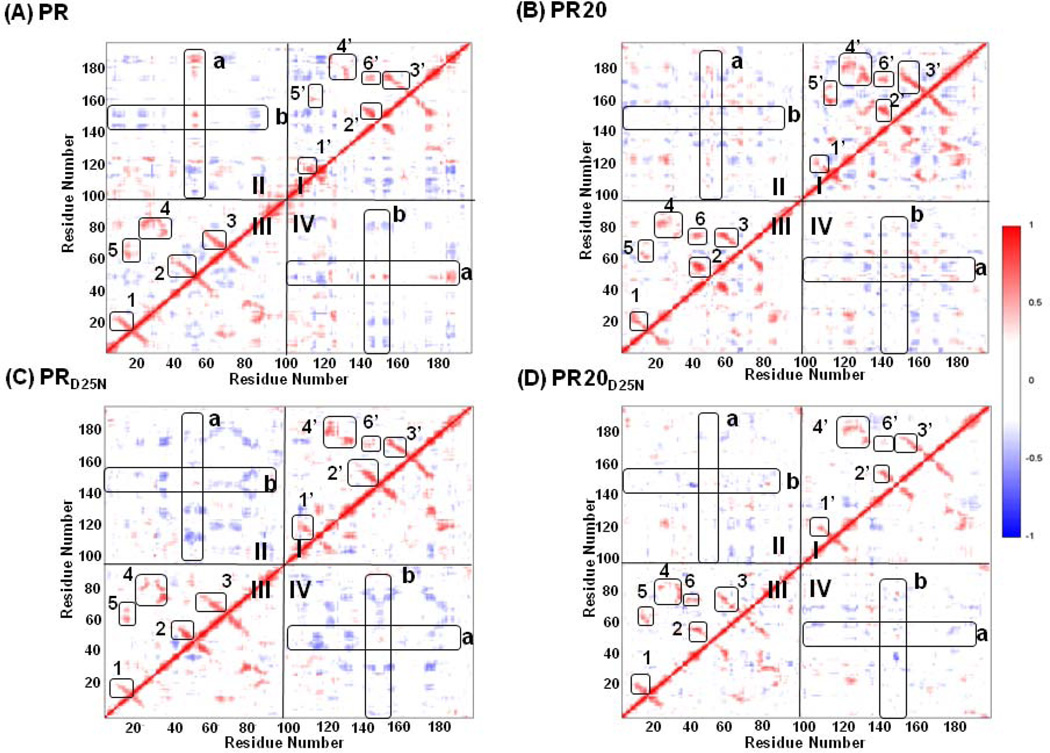
Dynamic fluctuations in the protein conformation are important in regulating protein function; and mutations can alter the protein dynamics [53; 54]. Therefore; the correlation coefficients of the atomic fluctuations were analyzed for each residue over time in order to identify the correlated motions and any change between the MD simulations for wild-type PR and PR20. Dynamical cross-correlation maps (DCCM) were drawn for each simulation (Figure 7). Assuming a null hypothesis of uncorrelated motions; the Student T-test indicates statistical significance for correlations with a magnitude larger than 0.063. Correlation coefficients of higher than 0.25 or lower than −0.25 are shown in the DCCM maps [46]; and the peaks for correlated motions are labeled in the maps. The peaks of correlated motions identified in the MD simulations of wild type PR and PR20 (Figure 7A and 7B) are notably larger than for the corresponding simulations of the inactive mutants incorporating D25N (Figure 7C and 7D). Our MD simulations agree with the NMR study showing that the single mutation of D25N altered the dynamic properties of the PR [22]. However; introducing the D25N mutation had no significant effect on the atomic positions or hydrogen bond interactions when comparing the crystal structures of PR and PRD25N dimers [22].
Figure 7.
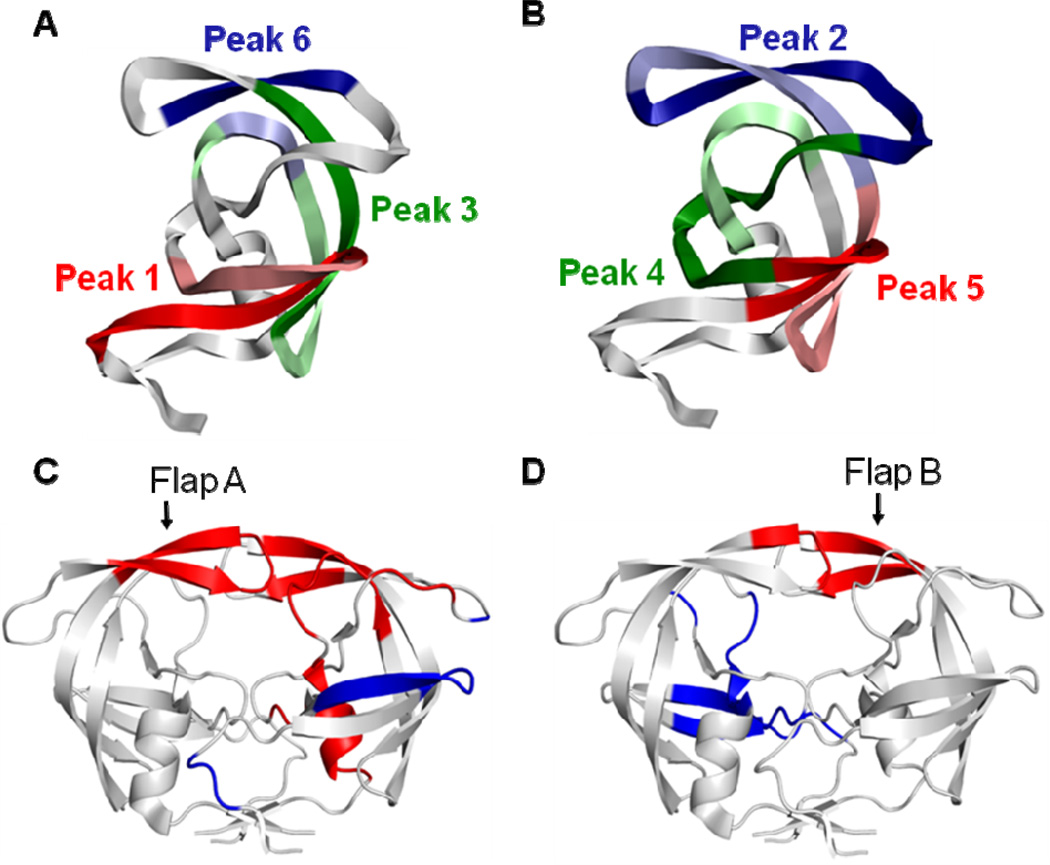
Dynamic Cross Correlation Maps show correlated motions in dimers: (A) PRMD; (B) PR20MD; (C) PRD25NMD and (D) PR20D25NMD. Cross-correlation coefficients Cij larger than 0.25 and smaller than −0.25 are shown in maps with the intensity represented as follows: red squares 0.25 <Cij< 1; blue squares −1 <Cij< −0.25. Quadrants I and III show intra-subunit correlated motions. Boxes 1; 2; and 3 correspond to surface turns between two beta-strands. Box 4 corresponds to two interacting loops within the active site cavity. Boxes 5 and 6 correspond to interactions between the flap residues and the surface loops. Quadrant II and the symmetry related quadrant IV show inter-subunit correlated motions. The boxes in these quadrants indicate the flap residues from the two monomers. The two flaps in both PR20 and PR20D25N show less correlation than is seen for the wild type protein.
Correlated motions identified in quadrants I and III of DCCM represent intra-subunit relationships and are consistent with observations from previous MD simulations [55; 56]. The labeled peaks correspond to structural elements; such as antiparallel beta strands; as indicated in Figure 8. The secondary structure patterns and the interface contacts have mainly positive correlations within each monomer; while anti-correlations exist between these regions. The major regions with positive correlations are illustrated in Figure 8A and 8B for the wild type PRMD. The peaks of correlated motions observed in the two monomers are similar but have different intensities; suggesting that each monomer has a similar pattern of dynamic motions. The second monomer of PR20MD (Figure 7b) shows the highest correlations; and more positive correlation than in the analysis of the simulation for wild type enzyme (PRMD) (Figure 7a); which is consistent with experimental studies suggesting a more stable monomer in PR20 compared to PR [15].
Figure 8.
Correlated motions mapped onto the protease structure. (A) and (B): Intrasubunit correlations from quadrants I and III of Figure 7 are shown in two panels to minimize overlap of residues. Six positively correlated peaks are labeled in the intra-subunit quadrants I and III of all subunits except that peak 5’ is absent from the second subunit of PR20D25NMD. Each peak defines two regions of residues represented as dark and light colors. The corresponding residues and colors are listed inside the parentheses: (A) Peak 1 (6–17 red and 15–25 light red); Peak 3 (56–67 green and 67–81 indicated in light green-light blue-light green due to overlap with residues of Peak 6); and Peak 6 (43–48 blue and 76–79 light blue). (B) Peak 2 (37–52 blue and 51–63 light blue); Peak 4 (21–36 green and 76–87 light green); and Peak 5 (12–21 red and 61–71 light red). (C) and (D): The peaks for intersubunit correlated motions in PRMD are also illustrated in two panels to avoid overlaps. Higher peaks occur for residues showing correlated motions with the two flaps (residues 45–55 in each subunit). Positively correlated motions are colored red; and anti-correlated motions are colored blue. (C) Residues 24’–25’; 35’–37’; 46’–48’; 53’–56’; 76’–82’; 84’–85’ and 87’–92’ have positively correlated motions with the flap residues 45–55 in the first subunit; and residues 4’–6’; 15’–22’ and 39’ show anti-correlated motions. (D) Residues 50–53 show positively correlated motion with the second flap (residues 45’–55’); and residues 7–13; 20–26; 34–35 and 81–86 have anti-correlated motions.
Correlated inter-subunit motions were identified in quadrants II and IV of the DCCM (Figure 7). Less correlation was observed between the motions of the two subunits compared to within each monomer. The inter-subunit quadrants have mostly negative correlations; suggesting that motions between two monomers have opposing directions. Correlated motions of the flaps with the active site cavity and peripheral residues have been reported in different MD simulations of wild-type PR [56; 57]. The major correlations of residues with the two flaps of PR are illustrated in Figure 8C and 8D. When the DCCMs are compared for the four MD simulations; the correlation coefficients in the inter-subunit quadrant are generally weaker in PR20MD and PR20D25NMD than in PRMD and PRD25NMD. In particular; the anti-correlated motions of the flaps (residues 45–55 in both subunits of the dimer) in the inter-subunit quadrant are weaker or disappear in PR20MD and PR20D25NMD; respectively. These motions suggest that the two flaps tend to fluctuate more independently in the PR20 mutant than in the wild-type enzyme; giving rise to asymmetrical conformations; as observed in the X-ray crystal structures. In summary; correlated motions are well preserved in each monomer; however; the cross communications between the two monomers are impaired in PR20 relative to wild type PR dimers.
DISCUSSION
The variant PR20 derived from a clinical isolate is highly resistant to the tested clinical inhibitors; which show several orders of magnitude worse affinity for PR20 compared to the wild type enzyme. Moreover; in contrast to the wild type precursor; saquinavir and darunavir do not inhibit autoprocessing of the mutant precursor containing PR20. Previous crystallographic studies of the PR20 dimer showed a large variation in the conformation of the flaps in the ligand-free structures. Here; we report two new flap conformations; designated twisted and tucked; in the dimers of PR20D25N crystallized in the absence of inhibitor. Three highly drug resistant variants PRP51; MDR769 and PR20 were observed to have widely separated flaps in their dimer structures; suggesting a common mechanism for resistance to inhibitors [16; 21; 58]; and new inhibitors have been designed to target the wide open flaps [59]. Among the new structures reported here; PR20D25Ntwist has the highest separation of the flap tips in the dimer. The rotation of the tips of the flaps in PR20D25Ntwist might further contribute to weaken the affinity for DRV and SQV. PR20D25Ntuck exhibits a unique flap conformation; which has not been reported previously for crystal structures of HIV PR. The tucked flap enters the active site cavity and can hinder the binding of substrates or inhibitors. Hence; this tucked flap provides a novel mechanism to lower the affinity for inhibitors.
The conformational dynamics of proteins contributes to their biological function; and correlated motions of domains regulate biological function [60; 61]. In the PR dimer; the substrate binding cavity is constructed by two identical monomers; and the cooperative opening and reclosing of the two flaps is critical during proteolysis of the natural substrate and binding of an inhibitor. The flaps exhibit diverse conformations in the crystal structures of ligand-free PR20 [16]. The new crystal structures reported here for ligand-free dimers of PR20D25N show additional conformational variation; including an unusual tucked form; where one flap is tucked inside the active site cavity. Isothermal titration calorimetry suggests that mutations in PR20 increase the stability of the monomer compared to wild type PR [15].
We performed 10ns MD simulations to analyze the dynamics of PR20 and wild type PR starting from the closed conformation for both the active protein and a commonly used inactive model system (D25N mutation). Cluster analysis of the MD simulations shows that the mutations in PR20 influence the conformational dynamics of the flaps. The individual monomers in the PR20 and wild type PR dimers exhibit similar conformational dynamics and correlated motions. The correlated motions between the two subunits of the dimer differ in the PR20 mutant and wild type enzyme. PR20 lacks strongly correlated motions between the flap and the other subunit in the dimer; which will tend to destabilize the flaps and promote asymmetrical structures; consistent with the diverse conformations observed in crystal structures of this mutant and the results from isothermal calorimetry. A 50ns simulation starting from the open conformation of the PR20 dimer suggested that apo-PR20 populates the open conformation more frequently than does the wild type enzyme because the loops were more variable in the PR20 than the wild type [51]. Neither study addresses the probability and time constants of the transition between open and closed conformations.
In summary; the mutations in PR20 induce subtle rearrangements in the structure that alter the conformational ensemble of the flaps. These changes are likely to influence the proteolytic activity and inhibitor affinity of the mutant enzyme; contributing to the high level of drug resistance exhibited by PR20. Importantly; the discovery of new flap conformations in crystal structures and MD simulations may hint at designs for novel antiviral inhibitors that capture the variant flap conformations of the resistant mutants.
Highlights.
Highly resistant HIV protease with 20 mutations was analyzed.
Crystal structures were solved for the protease mutant without inhibitor.
Molecular dynamics simulations were run on mutant and wild type protease.
Structural analysis revealed diverse flap conformations.
Fewer correlated intersubunit motions of mutant suggest role of 20 mutations.
Acknowledgments
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number U01GM062920 (I.T.W. & R.W.H.) and by the Georgia State University Molecular Basis of Disease Fellowship and the Georgia State University Research Program Enhancement Award in Bioinformatics (H.-C.S.). X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamline 22ID at the Advanced Photon Source; Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy; Office of Science; Office of Basic Energy Sciences; under Contract W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Konvalinka J, Krausslich HG, Muller B. Retroviral proteases and their roles in virion maturation. Virology. 2015;479–480C:403–417. doi: 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Wlodawer A. Structure-based design of AIDS drugs and the development of resistance. Vox Sang. 2002;83(Suppl 1):23–26. doi: 10.1111/j.1423-0410.2002.tb05261.x. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh AK, Anderson DD, Weber IT, Mitsuya H. Enhancing protein backbone binding--a fruitful concept for combating drug-resistant HIV. Angew. Chem. Int. Ed. Engl. 2012;51:1778–1802. doi: 10.1002/anie.201102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volarath P, Harrison RW, Weber IT. Structure based drug design for HIV protease: from molecular modeling to cheminformatics. Curr. Top. Med. Chem. 2007;7:1030–1038. doi: 10.2174/156802607780906744. [DOI] [PubMed] [Google Scholar]
- 5.Acevedo O, Ambrose Z, Flaherty PT, Aamer H, Jain P, Sambasivarao SV. Identification of HIV inhibitors guided by free energy perturbation calculations. Curr. Pharm. Des. 2012;18:1199–1216. doi: 10.2174/138161212799436421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornak V, Simmerling C. Targeting structural flexibility in HIV-1 protease inhibitor binding. Drug Discov. Today. 2007;12:132–138. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Titanji BK, Aasa-Chapman M, Pillay D, Jolly C. Protease inhibitors effectively block cell-to-cell spread of HIV-1 between T cells. Retrovirology. 2013;10:161. doi: 10.1186/1742-4690-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, Quer J, et al. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan PR, Hogg RS, Dong WW, Yip B, Wynhoven B, Woodward J, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. J. Infect. Dis. 2005;191:339–347. doi: 10.1086/427192. [DOI] [PubMed] [Google Scholar]
- 10.Weiser SD, Guzman D, Riley ED, Clark R, Bangsberg DR. Higher rates of viral suppression with nonnucleoside reverse transcriptase inhibitors compared to single protease inhibitors are not explained by better adherence. HIV Clin. Trials. 2004;5:278–287. doi: 10.1310/LNHD-K1R7-HQP5-HJCQ. [DOI] [PubMed] [Google Scholar]
- 11.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, et al. 2014 Update of the drug resistance mutations in HIV-1. Top Antivir. Med. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 12.Weber IT, Kneller DW, Wong-Sam A. Highly resistant HIV-1 proteases and strategies for their inhibition. Future Med. Chem. 2015;7:1023–1038. doi: 10.4155/fmc.15.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dierynck I, De Wit M, Gustin E, Keuleers I, Vandersmissen J, Hallenberger S, et al. Binding kinetics of darunavir to human immunodeficiency virus type 1 protease explain the potent antiviral activity and high genetic barrier. J. Virol. 2007;81:13845–13851. doi: 10.1128/JVI.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis JM, Aniana A, Weber IT, Sayer JM. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9072–9077. doi: 10.1073/pnas.1102278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis JM, Tozser J, Roche J, Matuz K, Aniana A, Sayer JM. Enhanced Stability of Monomer Fold Correlates with Extreme Drug Resistance of HIV-1 Protease. Biochemistry. 2013;52:7678–7688. doi: 10.1021/bi400962r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agniswamy J, Shen CH, Aniana A, Sayer JM, Louis JM, Weber IT. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51:2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agniswamy J, Louis JM, Shen CH, Yashchuk S, Ghosh AK, Weber IT. Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20. J. Med. Chem. 2015 doi: 10.1021/acs.jmedchem.5b00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh AK, Yashchuk S, Mizuno A, Chakraborty N, Agniswamy J, Wang YF, et al. Design of gem-difluoro-bis-tetrahydrofuran as P2 ligand for HIV-1 protease inhibitors to improve brain penetration: synthesis; X-ray studies; and biological evaluation. Chem. Med. Chem. 2015;10:107–115. doi: 10.1002/cmdc.201402358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robbins AH, Coman RM, Bracho-Sanchez E, Fernandez MA, Gilliland CT, Li M, et al. Structure of the unbound form of HIV-1 subtype A protease: comparison with unbound forms of proteases from other HIV subtypes. Acta Crystallogr. D: Biol. Crystallogr. 2010;66:233–242. doi: 10.1107/S0907444909054298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meher BR, Wang Y. Exploring the drug resistance of V32I and M46L mutant HIV-1 protease to inhibitor TMC114: Flap dynamics and binding mechanism. J. Mol. Graph. Model. 2015;56:60–73. doi: 10.1016/j.jmgm.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, et al. "Wide-open" 1.3 A structure of a multidrug-resistant HIV-1 protease as a drug target. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active site D25N mutation on the structure; stability; and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison RW, Weber IT. Molecular dynamics simulations of HIV-1 protease with peptide substrate. Protein Eng. 1994;7:1353–1363. doi: 10.1093/protein/7.11.1353. [DOI] [PubMed] [Google Scholar]
- 24.Harrison RW. Stiffness and energy conservation in molecular dynamics: An improved integrator. J. Comput. Chem. 1993;14:1112–1122. [Google Scholar]
- 25.Stigliani JL, Bernardes-Genisson V, Bernadou J, Pratviel G. Cross-docking study on InhA inhibitors: a combination of Autodock Vina and PM6-DH2 simulations to retrieve bioactive conformations. Org. Biomol. Chem. 2012;10:6341–6349. doi: 10.1039/c2ob25602a. [DOI] [PubMed] [Google Scholar]
- 26.188.ammp: SPEC CPU2000 Benchmark Description. 2015 https://www.spec.org/cpu2000/CFP2000/188.ammp/docs/188.ammp.html. [Google Scholar]
- 27.Bagossi P, Zahuczky G, Tözsér J, Weber I, Harrison R. Improved Parameters for Generating Partial Charges: Correlation with Observed Dipole Moments. J. Mol. Model. 1999;5:143–152. [Google Scholar]
- 28.Weber IT, Harrison RW. Molecular mechanics calculations on HIV-1 protease with peptide substrates correlate with experimental data. Protein Eng. 1996;9:679–690. doi: 10.1093/protein/9.8.679. [DOI] [PubMed] [Google Scholar]
- 29.Weber IT, Harrison RW. Molecular mechanics analysis of drug-resistant mutants of HIV protease. Protein Eng. 1999;12:469–474. doi: 10.1093/protein/12.6.469. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Weber IT, Harrison RW. Molecular dynamics simulations of 14 HIV protease mutants in complexes with indinavir. J. Mol. Model. 2004;10:373–381. doi: 10.1007/s00894-004-0205-x. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;267:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Meth. Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystalogr. D: Biol. Crystallogr. 2004;60 doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM); a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D: Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. San Carlos; CA: DeLano Scientific; 2002. [Google Scholar]
- 39.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, et al. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 40.Fang B, Fu G, Agniswamy J, Harrison RW, Weber IT. Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling. Apoptosis. 2009;14:741–752. doi: 10.1007/s10495-009-0333-y. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Yang M, Hu G, Shi S, Yi C, Zhang Q. Insights into the functional role of protonation states in the HIV-1 protease-BEA369 complex: molecular dynamics simulations and free energy calculations. J. Mol. Model. 2009;15:1245–1252. doi: 10.1007/s00894-009-0452-y. [DOI] [PubMed] [Google Scholar]
- 42.Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. Febs J. 2010;277:3699–3714. doi: 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kar P, Knecht V. Energetic basis for drug resistance of HIV-1 protease mutants against amprenavir. J. Comput.Aided Mol. Des. 2012;26:215–232. doi: 10.1007/s10822-012-9550-5. [DOI] [PubMed] [Google Scholar]
- 44.Everitt B, Hothorn T. A handbook of statistical analyses using R. 2nd ed. Boca Raton: CRC Press; 2010. [Google Scholar]
- 45.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2014 [Google Scholar]
- 46.Harte WE, Jr, Swaminathan S, Mansuri MM, Martin JC, Rosenberg IE, Beveridge DL. Domain communication in the dynamical structure of human immunodeficiency virus 1 protease. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8864–8868. doi: 10.1073/pnas.87.22.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 2000;301:1207–1220. doi: 10.1006/jmbi.2000.4018. [DOI] [PubMed] [Google Scholar]
- 48.Heaslet H, Rosenfeld R, Giffin M, Lin YC, Tam K, Torbett BE, et al. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr. D: Biol. Crystallogr. 2007;63:866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 49.Perryman AL, Lin JH, McCammon JA. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lepsik M, Kriz Z, Havlas Z. Efficiency of a second-generation HIV-1 protease inhibitor studied by molecular dynamics and absolute binding free energy calculations. Proteins. 2004;57:279–293. doi: 10.1002/prot.20192. [DOI] [PubMed] [Google Scholar]
- 51.Bhakat S, Chetty S, Martin AJ, Soliman ME. Multi-drug Resistance Profile of PR20 HIV-1 Protease is attributed to Distorted Conformational and Drug Binding Landscape: Molecular Dynamics Insights. J. Biomol. Struct. Dyn. 2015:1–46. doi: 10.1080/07391102.2015.1018326. [DOI] [PubMed] [Google Scholar]
- 52.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 53.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kokkinidis M, Glykos NM, Fadouloglou VE. Protein flexibility and enzymatic catalysis. Adv. Protein Chem. Struct. Biol. 2012;87:181–218. doi: 10.1016/B978-0-12-398312-1.00007-X. [DOI] [PubMed] [Google Scholar]
- 55.Trylska J, Tozzini V, Chang CE, McCammon JA. HIV-1 protease substrate binding and product release pathways explored with coarse-grained molecular dynamics. Biophys. J. 2007;92:4179–4187. doi: 10.1529/biophysj.106.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piana S, Carloni P, Parrinello M. Role of conformational fluctuations in the enzymatic reaction of HIV-1 protease. J. Mol. Biol. 2002;319:567–583. doi: 10.1016/S0022-2836(02)00301-7. [DOI] [PubMed] [Google Scholar]
- 57.Kurt N, Scott WR, Schiffer CA, Haliloglu T. Cooperative fluctuations of unliganded and substrate-bound HIV-1 protease: a structure-based analysis on a variety of conformations from crystallography and molecular dynamics simulations. Proteins. 2003;51:409–422. doi: 10.1002/prot.10350. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Chang YC, Louis JM, Wang YF, Harrison RW, Weber IT. Structures of Darunavir-Resistant HIV-1 Protease Mutant Reveal Atypical Binding of Darunavir to Wide Open Flaps. A.C.S. Chem. Biol. 2014 doi: 10.1021/cb4008875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dewdney TG, Wang Y, Liu Z, Sharma SK, Reiter SJ, Brunzelle JS, et al. Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease. Bioorg. Med. Chem. 2013;21:7430–7434. doi: 10.1016/j.bmc.2013.09.045. [DOI] [PubMed] [Google Scholar]
- 60.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu. Rev. Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fenwick RB, Orellana L, Esteban-Martin S, Orozco M, Salvatella X. Correlated motions are a fundamental property of beta-sheets. Nat. Commun. 2014;5:4070. doi: 10.1038/ncomms5070. [DOI] [PubMed] [Google Scholar]