Abstract
The development of efficacious carriers is an important long-standing challenge in gene therapy. In the past few decades, tremendous progress has been made toward non-viral vectors for gene delivery including cationic lipids and polymers. However, there continues to be a need for clinically translatable polymer-based delivery carriers because they offer tunable degradation profiles and functional groups, diverse structures/morphologies, and scalability in preparation. Herein, we developed a library of 144 degradable polymers with varying amine and hydrophobic content via a facile method that involves thiobutyrolactone aminolysis and consequent thiol-(meth)acrylate or acrylamide addition in one-pot. The polymer platform was evaluated for pDNA and siRNA delivery to HeLa cells in vitro. Hydrophobically modified 5S, 2E1, 6CY1, 5CY2, and 2M1 grafted HEMATL polymers are capable of delivering pDNA depending on the chemical composition and the size of the polyplexes. Hydrophobically modified 5S and 2B grafted HEMATL and 5S grafted ATL polymers exhibit capability for siRNA delivery that approaches the efficacy of commercially available transfection reagents. Due to tunable functionality and scalable preparation, this synthetic approach may have broad applicability in the design of delivery materials for gene therapy.
Graphical Abstract
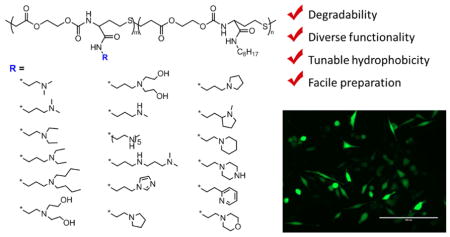
1. Introduction
Gene therapy has shown great potential for the treatment of a wide range of serious acquired and inherited diseases during the past three decades [1–3]. Polymer-based carriers are an essential component of this treatment strategy. Between 1990 and 2013, numerous gene therapy clinical trials commenced worldwide [4, 5], of which > 60% were related to cancer therapy. The first commercial gene therapy product was approved for the treatment of squamous cell carcinoma in China in 2003 [6]. The delivery of pDNA encoding for functional proteins to replace mutated or down-regulated genes remains a promising strategy to treat a variety of diseases [7]. More recently, delivery of shorter nucleic acids that invoke the RNA Interference (RNAi) pathway has been shown to be a powerful way to regulate gene expression post-transcriptionally. In particular, strategies that silence oncogenes using synthetic short interfering RNA (siRNA) or restore endogenous microRNA (miRNA) that function as tumor suppressors, represent next generation therapeutic approaches to treat cancer [8].
One of the practical challenges for both classical gene therapy and RNAi therapeutics is to efficiently deliver DNA or RNA into cells. A number of difficult barriers must be overcome to facilitate effective delivery. These barriers include protection from degradation, localization to the diseased tissue or tumor, cellular uptake into targeted cells, and intracellular release [9–12]. Viral vectors are established carriers for gene delivery because of their high efficiency [13]. However, safety concerns and production costs have restricted their utility. Non-viral vehicles for gene delivery have attracted much attention because of reduced immune response, low cost, and highly tunable diversity in structure [13–15]. Typically, cationic materials are used to bind negatively charged nucleic acids and facilitate cellular uptake. Cationic lipids represent one of the most extensively investigated non-viral vectors [16–18], and are commercially available for use as in vitro transfection reagents (e.g. Lipofectamine 2000 and RNAiMax). Cationic lipid nanoparticles have been used in human clinical trials [19]. The other representative non-viral delivery carrier, cationic polymers, have attracted increasing attention because of the flexibility in their synthesis and structural modifications, as well as the relatively higher stability of polyplexes (spherical complex of nucleic acids and cationic polymers) [15, 20, 21].
Some commonly used cationic polymers for non-viral gene delivery, such as poly(ethylene imine) (PEI), can generate high cytotoxicity because polymers with strong positive charges can induce hemolysis, apoptosis, or autophagy [22, 23]. Therefore, more biocompatible and biodegradable cationic polymers have been prepared for gene delivery in the past two decades. A large number of carriers have been synthesized and summarized in various reviews [14, 24–31]. In the context of research reported herein, we were particularly interested in degradable cationic polymers for gene delivery. For example, (oligo)PEI or poly((2-dimethylamino)ethyl methacrylate) (PDMAEMA) was grafted to degradable polymer backbones (e.g. poly(carbonate)s and poly(caprolactone)s) to utilize the pH buffering amino groups on PEI and PDMAEMA and increase the biocompatibility of the copolymers [32–34]. Polyesters, including poly(amino ester)s have been extensively studied as degradable polymer carriers for gene delivery [20, 35–45]. Polyamides, e.g. poly(ketal amidoamine) or polypeptides [46–50], polycarbonate-based polymers [25, 51–53], polyphosphoesters [54, 55], and bio-reducible poly(disulfide)s [33, 56–60] were also reported as gene delivery materials with varying physicochemical properties and distinct preparation procedures. However, classical synthesis of amino functional polyesters, polycarbonates, or polyamides sometimes involves multi-step protection/deprotection and/or ring-closure [25, 61, 62]. It has been shown that the development of combinatorial libraries of polymers is an effective way to discover efficacious nucleic acid carriers [63]. Because stringent polymerization conditions can limit the diversity of functional group incorporation, we aimed to synthesize a clinically-translatable platform with high structural diversity using a one-pot method.
In this report, we synthesized a poly(amino ester sulfide) library of 144 polymers with varying pKa and hydrophobic content using a one-pot method involving the reaction of amines with thiobutyrolactone (meth)acrylates or acrylamides. We were inspired by a recent report on an amine-thiol-ene polymerization strategy by Du Prez and coworkers [64], and decided to use a similar polymerization method to construct a polymer library. In this way, the resulting polymer library possesses a degradable backbone, diverse amine structures (modular apparent pKa), tunable hydrophobicity, and scalable preparation. In vitro screening of these polymers for pDNA and siRNA delivery to HeLa cells was performed to identify efficacious carriers. The binding of pDNA/siRNA with hit polymers and the size of the resulting polyplexes were comprehensively investigated in this paper to understand structure-function relationships. Due to tunable functionality and scalable preparation in one pot, this approach is a useful strategy to prepare polymer libraries for nucleic acid delivery.
2. Experimental
2.1 Materials
DL-Homocysteine thiolactone hydrochloride (CTH), acryloyl chloride, methacryloyl chloride, 2-hydroxyethyl methacrylate (HEMA), 4-nitrophenyl chloroformate (p-NPC), dimethylphenylphosphine (DPP) and all amines were purchased from Sigma-Aldrich and used as received. All organic solvents were purchased from Fisher Scientific and purified with a solvent purification system (Innovative Technology). Milli-Q water was used throughout the experiments. siRNA against luciferase (sense strand: 5′-GAUUAUGUCCGGUUAUGUA[dT][dT]-3′; anti-sense strand: 3′-UACAUAACCGGACAUAAUC[dT][dT]-5′), Dulbecco’s Modified Eagle Media (DMEM), and fetal bovine serum (FBS) were purchased from Sigma-Aldrich. gWiz pDNA (GFP) was purchased from Aldevron. OptiMEM was purchased from Life Technologies. Lipofectamine 2000 (LF2000) and RNAiMax was purchased from Invitrogen and used following the supplier’s recommended protocols.
2.2 Methods
The molecular weight of polymers was measured by Gel Permeation Chromatography (GPC) (Viscotek) equipped with RI detection and ViscoGEL I-series columns (Viscoteck I-MBLMW-3078) using DMF as the eluent at 0.75 mL/min and 45 °C. The instrument was calibrated with a series of 10 narrow polydispersity polystyrene standards (500 to 200,000 g/mol). The structure of monomers was characterized by 1H NMR (Varian, 400 MHz) in CDCl3. Flash chromatography was performed on a Teledyne Isco CombiFlash Rf-200i chromatography system.
2.3 Synthesis of monomers
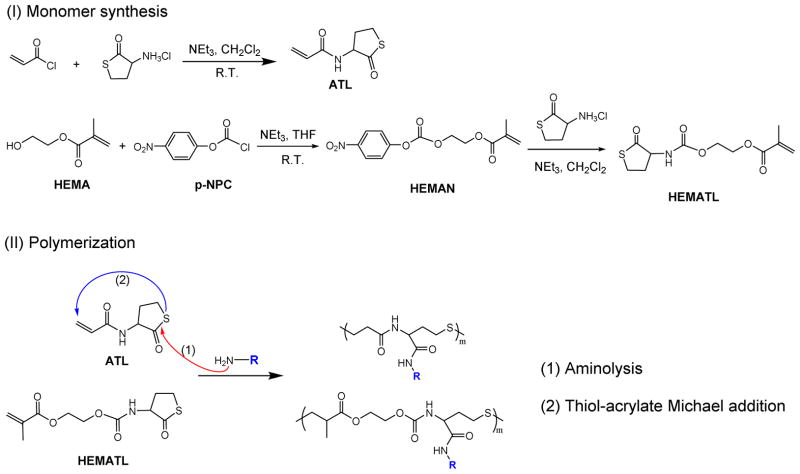
N-(2-oxotetrahydrothiophen-3-yl)acrylamide (ATL) was synthesized by the amidation of DL-homocysteine thiolactone hydrochloride with acryloyl chloride in dichloromethane (DCM) using triethylamine (TEA) as base (Scheme 1). 7.759 g (50 mmol) of CTH and 21.01 mL (150 mmol) of TEA were dissolved in 100 mL of DCM and the mixture was placed in a round bottom flask in an ice bath with stirring (600 rpm). 4.188 mL (50 mmol) of acryloyl chloride was added drop-wise into the mixture under N2 using an addition funnel. After 24 h reaction at r.t, the salt was removed and the filtrate was washed with 50 mL of NaHCO3 aqueous solution (10%) and 50 mL of HCl (0.1 N), dried with magnesium sulfate, and concentrated by rotary evaporation. ATL was purified by flash chromatography with a hexane/ethyl acetate gradient.
Scheme 1.
Synthesis of monomers ATL and HEMATL and preparation of functional poly(amino ester sulfide)s via combination of thiolbutyrolactone aminolysis and thiol-(meth)acrylate addition.
To synthesize 2-(((2-oxotetrahydrothiophen-3-yl)carbamoyl)oxy)ethyl methacrylate (HEMATL), the hydroxyl group in HEMA was activated by reaction with p-NPC, then coupled with the primary amine in CTH. 6.253 mL (50 mmol) of HEMA and 8.405 mL (60 mmol) of TEA were added into 50 mL of dry tetrahydrofuran (THF) at 0 °C, followed by addition of 10.50 grams (50 mmol) of p-NPC (dissolved in 30 mL THF) under N2. The mixture was kept stirring (600 rpm) at r.t. for 24 h. After removal of salt and THF, the raw intermediate was re-dissolved in 30 mL of DCM and added drop-wise into the mixture of 7.760 gram (50 mmol) of CTH and 15.40 mL (110 mmol) of TEA in 100 mL DCM. The reaction was kept at r.t. for 48 h. The pure HEMATL was isolated by flash chromatography using a hexane/ethyl acetate gradient.
2.4 Polymerization of monomers
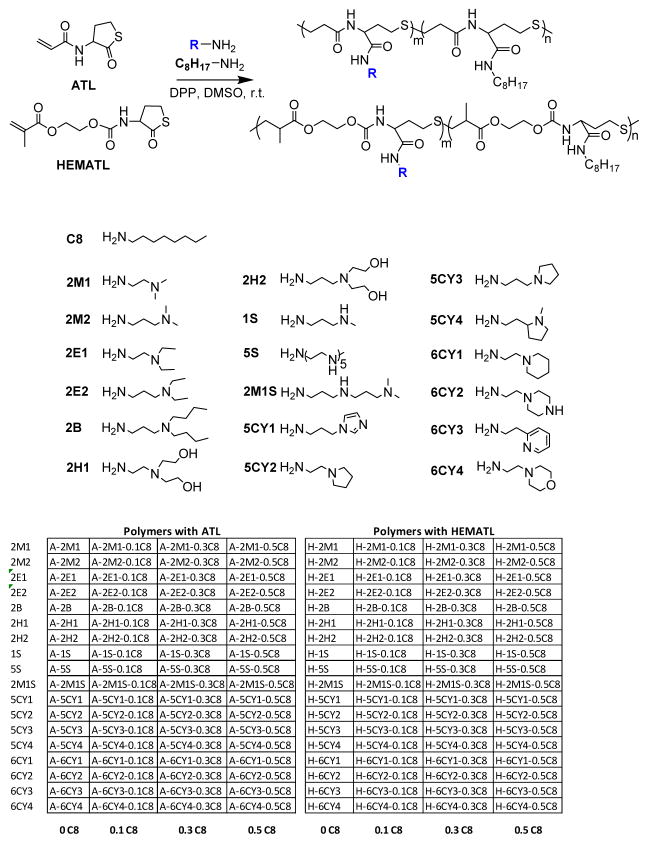
To optimize the polymerization, varying temperature, solvents and ratios of monomer and were applied in the preliminary polymerizations. Dimethylphenylphosphine (DPP) was evaluated as a catalyst for thiol-(meth)acrylate Michael addition [65]. The polymer library with varying amines and octylamine percentage was prepared at the optimized conditions. The concentration of monomers was 0.5 M in all polymerizations. As a typical example, 17.1 mg (0.1 mmol) ATL, 1.1 equivalent dimethylamino-1-propylamine (DMP) (11.4 mg, 0.11 mmol) and 0.05 equivalent DPP (0.7 mg, 0.005 mmol) were added to 0.2 mL of DMSO, and the mixture was kept stirring at r.t. for 24 h.
2.5 pDNA delivery with functional poly(amino ester sulfide)s
HeLa cells (ATCC) were seeded into opaque white 96-well plates (10,000 cells/well) and cultured in complete DMEM with 5% FBS (37 °C, 5% CO2) for in vitro pDNA transfection assays. After 24 hours, the media was replaced with fresh, FBS-containing media (200 μL/well). The polyplexes of pDNA and polymers were prepared at a polymer/pDNA ratio of 30:1 (wt/wt) in 10 mM phosphate buffer (pH 6.8) by vigorous pipette mixing in a 96-well plate. After incubation of 30 min at room temperature, 20 μL of polyplexes containing 200 ng of pDNA were added into each well. The cells were incubated with the polyplexes for 24 h, after which the medium was replaced with 100 μL PBS and the GFP fluorescence intensity was measured on a plate reader (Tecan Infinite M200 Pro). Transfections were performed in triplicate.
2.6 siRNA delivery with functional poly(amino ester sulfide)s
HeLa cells stably expressing luciferase (HeLa-Luc) were derived from HeLa cells (ATCC) by stable transfection of the Firefly Luciferase gene using Lentiviral infection followed by clonal selection. HeLa-Luc cells were seeded into opaque white 96-well plates (10,000 cells/well) and cultured in complete, phenol red free DMEM with 5% FBS (37 °C, 5% CO2) for in vitro siRNA transfection assays. After 24 hours, the media was replaced with fresh, FBS-containing media (200 μL/well). The polyplexes of siRNA and polymers were prepared at a polymer/siRNA ratio of 30:1 (wt/wt) in 10 mM citric acid/trisodium citrate buffer (pH 4.2) by vigorous mixing in a 96-well plate using pipette tips. 20 μL of polyplexes containing 50 ng of siRNA were added into each well. The cells were incubated with the polyplexes for 24 h, after which the cell viability and luciferase activity was analyzed using ONE-Glo + Tox assay kits (Promega). Transfections were performed in triplicate.
2.7 Fluorescence Microscopy
The complexes with selected polymers were added to HeLa cells using the same delivery protocol described above but using clear-bottom black plates. Fluorescence microcopy was performed on an AMG EVOS FL microscope.
2.8 pDNA/siRNA binding with poly(amino ester sulfide)s and polyplex size measurements
Polyplexes were prepared in the same way as that in the delivery assays for consistency. pDNA or siRNA binding was quantified using the Quant-iT RiboGreen Assay kit (Life Technologies). This assay measures free DNA/RNA and can therefore be used to quantify the binding efficiency of polyplexes. The size measurements were performed using dynamic light scattering (DLS, Malvern Zetasizer Nano ZS, He-Ne laser, λ = 632 nm). The number-weighted diameter (Dh) was the average result of 5 measurements.
3. Results and Discussion
3.1 Synthesis of N-(2-oxotetrahydrothiophen-3-yl)acrylamide (ATL) and 2-(((2-oxotetrahydrothiophen-3-yl)carbamoyl)oxy)ethyl methacrylate (HEMATL)
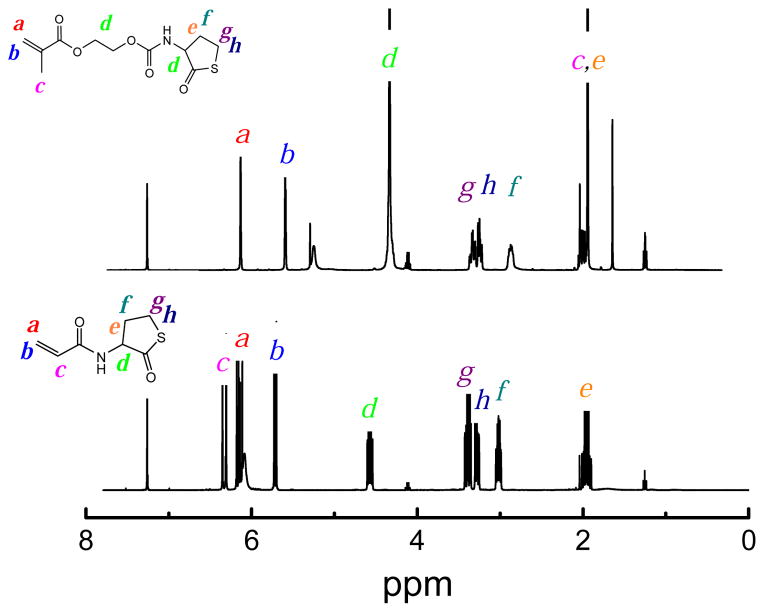
The one-pot polymerization method involving thiobutyrolactone aminolysis and consequent thiol-(meth)acrylate addition is an attractive route towards functional, degradable polymers [64]. We designed new monomers that contained both (meth)acrylate and thiolactone groups to produce polymers containing ester, urethane, and sulfide linkages in the polymer backbone. In particular, we investigated the effect of changing acrylate, methacrylate, and acrylamide functionalities, and the spacer length between the thiolactone amide and the (meth)acrylate. Monomers containing both (meth)acrylate and thiolactone groups were synthesized via the reaction of CTH and acryloyl chloride or HEMAN (a p-NPC activated derivative of HEMA) (Scheme 2). The structures were confirmed by 1H NMR spectra, as shown in Figure 1. The straightforward and easily scalable synthesis was achieved in 12.4 grams with ~72% yield and confirmed by 1H NMR (Figure 1).
Scheme 2.
Structure of amines and the percentage of octylamine in feed for the polymers.
Figure 1.
1H NMR spectra of ATL and HEMATL in CDCl3.
3.2 Preparation of a poly(amino ester sulfide) library
Before the combinatorial synthesis of the functional polymer library, the polymerization conditions were optimized with HEMATL and dimethylamino-1-propylamine (DMP) at varying temperature, solvent, and ratios of monomers and amines (Table S1, Figures S1 and S2). In general, the polymerization improved with increase of solvent polarity because more polar solvents are favorable for Michael addition reactions [65]. The aminolysis of thiolbutyrolactone could be completed in a few minutes [64]. The most polar organic solvent (DMSO) and DPP as a Michael addition catalyst was chosen for preparation of the polymer library. More trials in DMSO (Figure S2) show that polymerization is almost completed in 24 h and higher temperature is unfavorable. The addition of DPP promotes the polymerization of HEMATL and the amine at r.t (Figure S2a). Subsequently, the polymerization of the monomers was performed with 1.1 equivalent amines, 0.05 equivalent DPP, and a monomer concentration of 0.5 M in DMSO at r.t for 24 h. Figure 1 shows the GPC curve of a polymer that was prepared with HEMATL and DMP under these conditions and precipitated in diethyl ether.
3.3 pDNA delivery with poly(amino ester sulfide)s to HeLa cells
A library of 144 polymers was prepared under the optimized polymerization conditions with 0, 10, 30, and 50% octylamine (C8) by mole (Scheme 2). The polymers were diluted with DMSO and used in the preparation of polyplexes for delivery, binding, and DLS measurements to accelerate screening and identification of lead materials.
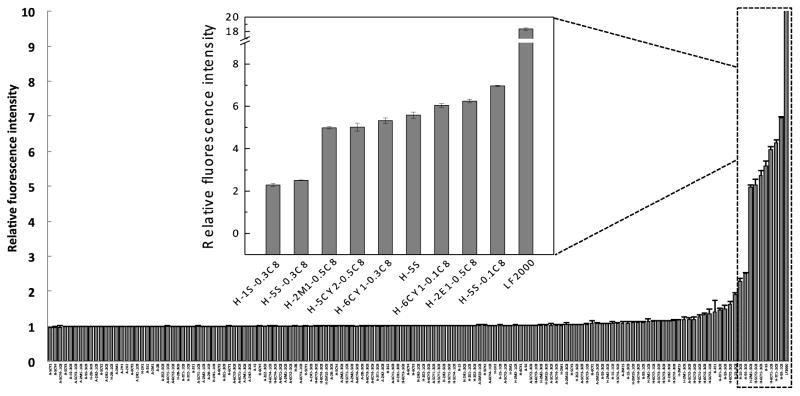
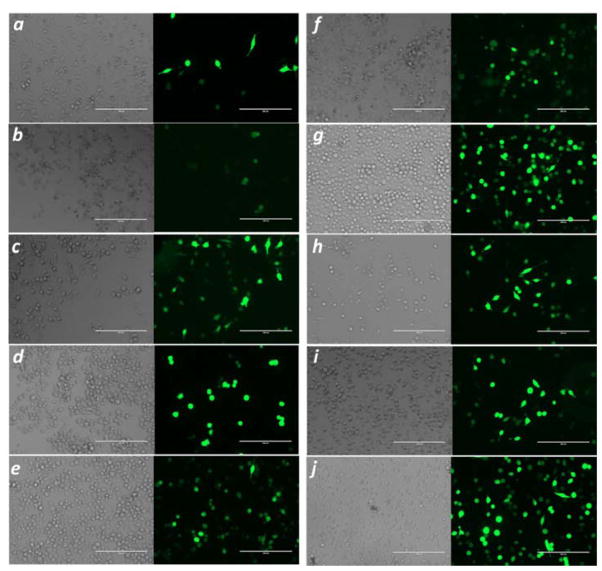
The successful delivery of pDNA (GFP) to HeLa cells induced the expression of green fluorescent protein leading to the increase of fluorescence intensity. Figure 3 plots fluorescence intensity after transfection with all 144 polyplexes. Nine polymers prepared with HEMATL and various amines produced a 2- to 7-fold increase in fluorescence intensity (insert in Figure 3). The transfection activity of those polyplexes was verified by the fluorescence microscopy (Figure 4). Side-by-side bright field and fluorescence images confirm the expression of GFP, which matches the average, relative expression measured in the plate reader.
Figure 3.
Relative fluorescence intensity after treatment with pDNA (GFP)-polymer polyplexes for 24 h. LF2000 was used as a positive control for pDNA delivery. The fluorescence intensity was normalized to untreated cells.
Figure 4.
Fluorescence and bright field images of cells after 24 hour-treatment with complexes of pDNA and H-1S-0.3C8 (a), H-5S-0.3C8 (b), H-2M1-0.5C8 (c), H-5CY2-0.5C8 (d), H-6CY1-0.3C8 (e), H-5S (f), H-6CY1-0.1C8 (g), H-2E1-0.5C8 (h), H-5S-0.1C8 (i), and LF2000 (j). The scale bar is 200 μm.
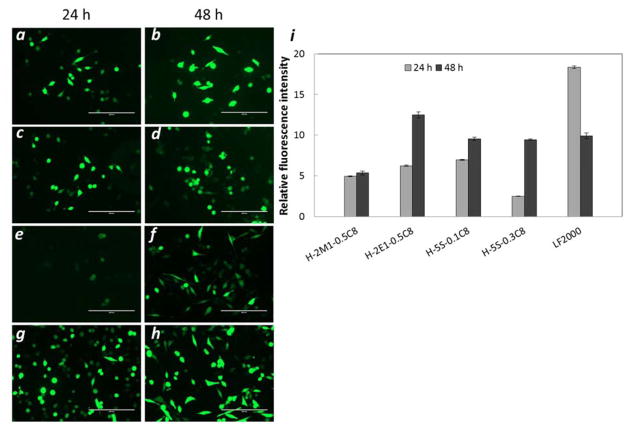
To further examine the delivery efficacy, a 48 hour-incubation was applied for H-2E1-0.5C8, H-5S-0.1C8, and H-5S-0.3C8. Figure 5 shows an increase in relative fluorescence intensity and brighter green fluorescence in microscopy images, indicating greater transfection to cells under these conditions. It is worth noting that longer incubation with LF2000-pDNA complexes led to ~2 fold decrease in fluorescence intensity, perhaps due to higher toxicity under these conditions of longer incubation time.
Figure 5.
Fluorescence images of cells after 24 or 48 hour treatment with polyplexes of pDNA and H-2E1-0.5C8 (a, b), H-5S-0.1C8 (c, d), H-5S-0.3C8 (e, f), and LF2000 (g, h), and relative fluorescence intensity normalized to untreated cells (i). The scale bar is 200 μm.
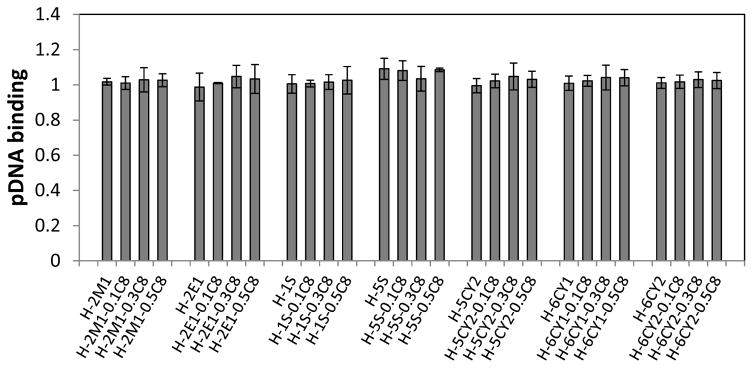
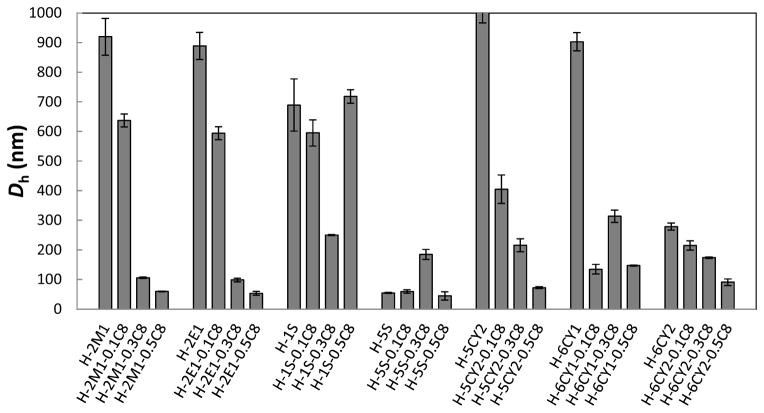
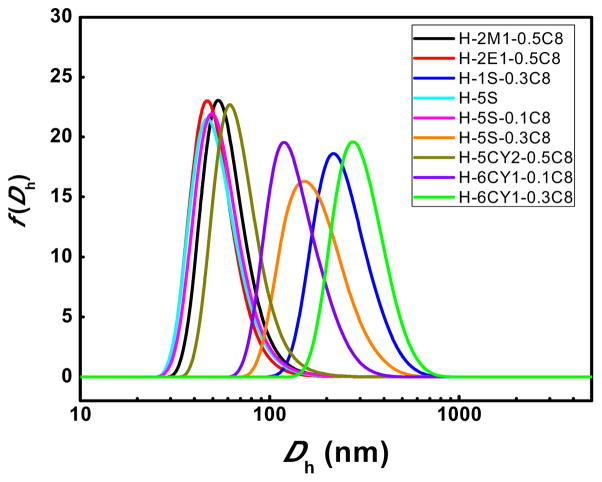
As shown in Figure 3, HEMATL-based polymers modified with C8, combined with dimethylamine, diethylamine, or ethyleneimine exhibited distinct delivery activity for pDNA. To better understand these results, the binding ability of the polymers to pDNA and the size of polyplexes were measured (Figures 6 and 7). All of the active polymers efficiently bound pDNA, likely due to the high polymer to pDNA weight ratio (30:1) and the cationic nature of the active polymer structures enabling efficient electrostatic interactions with the anionic phosphate backbone of the pDNA. Characterization of these polyplexes by DLS revealed that the hydrodynamic diameter varied greatly from 40 to 1000 nm depending on the polymer composition (Figure 7). For most of the polymer series, the size of polyplexes with pDNA decreases with an increase of octylamine percentage. That is most likely due to the hydrophobic octyl side groups, which results in more compact nanoparticles. Furthermore, the resulting nanoparticles with a smaller size and condensed structure contributes to the successful delivery of pDNA into HeLa cells. Figure 8 shows the size distribution of the most efficacious polyplexes (insert in Figure 3). Most of the polyplexes are smaller than 300 nm. None of the nanoparticles larger than 320 nm transfected pDNA to HeLa cells. Therefore, relatively small size is a necessary but not sufficient parameter for delivery. Several polymers (e.g. some in 5S and 6CY2 polymer series) formed smaller nanoparticles with pDNA but their polyplexes did not show good transfection activities. Polymerization control limitations did not allow us to study the effect of MW carefully in this paper, but a prior study of PBAEs for pDNA delivery indicated that the higher MW chains in a polymer mixture are solely responsible for pDNA delivery, which may also be the case with these polymers [40]. To summarize, all active polymers effectively bound pDNA. No conclusions could be made with respect to binding and pDNA efficacy. Correlating the results of size measurements with activity, increasing hydrophobic content of the polymer yielded smaller, more compact polyplexes that were generally more active in pDNA delivery.
Figure 6.
pDNA binding with polymer series prepared with HEMATL and amines 2M1, 2E1, 1S, 5S, 5CY2, 6CY1, and 6CY2. The polyplexes were prepared under the same condition as in delivery assays. The binding fraction is shown, indicating no free pDNA at a polymer/pDNA ratio of 30:1 (wt/wt).
Figure 7.
The hydrodynamic diameters (Dh, number weighted) of pDNA polyplexes from Figure 6.
Figure 8.
Dh distribution of the most efficacious pDNA polyplexes.
3.4 siRNA delivery with poly(amino ester sulfide)s to HeLa cells
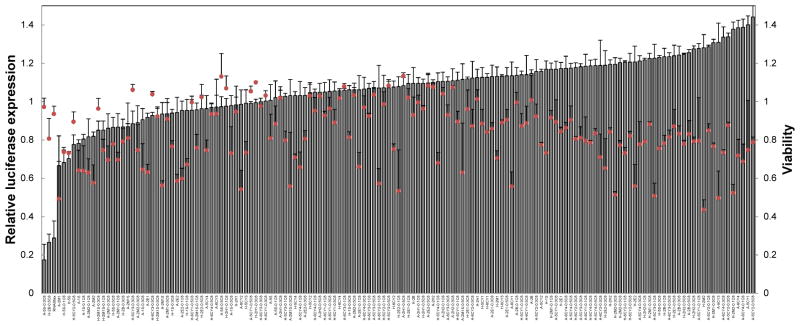
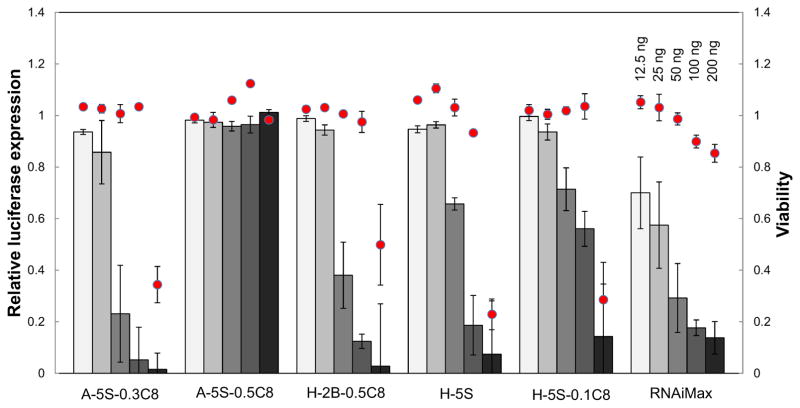
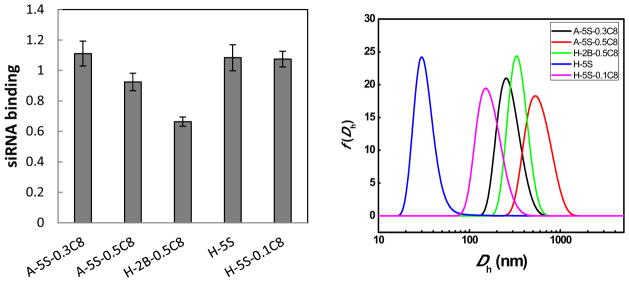
To extend the utilization of this degradable polymer platform, siRNA delivery capability was evaluated. HeLa cells, engineered to stably express a firefly luciferase reporter, were treated with polyplexes containing an siRNA against firefly luciferase (siLuc). As shown in Figure 9, a small number of siLuc polyplexes were capable of silencing luciferase expression at a dose of 50 ng siRNA per well. Focusing on the polymer hits, delivery efficacy was verified by a dose response experiment (Figure 10). Three polymers showed excellent dose response trends that approach (or exceed) the efficacy of RNAiMax at the siRNA doses above 50 ng. However, there was some cytotoxicity at the highest dose, an effect that could possibly be mitigated by decreasing the polymer:siRNA weight ratio. The binding with siRNA and the size of their polyplexes were measured (Figure 11). In comparison with pDNA delivery, the polymer hits do not necessarily show 100% binding with siRNA and the complex size is in a relatively wide range probably because the much shorter siRNA chain leads to less physical entanglement or van der Waals interactions between polymer chains and siRNA. Although most of the polymers were not effective, a few polymers approached the efficacy of RNAiMax. Further optimization of formulation and polymer structure is needed for better siRNA delivery in the future. Taken together, these results demonstrate the utility of polymer library synthesis and screening in the discovery and understanding of effective polyplexes for nucleic acid delivery.
Figure 9.
in vitro screening of the polymer library for luciferase siRNA delivery to HeLa cells. Gray bars and red circles indicate relative luciferase activity and cell viability, respectively.
Figure 10.
siRNA dose response curve for three active and one inactive polymer. RNAiMax was used as the positive control to benchmark the efficacy. The same polyplexes were prepared as those in Figure 9 and different amount was added to the cells depending on the desired dosage. The dose range varied from 12.5 to 200 ng (4.55 to 53.3 nM) per well. Red dots denote the cell viability and grey bars indicate relative luciferase expression compared to untreated cells.
Figure 11.
siRNA binding and the sizes of the siRNA polyplexes.
4. Conclusions
We developed a degradable functional polymer library with varying amines and hydrophobic degrees via a one-pot, combinatorial approach. 144 poly(amino ester sulfide)s were synthesized by the one-pot combination of thiolbutyrolactone aminolysis and the consequent thiol-(meth)acrylate addition at room temperature. Polyplexes were prepared by direct pipette tip mixing and applied to HeLa cells to screen pDNA or siRNA delivery capability of the degradable polymer platform. In vitro delivery experiments and fluorescence microscopy show that hydrophobically modified 5S, 2E1, 6CY1, 5CY2, and 2M1 grafted HEMATL polymers are capable of delivering pDNA with 24 hour transfection and the reporter gene expression increases with longer incubation (48 hrs). Binding assays and DLS measurements indicate that the polymer series with similar chemical structure have identical binding efficacy but different polyplex sizes with pDNA which affects delivery. Small polyplexes (typically <300 nm in diameter) are favorable for delivery. But only small size is not sufficient for successful pDNA delivery; certain chemical composition (pKa and hydrophobicity) is also required. In addition, hydrophobically modified 5S and 2B grafted HEMATL and 5S grafted ATL polymers exhibit capability for siRNA delivery that approaches the efficacy of commercially available transfection reagents. It is worth noting that the ATL polymers contain amide linkages that are generally less degradable than ester linkages under physiological conditions. To increase the chemical diversity, ATL was explored and some ATL-based polymers were capable of delivering siRNA to HeLa cells. Due to tunable functionality and scalable preparation, this synthetic approach may have broad applicability in the design of delivery materials for gene therapy. Future exploration will focus on improving polymer structures and formulation procedures with the aim of enhancing gene delivery even further.
Supplementary Material
Figure S1. GPC curves of polymers (1–14) prepared in THF (a) and dioxane or acetonitrile (b). The apparent Mw of peaks with higher molecular weight are indicated in the figure.
Figure S2. GPC curves of polymers (15–20) prepared in DMSO at 25 °C (a), 45 °C (b), and 90 °C (c) with polymerization time of 24, 48, 72, and 96 h. The apparent Mw of peaks with higher molecular weight are indicated in the figure.
Table S1. Conditions for polymerization of HEMATL and DMP.
Figure 2.
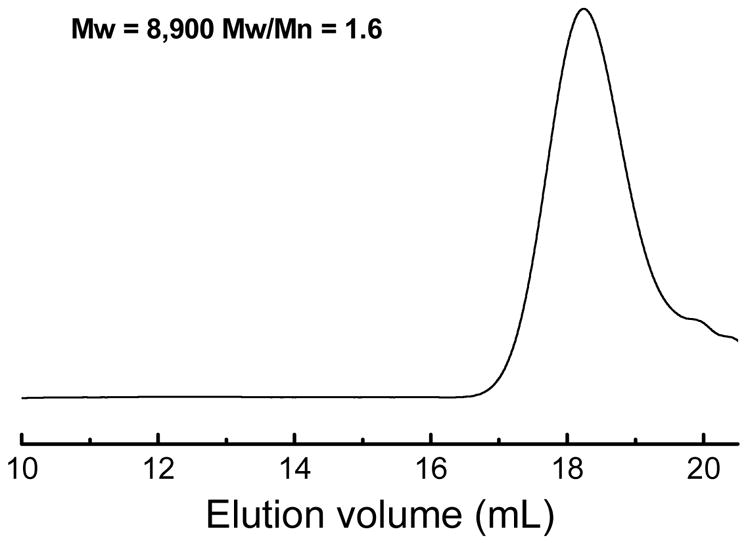
GPC curve of the HEMATL-DMP polymer after precipitation in diethyl ether.
Highlights.
We developed a combinatorial library of 144 degradable polymers with varying amine and hydrophobic content via a facile method that involves thiobutyrolactone aminolysis and consequent thiol-(meth)acrylate or acrylamide addition in one-pot.
Hydrophobically modified 5S, 2E1, 6CY1, 5CY2, and 2M1 grafted HEMATL polymers were capable of delivering pDNA depending on the chemical composition and the size of the polyplexes.
Hydrophobically modified 5S and 2B grafted HEMATL and 5S grafted ATL polymers exhibited capability for siRNA delivery that approaches the efficacy of commercially available transfection reagents.
Due to tunable functionality and scalable preparation, this synthetic approach may have broad applicability in the design of delivery materials for gene therapy.
Acknowledgments
The authors dedicate this article to honor Professor Krzysztof Matyjaszewski on occasion of his 65th birthday, and wish him continued personal happiness and research success. We gratefully acknowledge financial support from the Cancer Prevention and Research Institute of Texas (CPRIT) (Grant R1212), the Welch Foundation (Grant I-1855), the American Cancer Society (Grant ACS-IRG-02-196), the UTSW Friends of the Comprehensive Cancer Center 2014 Award in Cancer Research, and the UTSW Translational Pilot Program via the National Center for Advancing Translational Sciences of the National Institutes of Health (Grant UL1TR001105). The content is solely the responsibility of the authors and does not necessarily represent the official views of the CPRIT, Welch Foundation, ACS, or NIH. The authors also thank Professor Jinming Gao (UTSW) for providing access to the DLS instrument.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gardlik R, Palffy R, Hodosy J, Lukacs J, Turna J, Celec P. Vectors and delivery systems in gene therapy. Med Sci Moni. 2005;11:RA110–RA21. [PubMed] [Google Scholar]
- 2.Friedmann T. Human gene therapy - An immature genie, but certainly out of the bottle. Nat Med. 1996;2:144–7. doi: 10.1038/nm0296-144. [DOI] [PubMed] [Google Scholar]
- 3.Miller AD. Human gene-therapy comes of age. Nature. 1992;357:455–60. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 4.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, et al. T Lymphocyte-directed gene therapy for ADA–SCID: initial trial results after 4 years. Science. 1995;270:475–80. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 5.Wirth T, Parker N, Yla-Herttuala S. History of gene therapy. Gene. 2013;525:162–9. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- 6.Wilson JM. Gendicine: The first commercial gene therapy product. Hum Gene Ther. 2005;16:1014–4. doi: 10.1089/hum.2005.16.1014. [DOI] [PubMed] [Google Scholar]
- 7.Stribley JM, Rehman KS, Niu HR, Christman GM. Gene therapy and reproductive medicine. Fertil Steril. 2002;77:645–57. doi: 10.1016/s0015-0282(01)03233-2. [DOI] [PubMed] [Google Scholar]
- 8.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–39. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–7. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 10.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–77. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 11.Clarke DTW, McMillan NAJ. Gene delivery: Cell-specific therapy on target. Nat Nanotechnol. 2014;9:568–9. doi: 10.1038/nnano.2014.155. [DOI] [PubMed] [Google Scholar]
- 12.Ibraheem D, Elaissari A, Fessi H. Gene therapy and DNA delivery systems. Int J Pharm. 2014;459:70–83. doi: 10.1016/j.ijpharm.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 13.Boulaiz H, Marchal JA, Prados J, Melguizo C, Aranega A. Non-viral and viral vectors for gene therapy. Cell Mol Biol. 2005;51:3–22. [PubMed] [Google Scholar]
- 14.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–55. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 15.Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125:5316–23. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 16.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: A highly efficient, lipid-mediated DNA-transfection prodedure. Proc Natl Acad Sci USA. 1987;84:7413–7. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN. Improved DNA: Liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol. 1997;15:647–52. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 18.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–9. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. New Engl J Med. 2013;369:819–29. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat Mater. 2012;11:82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Dosari MS, Gao X. Nonviral gene delivery: Principle, limitations, and recent progress. AAPS J. 2009;11:671–81. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Ilarduya CT, Sun Y, Duezguenes N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010;40:159–70. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Lv HT, Zhang SB, Wang B, Cui SH, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–9. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Islam MA, Park T-E, Singh B, Maharjan S, Firdous J, Cho M-H, et al. Major degradable polycations as carriers for DNA and siRNA. J Control Release. 2014;193:74–89. doi: 10.1016/j.jconrel.2014.05.055. [DOI] [PubMed] [Google Scholar]
- 25.Tempelaar S, Mespouille L, Coulembier O, Dubois P, Dove AP. Synthesis and post-polymerisation modifications of aliphatic poly(carbonate)s prepared by ring-opening polymerisation. Chem Soc Rev. 2013;42:1312–36. doi: 10.1039/c2cs35268k. [DOI] [PubMed] [Google Scholar]
- 26.Teasdale I, Brueggemann O. Polyphosphazenes: Multifunctional, biodegradable vehicles for drug and gene delivery. Polymers. 2013;5:161–87. doi: 10.3390/polym5010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Ko NR, Oh JK. Recent advances in stimuli-responsive degradable block copolymer micelles: synthesis and controlled drug delivery applications. Chem Commun. 2012;48:7542–52. doi: 10.1039/c2cc32408c. [DOI] [PubMed] [Google Scholar]
- 28.Samal SK, Dash M, Van Vlierberghe S, Kaplan DL, Chiellini E, van Blitterswijk C, et al. Cationic polymers and their therapeutic potential. Chem Soc Rev. 2012;41:7147–94. doi: 10.1039/c2cs35094g. [DOI] [PubMed] [Google Scholar]
- 29.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliver Rev. 2012;64:61–71. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 30.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliver Rev. 2012;64:206–12. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Siegwart DJ, Oh JK, Matyjaszewski K. ATRP in the design of functional materials for biomedical applications. Prog Polym Sci. 2012;37:18–37. doi: 10.1016/j.progpolymsci.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng M, Librizzi D, Kilic A, Liu Y, Renz H, Merkel OM, et al. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials. 2012;33:6551–8. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 33.Dong X, Tian H, Chen L, Chen J, Chen X. Biodegradable mPEG-b-P(MCC-g-OEI) copolymers for efficient gene delivery. J Control Release. 2011;152:135–42. doi: 10.1016/j.jconrel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 34.Wang C-F, Lin Y-X, Jiang T, He F, Zhuo R-X. Polyethylenimine-grafted polycarbonates as biodegradable polycations for gene delivery. Biomaterials. 2009;30:4824–32. doi: 10.1016/j.biomaterials.2009.05.053. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Zou J, Cheng C. Synthesis and biomedical applications of functional poly(alpha-hydroxyl acid)s. Polym Chem. 2014;5:5854–72. [Google Scholar]
- 36.Keeney M, Onyiah S, Zhang Z, Tong X, Han L-H, Yang F. Modulating polymer chemistry to enhance non-viral gene delivery inside hydrogels with tunable matrix stiffness. Biomaterials. 2013;34:9657–65. doi: 10.1016/j.biomaterials.2013.08.050. [DOI] [PubMed] [Google Scholar]
- 37.Park H-J, Lee J, Kim M-J, Kang TJ, Jeong Y, Um SH, et al. Sonic hedgehog intradermal gene therapy using a biodegradable poly (beta-amino esters) nanoparticle to enhance wound healing. Biomaterials. 2012;33:9148–56. doi: 10.1016/j.biomaterials.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Fields RJ, Cheng CJ, Quijano E, Weller C, Kristofik N, Nha D, et al. Surface modified poly(beta amino ester)-containing nanoparticles for plasmid DNA delivery. J Control Release. 2012;164:41–8. doi: 10.1016/j.jconrel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endres T, Zheng M, Beck-Broichsitter M, Samsonova O, Debus H, Kissel T. Optimising the self-assembly of siRNA loaded PEG-PCL-lPEI nano-carriers employing different preparation techniques. J Control Release. 2012;160:583–91. doi: 10.1016/j.jconrel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(beta-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33:3594–603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on Poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–10. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao C-Q, Du J-Z, Sun T-M, Yao Y-D, Zhang P-Z, Song E-W, et al. A biodegradable amphiphilic and cationic triblock copolymer for the delivery of siRNA targeting the acid ceramidase gene for cancer therapy. Biomaterials. 2011;32:3124–33. doi: 10.1016/j.biomaterials.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Inoue S, Ding H, Portilla-Arias J, Hu J, Konda B, Fujita M, et al. Polymalic acid-based nanobiopolymer provides efficient systemic breast cancer treatment by inhibiting both HER2/neu receptor synthesis and activity. Cancer Res. 2011;71:1454–64. doi: 10.1158/0008-5472.CAN-10-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu C, Jung S, Luo S, Meng F, Zhu X, Park TG, et al. Co-delivery of siRNA and paclitaxel into cancer cells by biodegradable cationic micelles based on PDMAEMA-PCL-PDMAEMA triblock copolymers. Biomaterials. 2010;31:2408–16. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 45.Xiong X-B, Uludag H, Lavasanifar A. Biodegradable amphiphilic poly(ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomaterials. 2009;30:242–53. doi: 10.1016/j.biomaterials.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 46.Deng C, Wu J, Cheng R, Meng F, Klok H-A, Zhong Z. Functional polypeptide and hybrid materials: Precision synthesis via alpha-amino acid N-carboxyanhydride polymerization and emerging biomedical applications. Prog Polym Sci. 2014;39:330–64. [Google Scholar]
- 47.Barrett SE, Burke RS, Abrams MT, Bason C, Busuek M, Carlini E, et al. Development of a liver-targeted siRNA delivery platform with a broad therapeutic window utilizing biodegradable polypeptide-based polymer conjugates. J Control Release. 2014;183:124–37. doi: 10.1016/j.jconrel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 48.Zeng H, Little HC, Tiambeng TN, Williams GA, Guan Z. Multifunctional dendronized peptide polymer platform for safe and effective siRNA delivery. J Am Chem Soc. 2013;135:4962–5. doi: 10.1021/ja400986u. [DOI] [PubMed] [Google Scholar]
- 49.Lim H, Noh J, Kim Y, Kim H, Kim J, Khang G, et al. Acid-degradable cationic poly(ketal amidoamine) for enhanced RNA interference in vitro and in vivo. Biomacromolecules. 2013;14:240–7. doi: 10.1021/bm301669e. [DOI] [PubMed] [Google Scholar]
- 50.Kadlecova Z, Rajendra Y, Matasci M, Baldi L, Hacker DL, Wurm FM, et al. DNA delivery with hyperbranched polylysine: A comparative study with linear and dendritic polylysine. J Control Release. 2013;169:276–88. doi: 10.1016/j.jconrel.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 51.Feng J, Zhuo R-X, Zhang X-Z. Construction of functional aliphatic polycarbonates for biomedical applications. Prog Polym Sci. 2012;37:211–36. [Google Scholar]
- 52.Yang C, Ong ZY, Yang Y-Y, Ee PLR, Hedrick JL. Novel biodegradable block copolymers of poly(ethylene glycol) (PEG) and cationic polycarbonate: Effects of PEG configuration on gene delivery. Macromol Rapid Comm. 2011;32:1826–33. doi: 10.1002/marc.201100350. [DOI] [PubMed] [Google Scholar]
- 53.Ong ZY, Fukushima K, Coady DJ, Yang Y-Y, Ee PLR, Hedrick JL. Rational design of biodegradable cationic polycarbonates for gene delivery. J Control Release. 2011;152:120–6. doi: 10.1016/j.jconrel.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 54.Hao Y, He J, Zhang M, Tao Y, Liu J, Ni P. Synthesis and characterization of novel brush copolymers with biodegradable polyphosphoester side chains for gene delivery. J Polym Sci Pol Chem. 2013;51:2150–60. [Google Scholar]
- 55.Zhao Z, Wang J, Mao HQ, Leong KW. Polyphosphoesters in drug and gene delivery. Adv Drug Deliver Rev. 2003;55:483–99. doi: 10.1016/s0169-409x(03)00040-1. [DOI] [PubMed] [Google Scholar]
- 56.Phillips DJ, Gibson MI. Biodegradable poly(disulfide)s derived from RAFT polymerization: Monomer scope, glutathione degradation, and tunable thermal responses. Biomacromolecules. 2012;13:3200–8. doi: 10.1021/bm300989s. [DOI] [PubMed] [Google Scholar]
- 57.Beloor J, Choi CS, Nam HY, Park M, Kim SH, Jackson A, et al. Arginine-engrafted biodegradable polymer for the systemic delivery of therapeutic siRNA. Biomaterials. 2012;33:1640–50. doi: 10.1016/j.biomaterials.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 58.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–14. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ou M, Wang X-L, Xu R, Chang C-W, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjugate Chem. 2008;19:626–33. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YS, Kim SW. Bioreducible polymers for therapeutic gene delivery. J Control Release. 2014;190:424–39. doi: 10.1016/j.jconrel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan YF, Siegwart DJ. Scalable synthesis and derivation of functional polyesters bearing ene and epoxide side chains. Polym Chem. 2014;5:1362–71. [Google Scholar]
- 62.Taniguchi I, Mayes AM, Chan EWL, Griffith LG. A chemoselective approach to grafting biodegradable polyesters. Macromolecules. 2005;38:216–9. [Google Scholar]
- 63.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc Natl Acad Sci USA. 2011;108:12996–3001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Espeel P, Goethals F, Driessen F, Nguyen L-TT, Du Prez FE. One-pot, additive-free preparation of functionalized polyurethanes via amine-thiol-ene conjugation. Polym Chem. 2013;4:2449–56. [Google Scholar]
- 65.Li G-Z, Randev RK, Soeriyadi AH, Rees G, Boyer C, Tong Z, et al. Investigation into thiol-(meth)acrylate Michael addition reactions using amine and phosphine catalysts. Polym Chem. 2010;1:1196–204. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. GPC curves of polymers (1–14) prepared in THF (a) and dioxane or acetonitrile (b). The apparent Mw of peaks with higher molecular weight are indicated in the figure.
Figure S2. GPC curves of polymers (15–20) prepared in DMSO at 25 °C (a), 45 °C (b), and 90 °C (c) with polymerization time of 24, 48, 72, and 96 h. The apparent Mw of peaks with higher molecular weight are indicated in the figure.
Table S1. Conditions for polymerization of HEMATL and DMP.