Abstract
Shortly after the agricultural revolution, the domestication of bacteria, yeasts, and molds, played an essential role in enhancing the stability, quality, flavor, and texture of food products. These domestication events were likely the result of human food production practices that entailed the continual recycling of isolated microbial communities in the presence of abundant agricultural food sources. We suggest that within these novel agrarian food niches the metabolic requirements of those microbes became regular and predictable resulting in rapid genomic specialization through such mechanisms as pseudogenization, genome decay, interspecific hybridization, gene duplication, and horizontal gene transfer. The ultimate result was domesticated strains of microorganisms with enhanced fermentative capacities.
Introduction
Domestication refers to the genetic modification of a species by breeding it in isolation from it ancestral population in an effort to enhance its utility to humans [1]. The domestication of plants and animals lay at the core of the Neolithic Agricultural Revolution, a transition period that witnessed a rise of settled societies, an acceleration of technological innovations, and the establishment of organized systems of governance [1]. During this time, early farmers began selectively breeding plants and animals to become increasingly reliable and nutritive sources of sustenance. For instance, many crops were bred to increase the size and number of seeds, the loss of seed shattering, and a minimization of seed dormancy [2,3]. Similarly, favor was given to livestock which displayed increased passivity or docility, reductions in teeth size and number, alterations to body morphology, and reductions in brain size [4,5]. The genomic basis underlying many of these developmentally related phenotypes has been intensely studied due to their anthropological significance, applied agricultural importance, and suitability as a model system for evolutionary, genetic, and medical studies [2,6–9].
Less appreciated is the fact that a wealth of archaeological, molecular, and genetic evidence supports the parallel domestication of microbes along with that of plants and animals. Traditional artisanal food production practices such as back-slopping (the serial reinoculation of new foods with material from previous products) resulted in the continuous and long-term passage of isolated populations of microbes under specialized environmental conditions, leading to adaptation and genetic differentiation. The domestication of bacteria, yeasts, and molds was likely the unwitting result of Neolithic humans harnessing the metabolic capabilities of microbes in an effort to control the digestibility, palatability, and longevity of their newly abundant foods [10]. For example, cheese and yogurt appear very early on [11,12] and rely on the cooption of select microbes to break down lactose into lactic acid thereby making milk both more digestible and resistant to spoilage. Fruits and grain were similarly transformed and preserved by microbes, with evidence for wine, beer and bread dating back to at least 9,000 years ago [13,14]. The impact of this relationship is still observable in the form of traditional fermented food products such as wine, beer, cheese, bread, kefir, yogurt, shoyu, miso, and tempeh.
Until relatively recently, substantially less focus has centered on the genetics and genomics of microbial domestication in comparison with plant and animal domestication models, despite the crucial role microbes assume in food preservation, nutritional quality, consistency and flavor [15,16]. In this review, we focus on recent progress made toward (i) elucidating the origins of domesticated microbes in the fermented food environment, (ii) understanding the impact of domestication on genome architecture and function, and (iii) discovering of the complex microbial community dynamics responsible for a variety of fermented food products.
The Origin of Domesticated Microbes
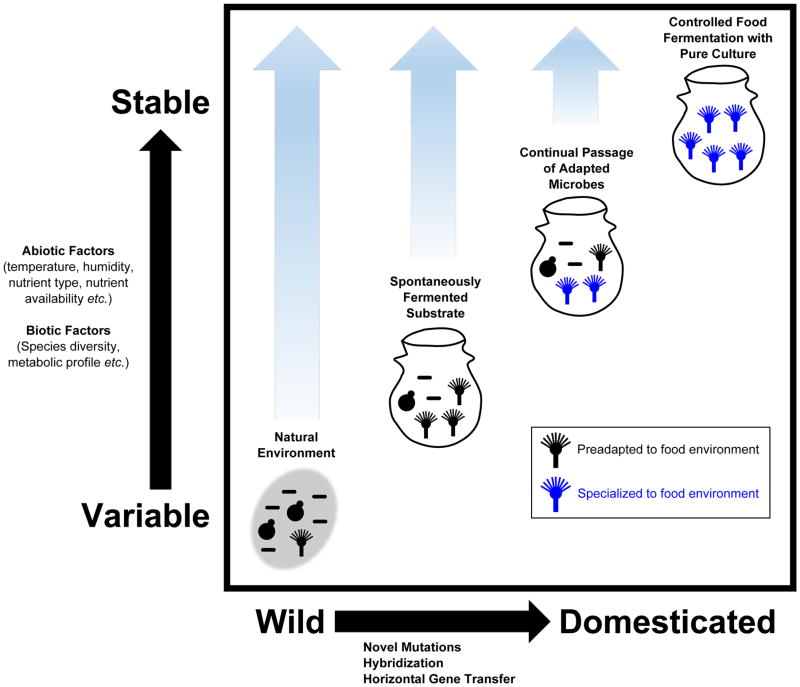
Much like domesticated plants and animals, it has been proposed that wild bacteria, yeast, and mold were “tamed” into the industrial organisms we use today [15,16]. The shift from variable and complex natural environments to more stable and relatively simple agrarian substrates favored specialized adaptations in these microbial isolates (Figure 1). Properly identifying the progenitors of domesticated species is vital to comparative genomics studies and to searching for target genes affected by artificial selection. In plant and animal examples the progenitor species are typically easily identifiable because of highly similar morphologies (with some exceptions such as maize/teosinte [17]) and overlapping geographical histories [18]. In contrast, identifying the progenitors of domesticated microbes it is often more challenging because phenotypic relationships between microbial species are not always apparent, and many microbial species are globally distributed [19]. Nevertheless, several studies have used genomic data to identify the source of domesticated microbial species.
Figure 1. Model of the progression toward microbial domestication.
In this model, a species of filamentous fungi has been domesticated (likewise, bacteria or yeast could have alternatively been depicted). The Y axis represents abiotic and biotic variability while the X axis reflects the progression from wild to domesticate. The natural environment is typically more variable than the food environment (depicted by stylized fermentation vessel) and the range of relative variability is depicted by light blue arrows in each stage. In the “spontaneously fermented substrate” stage (e.g. kimchi and cocoa), microbes preadapted to the food substrate and fermentation environment dominate the microbial community. Long-term continual passage of preadapted microbes (i.e. either through intentional back-slopping or through persistence in the food processing environment) in the more controlled food environment promotes adaptation and specialization (blue filamentous fungi in this case) which can be achieved through various mechanisms including novel mutations, hybridization, and horizontal gene transfer. Finally, in particular cases, such as industrial food production, abiotic and biotic factors are carefully regulated, and pure cultures of the domesticated microbe are used to ensure the quality, homogeneity, and safety of food product.
Lactic acid bacteria (LAB) were likely the first microbes used in food fermentation by humans and are known to enhance food flavor and texture, while also functioning to prevent spoilage by producing antimicrobial peptides and by lowering the pH of the food environment [20]. Characterized by their ability to convert hexose sugars to lactic acid [21], LAB remain a diverse and prominent starter culture for many types of fermented foods and, reflecting their industrial importance, many LAB genomes have been sequenced [22–24]. Plants represent the original habitat of some LAB species used in dairy fermentation, yet isolates of plant and dairy derived Lactococcus lactis display divergent metabolic and growth characteristics on plant derived carbohydrates [25]. This likely reflects the vastly different classes of carbohydrates found in plant and dairy sources and suggests that some dairy derived LAB lost ancestral metabolic pathways during specialization. Additionally, dairy strains show signatures of genome reduction and pseudogenization in loci involved in the metabolism of plant carbohydrates [26].
Yeasts are perhaps the most well-known domesticated microbes and are at the workhorse of fermentation during the production of beer, wine, and bread [27,28]. Species of Saccharomyces (mainly S. cerevisiae) are particularly well suited for food fermentation because they do not secrete toxic secondary metabolites, but they do produce high levels of alcohol as well as desirable flavor molecules including esters and phenols [16]. Lager-type beer originated in Bavaria around the 15th century and relies on a specialized, cryotolerant strain of Saccharomyces (S. pastorianus) that is especially amenable to cooler production temperatures of that region. Although it has been surmised that S. pastorianus was an interspecies hybrid of S. cerevisiae and a cold tolerant species, identifying the unknown species remained a mystery for several decades [29,30]. In 2011, Libkind and colleagues isolated two crytolerant yeast species from Patagonian forests, sequenced their genomes, and in doing so, identified the S. pastorianus missing hybridization donor species, which they named S. eubayanus [31]. Subsequent work has identified S. eubayanus from sources in North America [32] as well as in Asia [33] and strongly suggests an East Asian, rather than Patagonian, origin [33,34].
In addition to bacteria and yeast, molds have also served as an essential catalyst in fermented products [15]. Species of Rhizopus are used in the production of different alcoholic drinks as well as tempeh, Monascus purpureus is used to make red yeast rice, Penicillium species are used during cheese making, and Aspergillus species are utilized during the production of traditional alcoholic drinks, sauces, and condiments. A. oryzae is used in the saccharification of rice during sake production where the newly created sugars are then fermented to alcohol by S. cerevisiae [35]. It is well established that A. oryzae was domesticated from the aflatoxin producing agricultural pest, A. flavus [36], but specific details concerning the evolution of A. oryzae remained vague. To elucidate the origins of A. oryzae, Gibbons et al. [37] sequenced the whole genomes of a diverse collection of 14 A. flavus and A. oryzae isolates. Population, phylogenetic, and functional analysis confirmed that A. oryzae was likely the product of a single domestication event from an atoxigenic lineage of A. flavus [36,38,39], which may have been selected by sake makers because of its cooperation with yeast [37].
Genome Optimization of Domesticated Microbes
In their natural environment, microbes contend with labile abiotic conditions and intense competition for nutrients that are often heterogeneously distributed and intermittently available (Figure 1). In contrast, the human designated food milieu represents a stable, abundant, simplified, and less competitive niche in which artificial selection could rapidly drive microbial genome optimization. In an elegant experiment highlighting the speed at which selection may work in microbial systems, Bachmann et al. continually propagated a plant derived isolate of L. lactis on milk [26]. After only 1,000 generations the authors observed increased growth rates, increased acidification, and transcriptional profiles similar to dairy derived isolates of L. lactis. Below, we discuss several different mechanisms of genome optimization to the food environment.
The transition from generalist to specialist can lead to pseudogenization driven by relaxed selection for genes no longer useful or, in some cases, positive selection against genes now detrimental in the new environment [40]. For example, A. oryzae and A. sojae have accumulated various inactivating mutations in the cyclopiazonic acid and aflatoxin gene clusters [37,41–46], and the shochu brewing species A. kawachii, has lost a 21 Kb region of the polyketide synthase gene that drives the production of ochratoxin A [47]. Extensive loss of genes associated with primary and secondary metabolism was also observed in the M. purureus genome in comparison with other Eurotiales species [48]. In the food environment, metabolic defense mechanisms may be energetically inefficient or detrimental to interdependent microbial relationships. In S. pastorianus, both copies of the SUL1 sulfate transporters have become inactivated in favor of retaining the function of the two SUL2 genes which are more efficient under fermentation conditions [31]. LAB species have experienced comparable fates [24], exemplified in the Streprococcus thermophilus genome where more than 10% of genes, many associated with pathogenicity and carbon metabolism, have been pseudgenized or lost [49,50].
Copy number variation (CNV) is a rapid source of genotypic and phenotypic variation, is an effective strategy to alter levels of transcription and translation [49], and allows for quick adaptation to new environments in domesticated microbes. For example, A. oryzae is valued for its ability to digest rice starches and while growing on rice, the alpha-amylase gene appears as the most highly expressed gene and protein of the A. oryzae genome, but not in A. flavus [37]. Examination of the species’ respective genomes reveals the alpha-amylase gene is present in two or more copies in A. oryzae but found as only a single copy in A. flavus genomes [51]. In S. cerevisiae a number of studies observed an increased number of hexose transporter genes in strains from low glucose environments which resulted in heightened expression and increased glucose transport into the cell [52–54]. Additionally, diverse industrial strains exhibited adaptive CNV in genes functioning to assimilate different amino acids as their primary source of nitrogen [55].
Adaptive genetic variation can also be acquired via horizontal gene transfer (HGT), defined as “the non-genealogical transmission of genetic material from one organism to another” [56]. HGT is common in prokaryotes and a collection of studies over the last several years suggest it is also widely prevalent in microbial eukaryotes [57,58] and can facilitate rapid adaptation to novel food niches [59]. As HGT in industrial prokaryotes has been extensively reviewed elsewhere [59–61], here we focus on several examples of HGT events in domesticated microbial eukaryotes.
S. cerevisiae has been the recipient of several laterally transferred loci from bacteria [62–64], as well as other yeasts. Bolstered by phylogenetic and syntenic support, analysis of the wine making isolate S. cerevisiae EC1118 genome revealed several loci that appear to have been laterally transferred from the wine spoilage yeast species Zygosaccharomyces bailii [65]. Remarkably, many of these genes are relevant to the wine making process and are found almost entirely in isolates used to make wine. A. niger, used for its polysaccharide metabolizing capacities, recently acquired a 72 Kb locus from A. oryzae that contained the gene encoding for the starch degrading enzyme alpha-amylase [66]. The cheese environment was the stage for a massive lateral transfer of a genomic region containing nearly 250 genes between Penicillium camemberi and P. roqueforti [67]. The HGT region contained genes involved in conidiation and secondary metabolite production, and may have conferred a competitive advantage in the multi-microbe cheese environment. Importantly, the occurrence of HGT in domesticated microbes underscores the complex community dynamics and close organismal associations that take place in the production of most fermented foods [68,69]. As more genomes become publicly available, it is likely that additional cases of HGT will be discovered between domesticated microbes and their ecological neighbors.
Microbial Community Dynamics of Fermented Food
The interspecific interactions central to both HGT and hybridization events were the result of taxonomically heterogeneous fermentation environments. Such mixed environments were effectively unavoidable prior to the advent of the pure culture and sanitary techniques of the mid-nineteenth century, and all instances of historic, microbial domestication occurred within the context of broader microbial communities.
Traditional fermentations, which depend upon autochthonous (spontaneous) elements, are the simplest example of microbial communities being employed in human food production. Kimchi and cocoa are examples of food products derived entirely from spontaneous fermentations. Kimchi is a Korean food product resulting from the fermentation of raw vegetables in closed, un-sanitized vessels and relies solely upon the heterogeneous and variable collection of microbes present in the starting materials. While kimchi fermentations inevitably end up being predominated by just a few strains of LAB, the dominate genera can vary between given fermentations (reviewed in [70]). Moreover, in dongchimi (watery kimchi) metagenomic studies have shown the dominating microbes to vary dynamically with changing abiotic fermentation conditions (temperature, substrate, pH, free sugar etc.) [71–73]. Like kimchi, the fermentation of cocoa beans is also purely spontaneous but more dynamic and complex as it involves native LAB, dozens of yeast species, and acetic acid bacteria. However, cocoa fermentations appear to differ from those of kimchi in that individual fermentations show consistently similar bacterial profiles and progressions in the predominating species of wild yeast (reviewed in [15,16]). Therefore while the dominating microbial profiles within separate kimchi fermentations may be the result of both variation within the starting materials and their associated microbiomes, both kimchi and coca fermentations seem to derive a certain degree of batch-to-batch consistency through the human management of the food production milieu even in the absence of any inoculum (Figure 1).
Similar microbial progressions are present in the spontaneously fermented style of Belgian beer called lambic which is fermented by native microbes from both the local air and harbored in wooden fermentation and ageing vessels. Successive molecular characterization of the evolving microbiome over the course of lambic fermentations have identified qualitatively distinct, semi-overlapping phases dominated at first by a variety of enterobacteria and non-Saccharomyces yeasts followed by increasing levels of Saccharomyces including S. cerevisiae and S. bayanus [74,75], then LAB, and finally by Brettanomyces bruxellensis [75,76]. Similar microbial communities and progressions have been documented in other types of spontaneously fermented beer [28,77] and in traditionally produced kimoto-style sake [78]. Again, while no overt inoculation of microbes occurs during the production of these beverages, there are nonetheless microbes resident to the brew house environment which take part in the fermentation process and show signatures of adaptation [28,77] [79].
In contrast to such liquid and semi-solid growth conditions, fermented foods can also harbor microbial communities in the form of biofilms. Despite, their diverse structure, formation, and community composition, biofilms embody the predominate form of microbial life and confer significant fitness advantages in competitive environments. In pure-culture domestication environments (including food and laboratory) the loss of biofilm formation has been observed, reflecting its dispensability in low stress [80,81]. Cheese rinds, kombucha, and vinegar all rely on the properties of biofilms or pellicles to produce and preserve their associated food products [68]. The communities present on the exposed cheese rind are distinct from those found in the core of the cheese and are dominated by molds, yeasts, and aerobic bacteria but their interactions are equally complex and affected by milk processing and geographic location [82]. In contrast, the composition of the microbial communities of the rind appears “strikingly similar” between geographic location and harbor co-evolving communities of microbes [83]. This then suggests that the domestication signatures seen in some cheese microbes [67,84] likely arose under similar communal conditions.
Overall, the implication appears to be that the diversity of native, regional microbial communities can become wholly subsumed by the powerfully selective environment created by the food milieu. Once constrained by these human contrived niche environments, preadapted genera rapidly predominate and lay the essential ground work upon which the domestication process can further build (Figure 1).
Future Directions
Identifying the progenitor species or lineages of domesticated microbes remains a challenging and largely unexplored area of research that is fundamental to the reconstruction of genotypic and phenotypic evolution. To this end, extensive sampling combined with population genomic and phylogenomic approaches can be used to resolve the origin and number of domestication events [33,37,85,86]. Moreover, metagenomic sequencing of ecological niches that share (i) abiotic characteristics similar to the fermented food environment in question (i.e. those potentially promoting preadaptations such as temperature tolerance, flavor molecule production, carbon metabolism, and spoilage control [16]), and (ii) geographical proximity to the putative origins of the specific fermented food (e.g. cocoa producing regions), offers a useful approach for better understanding how preadapted species initially entered the food milieu. Advances in ancient DNA metagenomics also affords promising potential for identifying progenitors from preserved food [87]. Understanding the history of microbial domestication provides many instructive ethnomicrobiological and evolutionary insights, but also yields the promise of novel industrial applications. For example, the identification of the lager yeast progenitor species, S. eubayanus, has enabled the generation of new S. pastorianus strains [88,89].
Cheaper and more efficient DNA sequencing and genotyping technologies have substantially improved our ability to finely catalog genetic variation at single nucleotide resolution and to identify the genomic underpinnings of phenotypes. These approaches have recently been applied to yeast to map quantitative trait loci (QTL) responsible for industrially important traits such as sulfite resistance [90], aromatic compound production [91], flocculation [92], and thermotolerance [93]. Future work on other domesticated microbes would first have to focus on establishing conditions to induce sexual reproduction in cryptically sexual food-related eukaryotic microbes [94], as has been initiated in Aspergillus oryzae [95,96] and Penicillium roqueforti [97,98]. This would enable QTL analysis, and is promising for breeding strains with combinations of desirable traits.
For primarily (or solely) asexually reproducing species, experimental evolution followed by whole-genome sequencing (i.e. an evolve and resequence approach) would be a powerful strategy to select for desired phenotypes and then to track the mutational landscape in real time. Microbes offer rapid generation times, streamlined genomes, established phenotypic measurement assays, and long-term storage and viability, and are thus powerful models of experimental evolution to examine the speed of adaptation, rates of parallel evolution, and the spectrum of adaptive mutations in conditions similar to those experienced during domestication [26]. In recent years, experimental evolution systems have been developed around several species of filamentous fungi [99–102] and are reflective of the potential these approaches offer to rapidly select for desirable traits in domesticates and to model the impact of microbial domestication in progenitors.
Acknowledgments
We thank two anonymous reviewers for constructive suggestions on an earlier version of this review. We acknowledge financial support from Clark University (JGG) and NIDCD through NRSA F31 DC012991 (DCR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Lin Z, Li X, Shannon LM, Yeh CT, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer RS, DuVal AE, Jensen HR. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012;196:29–48. doi: 10.1111/j.1469-8137.2012.04253.x. [DOI] [PubMed] [Google Scholar]
- 4.Trut L, Oskina I, Kharlamova A. Animal evolution during domestication: the domesticated fox as a model. Bioessays. 2009;31:349–360. doi: 10.1002/bies.200800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkins AS, Wrangham RW, Fitch WT. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197:795–808. doi: 10.1534/genetics.114.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- 7.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 9.Wayne RK, vonHoldt BM. Evolutionary genomics of dog domestication. Mamm Genome. 2012;23:3–18. doi: 10.1007/s00335-011-9386-7. [DOI] [PubMed] [Google Scholar]
- 10.Ross RP, Morgan S, Hill C. Preservation and fermentation: past, present and future. Int J Food Microbiol. 2002;79:3–16. doi: 10.1016/s0168-1605(02)00174-5. [DOI] [PubMed] [Google Scholar]
- 11.Bogucki PI. Ceramic sieves of the linear pottery culture and their economic implications. Oxford Journal of Archaeology. 1984;3:15–30. [Google Scholar]
- 12.Salque M, Bogucki PI, Pyzel J, Sobkowiak-Tabaka I, Grygiel R, Szmyt M, Evershed RP. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature. 2013;493:522–525. doi: 10.1038/nature11698. [DOI] [PubMed] [Google Scholar]
- 13••.McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, Nunez A, Butrym ED, Richards MP, Wang CS, et al. Fermented beverages of pre- and proto-historic China. Proc Natl Acad Sci U S A. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. Chemical analysis of pottery from ancient China provides evidence for perhaps the first fermeted beverages dating 9,000 years ago. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsinelli M. Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol. 2003;57 (Suppl 1):S226–232. doi: 10.1007/s00239-003-0031-2. [DOI] [PubMed] [Google Scholar]
- 15.Douglas GL, Klaenhammer TR. Genomic evolution of domesticated microorganisms. Annu Rev Food Sci Technol. 2010;1:397–414. doi: 10.1146/annurev.food.102308.124134. [DOI] [PubMed] [Google Scholar]
- 16.Steensels J, Verstrepen KJ. Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu Rev Microbiol. 2014;68:61–80. doi: 10.1146/annurev-micro-091213-113025. [DOI] [PubMed] [Google Scholar]
- 17.Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 18.Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, Barton L, Climer Vigueira C, Denham T, Dobney K, et al. Current perspectives and the future of domestication studies. Proc Natl Acad Sci U S A. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 20.Price CE, Zeyniyev A, Kuipers OP, Kok J. From meadows to milk to mucosa - adaptation of Streptococcus and Lactococcus species to their nutritional environments. FEMS Microbiol Rev. 2012;36:949–971. doi: 10.1111/j.1574-6976.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- 21.Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007;189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bull MJ, Jolley KA, Bray JE, Aerts M, Vandamme P, Maiden MC, Marchesi JR, Mahenthiralingam E. The domestication of the probiotic bacterium Lactobacillus acidophilus. Sci Rep. 2014;4:7202. doi: 10.1038/srep07202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douillard FP, Ribbera A, Kant R, Pietila TE, Jarvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013;9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siezen RJ, Starrenburg MJ, Boekhorst J, Renckens B, Molenaar D, van Hylckama Vlieg JE. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl Environ Microbiol. 2008;74:424–436. doi: 10.1128/AEM.01850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Bachmann H, Starrenburg MJ, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2012;22:115–124. doi: 10.1101/gr.121285.111. An exceptional experimental evolution study examining the adaptation of a plant derived isolate of Lactococcus lactis to dairy in only 1,000 generations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 28••.Bokulich NA, Bamforth CW. The microbiology of malting and brewing. Microbiol Mol Biol Rev. 2013;77:157–172. doi: 10.1128/MMBR.00060-12. A comprehensive review of the major literature relevant to the microbiology of brewing yeast, fermentation management, and the microbial ecology of beer and brewing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn B, Sherlock G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008;18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, Nakamura N, Shimonaga T, Hattori M, Ashikari T. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009;16:115–129. doi: 10.1093/dnares/dsp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. An ecological survey and comparative genomic analysis that identified Saccharomyces eubayanus as the missing donor species of the lager yeast hybrid S. pastorianus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peris D, Sylvester K, Libkind D, Goncalves P, Sampaio JP, Alexander WG, Hittinger CT. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol Ecol. 2014;23:2031–2045. doi: 10.1111/mec.12702. [DOI] [PubMed] [Google Scholar]
- 33•.Bing J, Han PJ, Liu WQ, Wang QM, Bai FY. Evidence for a Far East Asian origin of lager beer yeast. Curr Biol. 2014;24:R380–381. doi: 10.1016/j.cub.2014.04.031. An ecological and phylogenetic study identifying S. eubayanus from the Tibetian plateau providing evidence that lager yeast originated from East Asia. [DOI] [PubMed] [Google Scholar]
- 34•.Wendland J. Lager yeast comes of age. Eukaryot Cell. 2014;13:1256–1265. doi: 10.1128/EC.00134-14. A timely minireview that is immensely useful in summarizing the many recent genomic discoveries and developments pertaining to the origin and optimiztion of cryotolerant brewing yeasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 2008;15:173–183. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 37••.Gibbons JG, Salichos L, Slot JC, Rinker DC, McGary KL, King JG, Klich MA, Tabb DL, McDonald WH, Rokas A. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr Biol. 2012;22:1403–1409. doi: 10.1016/j.cub.2012.05.033. A population, comparative, and functional genomic study providing strong evidence for the origin of Aspergillus oryzae and the adaptations that ensued during its adaptation to starch. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J. Whole genome comparison of Aspergillus flavus and A. oryzae. Medical Mycology. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 39.Geiser DM, Dorner JW, Horn BW, Taylor JW. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet Biol. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Grus WE, Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang PK, Horn BW, Dorner JW. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Chang PK, Matsushima K, Takahashi T, Yu J, Abe K, Bhatnagar D, Yuan GF, Koyama Y, Cleveland TE. Understanding nonaflatoxigenicity of Aspergillus sojae: a windfall of aflatoxin biosynthesis research. Appl Microbiol Biotechnol. 2007;76:977–984. doi: 10.1007/s00253-007-1116-4. [DOI] [PubMed] [Google Scholar]
- 43.Linz JE, Wee J, Roze LV. Aspergillus parasiticus SU-1 genome sequence, predicted chromosome structure, and comparative gene expression under aflatoxin-inducing conditions: evidence that differential expression contributes to species phenotype. Eukaryot Cell. 2014;13:1113–1123. doi: 10.1128/EC.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rank C, Klejnstrup ML, Petersen LM, Kildgaard S, Frisvad JC, Held Gotfredsen C, Ostenfeld Larsen T. Comparative Chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357) Metabolites. 2012;2:39–56. doi: 10.3390/metabo2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato A, Oshima K, Noguchi H, Ogawa M, Takahashi T, Oguma T, Koyama Y, Itoh T, Hattori M, Hanya Y. Draft genome sequencing and comparative analysis of Aspergillus sojae NBRC4239. DNA Res. 2011;18:165–176. doi: 10.1093/dnares/dsr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tominaga M, Lee YH, Hayashi R, Suzuki Y, Yamada O, Sakamoto K, Gotoh K, Akita O. Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl Environ Microbiol. 2006;72:484–490. doi: 10.1128/AEM.72.1.484-490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Futagami T, Mori K, Yamashita A, Wada S, Kajiwara Y, Takashita H, Omori T, Takegawa K, Tashiro K, Kuhara S, et al. Genome sequence of the white koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese distilled spirit shochu. Eukaryot Cell. 2011;10:1586–1587. doi: 10.1128/EC.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Liu B, Du X, Li P, Liang B, Cheng X, Du L, Huang D, Wang L, Wang S. Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Sci Rep. 2015;5:8331. doi: 10.1038/srep08331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goh YJ, Goin C, O’Flaherty S, Altermann E, Hutkins R. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact. 2011;10 (Suppl 1):S22. doi: 10.1186/1475-2859-10-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunter AJ, Jin B, Kelly JM. Independent duplications of alpha-amylase in different strains of Aspergillus oryzae. Fungal Genet Biol. 2011;48:438–444. doi: 10.1016/j.fgb.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 53.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Z, Li WH. Expansion of hexose transporter genes was associated with the evolution of aerobic fermentation in yeasts. Mol Biol Evol. 2011;28:131–142. doi: 10.1093/molbev/msq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibanez C, Perez-Torrado R, Chiva R, Guillamon JM, Barrio E, Querol A. Comparative genomic analysis of Saccharomyces cerevisiae yeasts isolated from fermentations of traditional beverages unveils different adaptive strategies. Int J Food Microbiol. 2014;171:129–135. doi: 10.1016/j.ijfoodmicro.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 56.Goldenfeld N, Woese C. Biology’s next revolution. Nature. 2007;445:369. doi: 10.1038/445369a. [DOI] [PubMed] [Google Scholar]
- 57.Richards TA, Talbot NJ. Horizontal gene transfer in osmotrophs: playing with public goods. Nat Rev Microbiol. 2013;11:720–727. doi: 10.1038/nrmicro3108. [DOI] [PubMed] [Google Scholar]
- 58.Fitzpatrick DA. Horizontal gene transfer in fungi. FEMS Microbiol Lett. 2012;329:1–8. doi: 10.1111/j.1574-6968.2011.02465.x. [DOI] [PubMed] [Google Scholar]
- 59.Rossi F, Rizzotti L, Felis GE, Torriani S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiology. 2014;42:232–243. doi: 10.1016/j.fm.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 60.Kelly BG, Vespermann A, Bolton DJ. Gene transfer events and their occurrence in selected environments. Food Chem Toxicol. 2009;47:978–983. doi: 10.1016/j.fct.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 61.van Reenen CA, Dicks LM. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: what are the possibilities? A review. Arch Microbiol. 2011;193:157–168. doi: 10.1007/s00203-010-0668-3. [DOI] [PubMed] [Google Scholar]
- 62.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hall C, Dietrich FS. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics. 2007;177:2293–2307. doi: 10.1534/genetics.107.074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, Cambon B, Legras JL, Wincker P, Casaregola S, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci U S A. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. A interesting example of adaptive horizontal gene transfer between divergent occupants of the wine fermentation environment S. cerrevisae and Zygosaccharomyces bailii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, Frisvad JC, Nielsen KF, Albang R, Albermann K, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21:885–897. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, Abraham AL, Ceppi M, Conseiller E, Debuchy R, et al. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat Commun. 2014;5:2876. doi: 10.1038/ncomms3876. An exceptional example of a massive hoizontally transferred genomic region between species of Penicillium associated with dairy and cheesemaking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe BE, Dutton RJ. Fermented Foods as Experimentally Tractable Microbial Ecosystems. Cell. 2015;161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 69.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 70.Jung JY, Lee SH, Jeon CO. Kimchi microflora: history, current status, and perspectives for industrial kimchi production. Appl Microbiol Biotechnol. 2014;98:2385–2393. doi: 10.1007/s00253-014-5513-1. [DOI] [PubMed] [Google Scholar]
- 71.Cho J, Lee D, Yang C, Jeon J, Kim J, Han H. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol Lett. 2006;257:262–267. doi: 10.1111/j.1574-6968.2006.00186.x. [DOI] [PubMed] [Google Scholar]
- 72•.Jeong SH, Jung JY, Lee SH, Jin HM, Jeon CO. Microbial succession and metabolite changes during fermentation of dongchimi, traditional Korean watery kimchi. Int J Food Microbiol. 2013;164:46–53. doi: 10.1016/j.ijfoodmicro.2013.03.016. A tidy study that examines the sucession of both microbial communities and metabolite levels over the entire course of the fermentation of a traditional style of kimchi. [DOI] [PubMed] [Google Scholar]
- 73.Kyung KH, Medina Pradas E, Kim SG, Lee YJ, Kim KH, Choi JJ, Cho JH, Chung CH, Barrangou R, Breidt F. Microbial ecology of watery kimchi. J Food Sci. 2015;80:M1031–1038. doi: 10.1111/1750-3841.12848. [DOI] [PubMed] [Google Scholar]
- 74.Martens H, Dawoud E, Verachtert H. Wort Enterobacteria and other Microbial Populations Involved during the First Month of Lambic Fermentation. Journal of the Institute of Brewing. 1991;97:435–439. [Google Scholar]
- 75•.Van Oevelen D, Spaepen M, Timmermans P, Verachtert H. Microbial Aspects of Spontaneous Wort Fermentation in the Production of Lambic and Gueuze. Journal of the Institute of Brewing. 1977;83:356–360. The seminal study to comprehensively characterize lambic fermentations over the two year matuartion period. [Google Scholar]
- 76•.Spitaels F, Wieme AD, Janssens M, Aerts M, Daniel HM, Van Landschoot A, De Vuyst L, Vandamme P. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS One. 2014;9:e95384. doi: 10.1371/journal.pone.0095384. An updated timecourse examination of lambic fermentation that uses MALDI-TOF MS and sequencing technologies to characterize microbial diversity both within the beer and within the brewign environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bokulich NA, Bamforth CW, Mills DA. Brewhouse-resident microbiota are responsible for multistage fermentation of American coolship ale. PLoS One. 2012;7:e35507. doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bokulich NA, Ohta M, Lee M, Mills DA. Indigenous bacteria and fungi drive traditional kimoto sake fermentations. Appl Environ Microbiol. 2014;80:5522–5529. doi: 10.1128/AEM.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Bokulich NA, Bergsveinson J, Ziola B, Mills DA. Mapping microbial ecosystems and spoilage-gene flow in breweries highlights patterns of contamination and resistance. Elife. 2015;4 doi: 10.7554/eLife.04634. A comprehensive examination of how microbial populations even in modern, industrialized contexts display a dynamism of their own and very suggestive of how microbially diverse premodern brewing environments must have been. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 82.Montel MC, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol. 2014;177:136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 83••.Wolfe BE, Button JE, Santarelli M, Dutton RJ. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–433. doi: 10.1016/j.cell.2014.05.041. Results of metagenomic sequencing of cheese rind communities reveal that their formation and assembly are geographically conserved, and when communal variability is observed it is attributable to differences in the cheese making process itself. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Passerini D, Beltramo C, Coddeville M, Quentin Y, Ritzenthaler P, Daveran-Mingot ML, Le Bourgeois P. Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS One. 2010;5:e15306. doi: 10.1371/journal.pone.0015306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gillot G, Jany JL, Coton M, Le Floch G, Debaets S, Ropars J, Lopez-Villavicencio M, Dupont J, Branca A, Giraud T, et al. Insights into Penicillium roqueforti Morphological and Genetic Diversity. PLoS One. 2015;10:e0129849. doi: 10.1371/journal.pone.0129849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Millar CD, Huynen L, Subramanian S, Mohandesan E, Lambert DM. New developments in ancient genomics. Trends Ecol Evol. 2008;23:386–393. doi: 10.1016/j.tree.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 88.Krogerus K, Magalhaes F, Vidgren V, Gibson B. New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol. 2015;42:769–778. doi: 10.1007/s10295-015-1597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89••.Hebly M, Brickwedde A, Bolat I, Driessen MR, de Hulster EA, van den Broek M, Pronk JT, Geertman JM, Daran JM, Daran-Lapujade P. S. cerevisiae x S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov005. A bonafide industrial application of domesticated microbial evolutionary genomics made possible by identifying the origin of S. pastorianus. [DOI] [PubMed] [Google Scholar]
- 90.Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P. QTL dissection of Lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One. 2014;9:e86298. doi: 10.1371/journal.pone.0086298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steyer D, Ambroset C, Brion C, Claudel P, Delobel P, Sanchez I, Erny C, Blondin B, Karst F, Legras JL. QTL mapping of the production of wine aroma compounds by yeast. BMC Genomics. 2012;13:573. doi: 10.1186/1471-2164-13-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilkening S, Lin G, Fritsch ES, Tekkedil MM, Anders S, Kuehn R, Nguyen M, Aiyar RS, Proctor M, Sakhanenko NA, et al. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics. 2014;196:853–865. doi: 10.1534/genetics.113.160291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Y, Foulquie-Moreno MR, Clement L, Erdei E, Tanghe A, Schaerlaekens K, Dumortier F, Thevelein JM. QTL analysis of high thermotolerance with superior and downgraded parental yeast strains reveals new minor QTLs and converges on novel causative alleles involved in RNA processing. PLoS Genet. 2013;9:e1003693. doi: 10.1371/journal.pgen.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuck U, Bohm J. Mating type genes and cryptic sexuality as tools for genetically manipulating industrial molds. Appl Microbiol Biotechnol. 2013;97:9609–9620. doi: 10.1007/s00253-013-5268-0. [DOI] [PubMed] [Google Scholar]
- 95.Wada R, Jin FJ, Koyama Y, Maruyama J, Kitamoto K. Efficient formation of heterokaryotic sclerotia in the filamentous fungus Aspergillus oryzae. Appl Microbiol Biotechnol. 2014;98:325–334. doi: 10.1007/s00253-013-5314-y. [DOI] [PubMed] [Google Scholar]
- 96.Wada R, Maruyama J, Yamaguchi H, Yamamoto N, Wagu Y, Paoletti M, Archer DB, Dyer PS, Kitamoto K. Presence and functionality of mating type genes in the supposedly asexual filamentous fungus Aspergillus oryzae. Appl Environ Microbiol. 2012;78:2819–2829. doi: 10.1128/AEM.07034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ropars J, Dupont J, Fontanillas E, Rodriguez de la Vega RC, Malagnac F, Coton M, Giraud T, Lopez-Villavicencio M. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PLoS One. 2012;7:e49665. doi: 10.1371/journal.pone.0049665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ropars J, Lopez-Villavicencio M, Dupont J, Snirc A, Gillot G, Coton M, Jany JL, Coton E, Giraud T. Induction of sexual reproduction and genetic diversity in the cheese fungus Penicillium roqueforti. Evol Appl. 2014;7:433–441. doi: 10.1111/eva.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Crecy E, Jaronski S, Lyons B, Lyons TJ, Keyhani NO. Directed evolution of a filamentous fungus for thermotolerance. BMC Biotechnol. 2009;9:74. doi: 10.1186/1472-6750-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeon J, Choi J, Lee GW, Dean RA, Lee YH. Experimental evolution reveals genome-wide spectrum and dynamics of mutations in the rice blast fungus, Magnaporthe oryzae. PLoS One. 2013;8:e65416. doi: 10.1371/journal.pone.0065416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schoustra SE, Debets AJ, Slakhorst M, Hoekstra RF. Reducing the cost of resistance; experimental evolution in the filamentous fungus Aspergillus nidulans. J Evol Biol. 2006;19:1115–1127. doi: 10.1111/j.1420-9101.2006.01102.x. [DOI] [PubMed] [Google Scholar]
- 102.Schoustra SE, Debets AJ, Slakhorst M, Hoekstra RF. Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet. 2007;3:e68. doi: 10.1371/journal.pgen.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]