To maneuver within their environment, aquatic organisms employ a variety of locomotive strategies. These diverse mechanisms offer inspiration in designing artificial microswimmers for applications ranging from directed drug delivery to accelerated environmental decontamination.[1–6] One challenge in adapting naturally-evolved designs for “smart” microswimmer systems lies in replicating their complex biomimetic form and function. Here, using a rapid 3D microprinting platform – microscale continuous optical printing (μCOP) – we engineered hydrogel microfish featuring biomimetic structures, locomotive capabilities, and functionalized nanoparticles. The μCOP system can print complex 3D structures within seconds at high resolution (~1 μm) across multiple orders of magnitude in scale. The 3D-printed microfish exhibit propulsion that is highly-efficient, chemically-powered, and magnetically-guidable. By incorporating polydiacetylene nanoparticles, we demonstrate the microfish’s utility in toxin-neutralization applications. The multiple capabilities integrated within these proof-of-concept microfish highlight the technical flexibility and broad applicability of our approach in engineering advanced functional bio-robotics for actuation, sensing, and detoxification.
With recent advances in nanoscience and nanomanufacturing technologies, the areas of biomimetic micro-robotics and nanomotors have seen rapid development in realizing functionalities mimicking natural organisms with self-propulsion.[1–6] Significant progress has been made over the past decade towards the design and fabrication of micro-/nanoswimmers with various locomotion mechanisms.[7–10] For instance, techniques for creating rolled-up nanostructures have led to the fabrication of tubular microjet engines and microdrillers.[11–14] Template-assisted electrochemical deposition has also been used to synthesize high-performance tubular micro-rockets and nanowire motors.[15,16] These microswimmers feature relatively simple spherical or cylindrical geometries, indicating the limitation of these techniques in creating complex 3D designs to mimic the sophisticated structure and function of their biological counterparts. Moreover, most microswimmers are composed of homogeneous inorganic materials and thus lack the capability of multiplexing sophisticated functionalities. While laser direct-writing enables the fabrication of microswimmers with more sophisticated 3D designs, such as helical structures used for targeted cargo transport,[17] the technique offers limited scalability due to the serial nature of its raster-scanning process, which is prohibitively time-consuming for large batch fabrication. Molding approaches have been used to create biomimetic polydimethylsiloxane (PDMS) microswimmers, propelled by either external magnetic fields or contractile cardiomyocytes.[18–21] However, the ferromagnetic materials or contractile cells must be post-deposited or seeded after the PDMS molding process, preventing more complex functionalization of the microswimmers. In addition, the near-millimeter size of these PDMS swimmers limits their applicability in micro- or nano-motion studies. The capability to fabricate complex architectures and miniaturize the dimension is highly desired for designing and customizing more functionalized, integrated and intelligent micromachines for different applications as well as investigating the fundamental aspects of microscale locomotion.[22] In light of these challenges, rapid 3D optical printing offers a promising alternative for efficiently manufacturing controllable microswimmers with complex 3D microscale structures composed of patterned heterogeneous materials as well as different functional components.[23–25]
In this work, we have developed a rapid 3D printing technology - microscale continuous optical printing (μCOP) - to engineer functionalized artificial microfish with diverse biomimetic structures and locomotive capabilities. The μCOP technology offers efficient fabrication speeds, provides improved feature resolution, and requires no harsh chemicals for fabrication. To 3D print artificial microfish with high-fidelity shapes and structures in an economical and scalable manner, we optimized this μCOP system to construct freely swimming microfish composed of polyethylene glycol diacrylate (PEGDA)-based hydrogels and functional nanoparticles as a proof of concept. By extending the μCOP technology to print advanced materials that incorporate three different types of functional nanoparticles – platinum (Pt) nanoparticles for chemically-mediated propulsion, iron oxide (Fe3O4) nanoparticles for magnetic guidance, and polydiacetylene (PDA) nanoparticles for toxin neutralization – we can create highly functionalized microfish using an approach that can be more broadly applied to engineering complex biomimetic micromotors with diverse custom-designed functions.
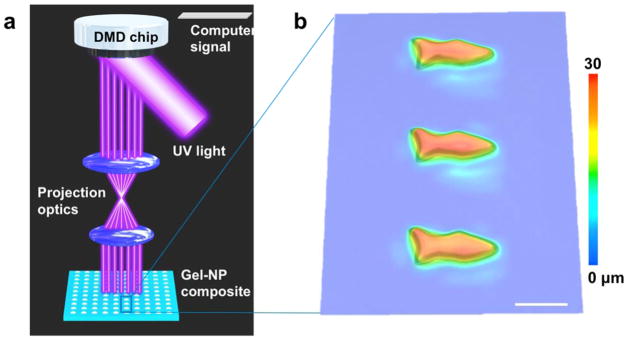
The setup of the μCOP platform is schematically illustrated in Figure 1a. The key component of the μCOP system is a digital micromirror device (DMD) chip composed of approximately two million micromirrors for continuous optical pattern generation. The DMD chip modulates the UV light and projects the optical pattern – dictated by the custom designed computer-aided design (CAD) model – onto the photopolymer solution. The 3D model can be defined using commercially available CAD software and sliced into a series of digital images. Through a computer interface coupled with in-house developed software, these images are automatically and continuously loaded one-by-one onto the DMD chip and then projected onto the photopolymerizable materials, which are loaded on a motorized stage. Areas illuminated by UV light crosslink within seconds, while leaving the dark regions uncrosslinked, forming a patterned layer in a specific polymerization plane. By iterating through the sequence of optical patterns in continuous synchrony with the movement of the sample stage during the required UV exposure time, we can construct polymer scaffolds with complex structures and submicron feature resolutions. PEGDA, a biocompatible hydrogel that has been widely used in medical implants, drug delivery devices, and tissue culture, was chosen as the photopolymerizable matrix material for the fabrication of the fish body and the encapsulation of functional nanoparticles.
Figure 1.
(a) Schematic illustration of the μCOP method to fabricate microfish. UV light illuminates the DMD mirrors, generating an optical pattern specified by the control computer. The pattern is projected through optics onto the photosensitive monomer solution to fabricate the fish layer-by-layer. (b) 3D microscopy image of an array of printed microfish. Scale bar, 100 μm.
Optimizing the swimming performance of microswimmers requires precise control over their shape and size. Our μCOP platform is capable of building a wide array of complex 3D geometries, allowing for efficient iteration through various microswimmer designs. Here we chose to print microfish of multiple biomimetic forms to demonstrate the potential for using this approach to systematically refine the microswimmer design to optimize its swimming performance. With this digitized and continuous optical printing technique, we are able to fabricate a large array of 3D microfish within mere seconds. In order to characterize the 3D morphology of the printed microfish, a 3D observation digital microscope (Keyence VHX1000) is used to capture the images of the microfish array from bottom to top at a step-size of 1 μm and to subsequently construct a 3D microscopy image that represents the height profile (Figure 1b). The thickness of the microfish is approximately 30 μm and the length of the microfish is 120 μm. These parameters can be readily varied during the 3D fabrication process by changing the digital masks and layer settings.
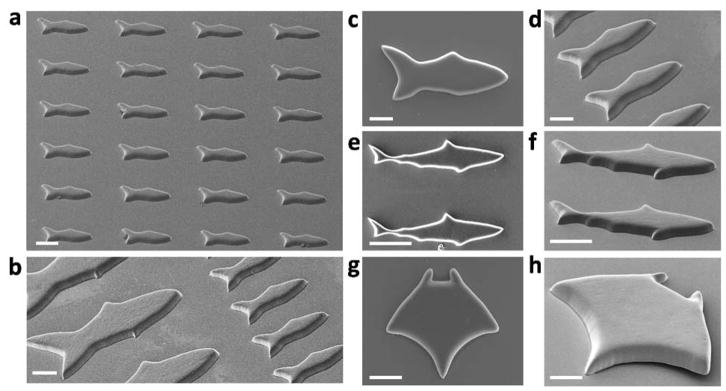
The digitized nature of the μCOP technique enables us to modulate the parameters as well as the designs of the microfish conveniently (Figure 2). Figure 2a illustrates that large arrays of microfish can be printed with excellent uniformity and precision. Multiple sizes (Figure 2b) and shapes (Figure 2c–h) of microfish can be produced on the same substrate, with the size of the microfish tunable from micrometer to nearly centimeter scale. Furthermore, the PEGDA material comprising the microfish bodies can be substituted for other photopolymerizable polymers as appropriate for different functions and applications. Importantly, the flexibility of this technique enables us to pattern or embed different materials at specific locations for custom-designed functions. For example, we are able to fabricate a microfish with magnetic Fe3O4 nanoparticles at the head and catalytic Pt nanoparticles at the tail (Figure 3). With these two types of functional nanoparticles, the swimming direction and speed of the microfish can be controlled readily.
Figure 2.
Assorted SEM images of the microfish. (a) A uniform array of microfish. (b) Microfish of various sizes and (c–h) fabricated fish of different designs: fish, shark, and manta ray, respectively. Scale bar, 50 μm.
Figure 3.
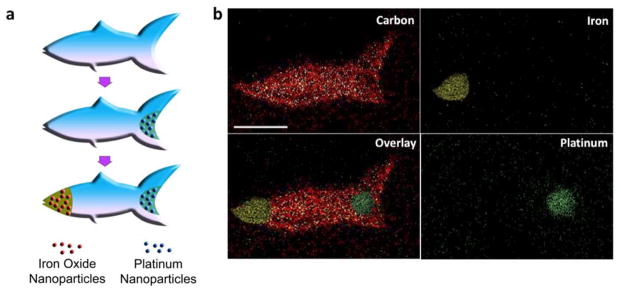
(a) Schematic illustration of the process of functionalizing a microfish for guided catalytic propulsion. Pt nanoparticles are first loaded into the tail of the fish for propulsion via catalytic decomposition of H2O2. Secondly, Fe3O4 nanoparticles are loaded into the head of the fish for magnetic control. (b) EDX spectroscopy images illustrating the iron-oxide head and platinum tail with respect to the PEGDA microfish body. Scale bar, 50 μm.
The fabrication process of the chemically-powered and magnetically-guided microfish is illustrated step by step in Figure 3a. First of all, the hydrogel body of the fish is fabricated with photopolymerizable solutions such as PEGDA in this particular case. Secondly, the solution is replaced with PEGDA containing catalytic Pt nanoparticles and only the tail of the microfish is polymerized to encapsulate the Pt nanoparticles, which enables efficient self-propulsion via decomposition of the peroxide fuel. Thirdly, the solution is replaced with PEGDA containing magnetic Fe3O4 nanoparticles and only the fish head is polymerized to encapsulate the magnetic Fe3O4 nanoparticles, which guide the motion of the microfish. Energy-dispersive X-ray (EDX) spectroscopy images show the successful functionalization of the microswimmers with the magnetic control and catalytic patch segments at different regions (Figure 3b), demonstrating the capability of the μCOP system to localize different functional nanoparticles at specific positions with high accuracy. Other nanoparticles can be further patterned on the microfish to achieve greater multiplexing and more complex functionalities.
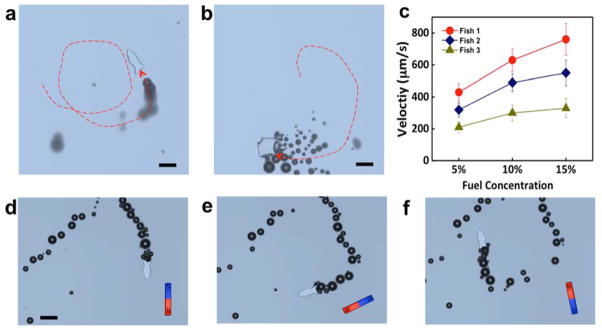
Pt nanoparticle encapsulation allows for efficient propulsion of the microfish, as illustrated in Figure 4. Hydrogen peroxide was used as the fuel source. The track lines of microfish motion, shown in Figure 4a and 4b and corresponding to Supplementary Movie 1 and 2, illustrate the thrust created by bubbles generated during peroxide decomposition at the tail ends of the fish, demonstrating the efficacy of the site-specific catalytic nanoparticle loading. The catalytically-generated oxygen bubbles expelled from microfish tail propel the microfish while fast and high-frequency bubble ejection leads to continuous motion of the microfish. Fundamentally, the propulsion force increases with the rate of the catalytic reaction that leads to faster bubble generation.[26] Therefore, increased peroxide concentration and loading of the Pt catalyst both lead to faster oxygen bubble generation for a more efficient propulsion. The microfish are able to achieve a speed of 780 μm s−1 in a 15 % peroxide solution. The Stokes’s drag force is highly related to the geometry of the microstructures.[27] Different fish geometries result in different speeds under identical fuel concentrations, owing to different Stokes’ drag at such small scales. In Figure 4c, Fish 1 and Fish 2 are both loaded with the same concentration of Pt nanoparticles, while Fish 1 has a shape of common fish and Fish 2 has shape of manta ray. The average speed of Fish 1 is 1.5 times higher than that of Fish 2, indicating that streamline geometry of microfish 1 can reduce drag forces for faster locomotion compared to the micro manta rays (Figure 4c). In general, our results indicate that the shape of the microfish can be optimized to achieve higher speeds. The speed can also be readily tuned by tailoring the loaded concentration of the catalytic Pt nanoparticles. In Figure 4c, Fish 1 and Fish 3 have the same shape of common fish, yet Fish 1 is loaded with twice the amount of Pt nanoparticles than that of Fish 3, yielding a higher speed under the same fuel concentrations.
Figure 4.
Track lines of motion of (a) a microfish and (b) a micro manta ray, over 3 seconds in the presence of 10 % hydrogen peroxide (highlighted from Supplementary Movie 1 and 2). (c) Speed profiles of microfish with different shape designs and different Pt nanoparticle concentrations at 5, 10, and 15% H2O2. Fish 1: common fish with 8.0×108 Pt particles per ml; Fish 2: manta ray with 8.0×108 Pt particles per ml; Fish 3: common fish with 4.0×108 Pt particles per ml. (d–f) Time-lapse images of the microfish movement in 15% H2O2, performing sharp turns with magnetic guidance (Supplementary Movie 3). Scale bar, 100 μm.
The encapsulation of Fe3O4 nanoparticles also allows for magnetic guidance. Such encapsulation of magnetic nanoparticles within the fish head provides a net magnetization, thus enabling magnetic alignment and guidance by a remote magnet during the chemical propulsion. The time-lapse images in Figure 4d–f, corresponding to Supplementary Movie 3, illustrate remote guidance of the microfish achieved by rotating a nearby magnet. The swimming microfish exhibits rapid alignment to the external magnetic field. Combined, the nanoparticle encapsulation methods can readily convert these polymeric microfish into efficient and controllable microswimmers for diverse applications.
To explore a potential application of the 3D printed microfish, we further incorporated functional toxin-neutralizing nanoparticles into the hydrogel matrix of the fish body as a proof-of-concept demonstration of its use in a detoxification context. Specifically, we used PDA nanoparticles, made from the self-assembly of 10,12-pentacosadiynoic acid (PCDA), for the detoxification of melittin.[24] The PDA nanoparticles have the structure of nanovesicles, with the surface made of a π-conjugated polymer. This unique surface structure can be used to mimic the biological membrane of a cell to effectively capture and neutralize pore-forming toxins via binding interactions between the nanoparticles and the toxins. The bound toxins will disrupt the ordered π-conjugated chain structure of the PDA nanoparticle surface, leading to fluorescence emission, which enables us to use these particles as toxin sensors as well. Change of the fluorescence intensity can serve as a good indicator of the detoxification efficiency during the experiment.
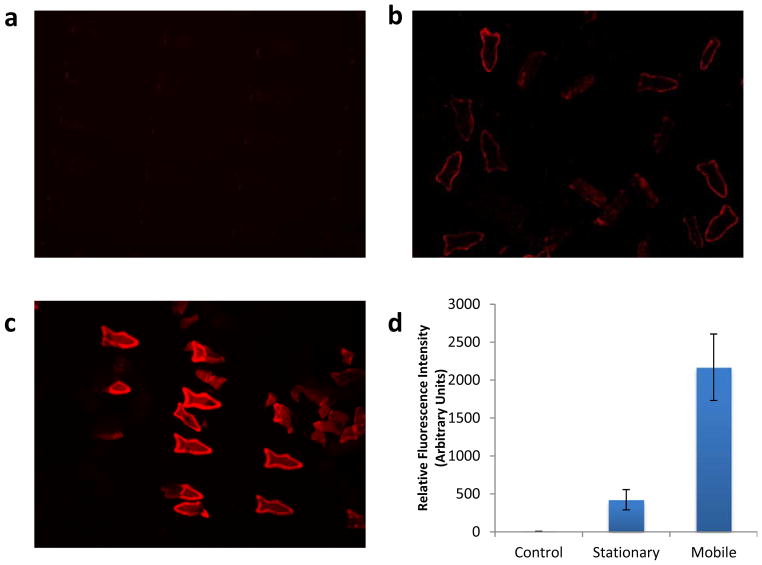
To evaluate the detoxification capability of the PDA nanoparticle enhanced microfish, we designed three test groups (Figure 5). In the control group, we incubated these functionalized microfish with 5% H2O2 solution at 37 °C for 10 mins, and no visible fluorescence was detected (Figure 5a). In the second group, we incubated the microfish in a 5% H2O2 solution containing 2.5 mg/ml melittin at 37 °C for 10 mins. No Pt nanoparticles were incorporated into this group, so the microfish were stationary during the incubation period. As shown in Figure 5b, a low intensity of fluorescence was observed. Comparing these two groups, we confirmed that the interaction between the melittin toxin and the PDA nanoparticles led to the red fluorescence emission. In the third group, Pt nanoparticles were incorporated within the tails of the functionalized microfish to make them mobile when exposed to the peroxide solution. These microfish were also incubated in a 5% H2O2 solution containing 2.5 mg ml−1 melittin at 37 °C for 10 mins. Figure 5c indicates that the motion of the microfish significantly enhanced the interaction between the melittin toxin and the PDA nanoparticles encapsulated in the microfish. The fluorescence intensity was further quantified to compare the detoxification efficacy (Figure 5d) using ImageJ, as previously described.[28] Statistical analysis was performed to compare sample means by ANOVA followed by Tukey’s post-hoc test, and p < 0.05 was considered statistically significant. These data indicate that there is a significant difference between the mobile and stationary microfish, highlighting the importance of the microswimmer movement for enhancing detoxification process.
Figure 5.
Fluorescent images demonstrating the detoxification capability of the microfish containing encapsulated PDA nanoparticles. (a) Control group of microfish incubated in 5% H2O2 without melittin toxin. (b) Stationary microfish incubated in 2.5 mg ml−1 melittin solution (5% H2O2). (c) Mobile microfish incubated in 2.5 mg/ml melittin solution (5% H2O2). (d) Relative fluorescence intensity measurements indicating the amount of melittin absorbed by the microfish. Statistical analysis indicates that there is significant difference between any two conditions among control, stationary or mobile, p<0.05.
The natural locomotion of aquatic and airborne species within fluids has motivated scientists and engineers who aim to develop devices and robotics with efficient locomotive properties for use in transportation, delivery and other applications.[29] Engineering nature-inspired structures and functionalities requires hierarchical design over several orders of magnitude, both in spatial and temporal domains. Biomimetic microswimmers have emerged over the past decade with the potential to be useful in biomedical applications or as rheological probes in vivo. Manufacturing such microswimmers with optimal design and swimming capacity is a critical step towards achieving these goals. We have demonstrated as proof of concept that functional microfish can be created by patterning biocompatible hydrogels with functionalized nanoparticles using our advanced 3D printing technology. With the μCOP system, diverse and complex 3D microswimmer designs can be rapidly fabricated in large, uniform arrays, demonstrating the enhanced scalability and high-throughput capacity of our approach when compared to conventional strategies. Biocompatible hydrogels are specifically chosen as the matrix material to encapsulate functional nanoparticles due to their wide use in medical implants, drug delivery devices, and tissue culture. PEG-based hydrogels, in particular, have proven extremely versatile for tissue engineering applications since they exhibit high biocompatibility and little or no immunogenicity. In addition, PEG-based hydrogels display tunable mechanical properties over a wide range for various tissues/organs. Importantly for patterning applications, PEG-based hydrogels are intrinsically resistant to protein adsorption and cell adhesion, thus providing biological “blank slates” upon which desired biofunctionality can be built.[30, 31] Last but not least, in the present work of microswimmers, PEGDA has demonstrated favorable stability of maintaining the fabricated shapes of the microfish as well as the encapsulation of nanoparticles over an extensive time period. The microfish can maintain the propulsion capability after more than two hours (longest swimming time tested) of swimming in the peroxide solution, and after the fabrication, they can be stored at room temperature for over one week before use.
The systematic engineering method reported here provides a general strategy for designing nature-inspired swimmers in fluidic environments. Beyond the example of microfish shown here, the digitized nature of the 3D μCOP system can be applied to fabricate virtually any other designs to optimize the locomotive performance of the microswimmers. With the μCOP system, we have demonstrated in the previous work that we can readily alternate the designs and fabricate a variety of structures with different materials encapsulating nanoparticles, biochemical molecules and even living cells.[24, 25, 32, 33] Applying this approach to the micromotor field, we have the capability to achieve diverse 3D microswimmer designs, ranging from simple cylindrical or tubular geometries to complex biomimetic architectures. We integrated platinum and iron oxide nanoparticles for guided chemical propulsion in hydrogen peroxide fuel, but other catalytic components, such as Ag, Ir, MnO2 or catalase, can also be integrated using the same approach for use with different chemical fuels.[34–36] Alternatively, new 3D printed micromachines that can harvest energy from their own surrounding environment, i.e., use a biological fluid or water as their fuel source, are also expected to be developed by our current manufacturing platform for diverse functionalities.[37, 38] Furthermore, as a proof-of-concept, we integrated PDA nanoparticles into the microfish and realized an improved detoxification platform as well as a real-time sensing tool. This strategy can be readily extended to incorporate multiple other functional nanoparticles into a fully-integrated microswimmer system as a powerful platform for applications including but not limited to drug delivery, personal therapeutics, and environmental conservation.
Experimental Section
3D printing of microfish
PEGDA (MW = 700 Da) was purchased from Sigma (USA). Photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate was synthesized according to the previous work.[30] PEGDA (40 wt%) in H2O with lithium phenyl-2,4,6-trimethylbenzoylphosphinate (1 wt%) was used as the matrix material. For the head of the microfish, magnetic Fe3O4 nanoparticles were diluted in the same matrix material with a concentration of 5 mg/ml; for the tail, catalytic Pt nanoparticles were diluted in the same matrix material with two different concentrations of 8.0×108 Pt particles per ml and 4.0×108 Pt particles per ml to evaluate the effect of Pt concentration on the swimming speed. For detoxification, PDA nanoparticles (3 mg ml−1) were loaded to the matrix material for the body of the microfish. The DMD chip used in the set-up is DLP-07 XGA from Texas Instruments.
Preparation of PDA nanoparticles
Polydiacetylene (PDA) nanoparticles were prepared according to previous work.[24] 10,12-pentacosadiynoic acid (PCDA) was purchased from Sigma (USA). PCDA was dissolved in dichloromethane and then filtered for purifying purpose. Dichloromethane was evaporated by heating after the filtration. Distilled water (6 ml) was added to purified PCDA (36mg), followed by probe sonication for 5 mins at ~80 °C. Then the self-assembled PDA nanoparticle solution was refrigerated at 4 °C overnight for stabilization. Finally, before 3D printing, the PDA nanoparticle solution was irradiated with ultraviolet light (285 nm) for 10 mins.
Characterization and propulsion experiments
The 3D microscopy images with height profile were obtained on microscope Keyence VHX1000. High-resolution electroscope microscopy images and EDX spectroscopy images were obtained on Phillips XL30 ESEM. In order to get self-propelled microfish, aqueous H2O2 solutions with concentrations ranging from 5% to 15% were used as chemical fuels. Videos were captured by an inverted optical microscope (Nikon Instrument Inc. Ti-S/L100), coupled with 10× and 4× objectives and a Hamamatsu digital camera C11440, using the NIS Elements AR 3.2 software. The speed of the microfish was obtained by tracking and analyzing their motion with the NIS Elements AR 3.2 software.
Detoxification experiments
Detoxification of melittin by the microfish was performed by loading the microfish to 100 μl of different solutions at 37 °C for 10 mins. For Figure 5a, the solution was 5% H2O2 without melittin toxin; for Figure 5b and 5c, the solution was the same with melittin (2.5 mg ml−1) in 5% H2O2. The microfish in Figure 5b were not encapsulated with Pt nanoparticles to keep them stationary (i.e., no movement). The detoxification efficiency was evaluated by fluorescence microscopy imaging of the microfish after the neutralization process. ImageJ (developed by National Institute of Health) was used to measure the relative fluorescence intensity according to the previous work.[28] The relative fluorescence intensity is calculated according to equation (1). Both the integrated density and area can be measured directly with ImageJ from the fluorescence microscopy images. Data are reported as mean ± standard deviation. Comparison of sample means was performed by ANOVA followed by Tukey’s post-hoc test (Origin software), p < 0.05 was considered statistically significant.
| (1) |
Supplementary Material
Acknowledgments
This project received support from the Defense Threat Reduction Agency-Joint Science and Technology Office for Chemical and Biological Defense (Grants no. HDTRA1-14-1-0064 and HDTRA1-13-1-0002), the National Science Foundation (Grants no. CMMI-1120795, CMMI-1332681) and National Institutes of Health (Grant no. EB012597, EB017876). R.D. and Z.W. acknowledge the China Scholarship Council (CSC) for the financial support. We thank Michael Galarnyk and John Warner for their assistance.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Wang J. Nanomachines: Fundamentals and Applications. John Wiley & Sons; 2013. [Google Scholar]
- 2.Ismagilov RF, Schwartz A, Bowden N, Whitesides GM. Angew Chemie. 2002;114:674. [Google Scholar]
- 3.Mallouk TE, Sen A. Sci Am. 2009;300:72. doi: 10.1038/scientificamerican0509-72. [DOI] [PubMed] [Google Scholar]
- 4.Ozin GA, Manners I, Fournier-Bidoz S, Arsenault A. Adv Mater. 2005;17:3011. doi: 10.1039/b414896g. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Manesh KM. Small. 2010;6:338. doi: 10.1002/smll.200901746. [DOI] [PubMed] [Google Scholar]
- 6.Mei Y, Solovev AA, Sanchez S, Schmidt OG. Chem Soc Rev. 2011;40:2109. doi: 10.1039/c0cs00078g. [DOI] [PubMed] [Google Scholar]
- 7.Loget G, Kuhn A. Nat Commun. 2011;2:535. doi: 10.1038/ncomms1550. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DA, Nolte RJM, van Hest JCM. Nat Chem. 2012;4:268. doi: 10.1038/nchem.1281. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Gao W. ACS Nano. 2012;6:5745. doi: 10.1021/nn3028997. [DOI] [PubMed] [Google Scholar]
- 10.Fischer P, Ghosh A. Nanoscale. 2011;3:557. doi: 10.1039/c0nr00566e. [DOI] [PubMed] [Google Scholar]
- 11.Mei Y, Huang G, Solovev AA, Ureña EB, Mönch I, Ding F, Reindl T, Fu RKY, Chu PK, Schmidt OG. Adv Mater. 2008;20:4085. [Google Scholar]
- 12.Balasubramanian S, Kagan D, Hu CJ, Campuzano S, Lobo-Castañon MJ, Lim N, Kang DY, Zimmerman M, Zhang L, Wang J. Angew Chem Int Ed Engl. 2011;50:4161. doi: 10.1002/anie.201100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solovev AA, Mei Y, Ureña EB, Huang G, Schmidt OG. Small. 2009;5:1688. doi: 10.1002/smll.200900021. [DOI] [PubMed] [Google Scholar]
- 14.Solovev AA, Xi W, Gracias DH, Harazim SM, Deneke C, Sanchez S, Schmidt OG. ACS Nano. 2012;6:1751. doi: 10.1021/nn204762w. [DOI] [PubMed] [Google Scholar]
- 15.Paxton W, Kistler K, Olmeda C. J Am Chem Soc. 2004;126:13424. doi: 10.1021/ja047697z. [DOI] [PubMed] [Google Scholar]
- 16.Guix M, Mayorga-Martinez C, Merkoçi A. Chem Rev. 2014;114:11862. doi: 10.1021/cr400273r. [DOI] [PubMed] [Google Scholar]
- 17.Tottori S, Zhang L, Qiu F, Krawczyk KK, Franco-Obregón A, Nelson BJ. Adv Mater. 2012;6:811. doi: 10.1002/adma.201103818. [DOI] [PubMed] [Google Scholar]
- 18.Stanton MM, Trichet-Paredes C, Sánchez S. Lab Chip. 2015;7:1634. doi: 10.1039/c5lc90019k. [DOI] [PubMed] [Google Scholar]
- 19.Qiu T, Lee TC, Mark AG, Morozov KI, Münster R, Mierka O, Turek S, Leshansky AM, Fischer P. Nat Commun. 2014;5:5119. doi: 10.1038/ncomms6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawroth JC, Lee H, Feinberg AW, Ripplinger CM, McCain ML, Grosberg A, Dabiri JO, Parker KK. Nat Biotechnol. 2012;30:792. doi: 10.1038/nbt.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams BJ, Anand SV, Rajagopalan J, Saif MTA. Nat Commun. 2014;5:3081. doi: 10.1038/ncomms4081. [DOI] [PubMed] [Google Scholar]
- 22.Parmar J, Ma X, Katuri J, Simmchen J, Stanton MM, Trichet-Paredes C, Soler L, Sanchez S. Sci Technol Adv Mater. 2015;16:014802. doi: 10.1088/1468-6996/16/1/014802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang AP, Qu X, Soman P, Hribar KC, Lee JW, Chen S, He S. Adv Mater. 2012;24:4266. doi: 10.1002/adma.201202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gou M, Qu X, Zhu W, Xiang M, Yang J, Zhang K, Wei Y, Chen S. Nat Commun. 2014;5:3774. doi: 10.1038/ncomms4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K, Zhu W, Qu X, Aaronson C, McCall W, Chen S, Sirbuly DJ. ACS Nano. 2014;8:9799. doi: 10.1021/nn503268f. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Huang G, Ye M, Li M, Liu R, Mei Y. Nanoscale. 2011;3:5083. doi: 10.1039/c1nr10840a. [DOI] [PubMed] [Google Scholar]
- 27.Dusenbery DB. Living at Micro Scale. Harvard University Press; Cambridge, Mass, USA: 2009. [Google Scholar]
- 28.Burgess A, Vigneron S. Proc Natl Acad Sci USA. 2010;107:12564. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ijspeert AJ. Science. 2014;346:196. doi: 10.1126/science.1254486. [DOI] [PubMed] [Google Scholar]
- 30.Qu X, Zhu W, Huang S, Li Y, Chien S, Zhang K, Chen S. Biomaterials. 2013;34:9812. doi: 10.1016/j.biomaterials.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha C, Soman P, Zhu W, Nikkhah M, Camci-Unal G, Chen S, Khademhosseini Ali. Biomater Sci. 2014;2:703. doi: 10.1039/C3BM60210A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suri S, Han LH, Zhang W, Singh A, Chen S, Schmidt CE. Biomed Microdevices. 2011;13:983. doi: 10.1007/s10544-011-9568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soman P, Chung PH, Zhang A, Chen S. Biotechnol Bioeng. 2013;110:3038. doi: 10.1002/bit.24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Zhao G, Pumera M. J Am Chem Soc. 2014;136:2719. doi: 10.1021/ja411705d. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez S, Solovev AA, Mei Y, Schmidt OG. J Am Chem Soc. 2010;132:13144. doi: 10.1021/ja104362r. [DOI] [PubMed] [Google Scholar]
- 36.Gao W, Pei A, Dong R, Wang J. J Am Chem Soc. 2014;136:2276. doi: 10.1021/ja413002e. [DOI] [PubMed] [Google Scholar]
- 37.Gao W, Uygun A, Wang J. J Am Chem Soc. 2012;134:897. doi: 10.1021/ja210874s. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Singh VV, Sattayasamitsathit S, Orozco J, Kaufmann K, Dong R, Gao W, Jurado-Sanchez B, Fedorak Y, Wang J. ACS Nano. 2014;8:11118. doi: 10.1021/nn505029k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.