Abstract
Background and Objectives:
Some 2 million tons of chicken meat is produced by Iran per annum, positioning Iran among the top producers in the region. This study aimed to evaluate the molecular epidemiology and genetic characteristics of Salmonella enterica Enteritidis in Iran.
Materials and Methods:
A representative selection of isolates (n=76), initially genotyped by a 7-locus MLVA typing system, was examined by the standard MLST genotyping.
Results and Conclusion:
All the MLVA typed isolates, classified into six types, were gathered under a single ST11 MLST type. This is an intriguing observation as much more genome heterogeneity was expected considering the extent of diversity in the host and geography origin of the examined isolates. ST11, on the other hand is not exclusively found in Iran as it is reported also from Brazil, Denmark, Japan and the United States. In explanation of these observations, ST11 might stand for a single probably ancestral clone of Salmonella enterica Enteritidis successfully scattered in all these geographically diverse countries. Further global investigation covering more isolates and methods like whole genome sequencing would be advisable.
Keywords: Salmonella enterica Enteritidis, MLST, MLVA, Genetic diversity
INTRODUCTION
Salmonella is considered a major public health problem on a global scale. It is estimated that 93.8 million cases of gastroenteritis due to Salmonella species with 155,000 deaths occur each year (1). Salmonella enterica including six subgroups (Ι, ΙΙ, ΙΙΙa, ΙΙΙb, ΙV, and VΙ) together with Salmonella bongori compose the Salmonella genus which accounts for more than 2600 serovars (2). Subgroup I defined as Salmonella enterica subsp. enterica, consisting of more than 1500 serovars (3), is the main cause of zoonotic food-borne diseases throughout the world (4). Among which, Salmonella enterica serovar Enteritidis is one of the most frequently reported reasons of human salmonellosis in developed countries. For nearly 2 decades, from 1970s through the mid 1990s this serotype experienced a significant increase, due to shelled eggs as its main transmission’s vehicle (5). In the first decade of 21st. century Salmonella Enteritidis along with Salmonella Typhimuium as the main etiological agents for gastroenteritis and diarrhea in Europe, affected over 75% of human cases of salmonellosis (4).
While typhoidal Salmonella serovars are host restricted (6), non-typhoidal Salmonella (NTS) are zoonotic agents, that are found in a wide range of animal reservoirs such as poultry, pigs, and cattle. These are considered as the common vehicle of human salmonellosis (7). Salmonella enterica Enteritidis as the leading NTS serotype is capable of persisting in the caecum or ovaries of chicken without presenting any clinical sign. In young chicken next to the probable condition of high mortality due to severe diarrhoea and dehydration, there is a greater risk of transforming to a carrier state in clinically recovered animals (8). Determining contamination routes, differentiating strains isolated in an outbreak from those of obtained in sporadic cases and collecting data about global distribution of a pathogen in a population has raised the importance of epidemiological studies to great extent (9). To conduct a precise epidemiological surveillance and outbreak studies, subtyping the microbial pathogens is critical to let closely related isolates be discriminated with an acceptable resolution. Differentiation investigations among Salmonella Enteritidis isolates is a challenging task, as this pathogen stands atop in list of genetically homogenous serotypes (10).
Regardless of Kaufmann-White scheme based on immunological classifications which can’t go beyond the serotyping level (11) other phenotypic approaches, like phage typing, biotyping and antibiotic susceptibility testing have a series of drawbacks. Phage typing demands specific typing phages, which makes its reproducibility a great concern. To be more specific, all these techniques lack the discriminatory power which is essential for epidemiological related Salmonella isolates (12). DNA-based techniques, including arbitrary primed PCR(13), pulsed-field gel electrophoresis (PFGE) and ribotyping (14) have resolved some of the problems, but still have shortcomings in their reproducibility and discriminatory abilities (15).
In recent years, multiple-locus variable-number of tandem repeat (MLVA) analysis (16) and multiple-locus sequence typing (MLST) (17) have been globally recognized as highly discriminative standard strategies in modern epidemiological studies of salmonellosis. MLVA genotyping has been successfully employed an effective tool for investigating strains that are epidemiologically related or unrelated in specific outbreaks (18). This method enhanced the characterization of subspecies amid of a complex epidemics risen by a single serovar with identical phages happening simultaneously in different geographical regions (19).
Multi locus sequence typing (MLST) was introduced to molecular technology for the first time in 1998 as an optimal approach to represent accurate, reproducible data as a solution to epidemiological survey of bacterial pathogen in one hand and evolutionary and population biology on the other (20). This technique is based on assigning the nucleotide sequences of a series of specific housekeeping, ribosomal, virulence-related genes. Data produced by MLST is very similar to those provided by multi locus enzyme electrophoresis MLEE but with greater precision, since it has the capability to detect individual nucleotide alterations rather than pheno-typical screening the expression of electrophoretic mobilities (EM) of multiple core metabolic enzymes.
Although, there is not abundant information about molecular epidemiology of Salmonella but in recent years it seems a new wave of progressive tendency spreading among the Iranian researchers (21–23) .
The objective of current study is MLST analysis of population structure of Salmonella Enteritidis for the first time in Iran to provide new insights into a global scale lineage evaluation.
MATERIALS AND METHODS
Selection of bacterial isolates.
A total of seventy-five Salmonella Enteritidis isolates incorporated in the study. Sixty-five chicken isolates were obtained from cloaca of slaughtered bird carcasses. The remaining chicken isolates were obtained from autopsied birds from Khorasan Razavi province. The human isolates were provided by fecal samples of sporadic clinical cases which were archived in Microbiology Department collection of Razi Vaccine and Serum Research Institute. To make a comparison analysis of the results, one S. Enteritidis type strain (ATCC13076) was included (Table 1).
Table 1.
Bacterial isolates, animal and human source and locations
| Isolate ID | RTTC Collection | Host | Province | City | Farm | Serotype |
|---|---|---|---|---|---|---|
| 1 | 1697-1 | poultry | Qazvin | Qazvin | A | S. enteritidis |
| 2 | 1697-2 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 3 | 1697-3 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 4 | 1697-4 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 5 | 1697-5 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 6 | 1697-6 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 7 | 1697-7 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 8 | 1697-8 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 9 | 1697-9 | poultry | Qazvin | Qazvin | S. enteritidis | |
| 10 | 1718-1 | poultry | Qazvin | Takestan | B | S. enteritidis |
| 11 | 1718-2 | poultry | Qazvin | Takestan | S. enteritidis | |
| 12 | 1718-3 | poultry | Qazvin | Takestan | S. enteritidis | |
| 13 | 1718-4 | poultry | Qazvin | Takestan | S. enteritidis | |
| 14 | 1718-5 | poultry | Qazvin | Takestan | S. enteritidis | |
| 15 | 1714-1 | poultry | Fars | Shiraz | C | S. enteritidis |
| 16 | 1714-9 | poultry | Fars | Shiraz | S. enteritidis | |
| 17 | 1714-20 | poultry | Fars | Shiraz | S. enteritidis | |
| 18 | 1714-21 | poultry | Fars | Shiraz | S. enteritidis | |
| 19 | 1714-23 | poultry | Fars | Shiraz | S. enteritidis | |
| 20 | 1714-24 | poultry | Fars | Shiraz | S. enteritidis | |
| 21 | 1714-25 | poultry | Fars | Shiraz | S. enteritidis | |
| 22 | 1714-38 | poultry | Fars | Shiraz | S. enteritidis | |
| 23 | 1714-39 | poultry | Fars | Shiraz | S. enteritidis | |
| 24 | 1714-54 | poultry | Fars | Shiraz | S. enteritidis | |
| 25 | 1714-55 | poultry | Fars | Shiraz | S. enteritidis | |
| 26 | 1714-59 | poultry | Fars | Shiraz | S. enteritidis | |
| 27 | 1714-60 | poultry | Fars | Shiraz | S. enteritidis | |
| 28 | 1714-61 | poultry | Fars | Shiraz | S. enteritidis | |
| 29 | 1709-6 | poultry | Zanjan | Abhar | D | S. enteritidis |
| 30 | 1714-26 | human | Fars | Shiraz | E | S. enteritidis |
| 31 | 1714-66 | human | Fars | Shiraz | F | S. enteritidis |
| 32 | 1714-67 | human | Fars | Shiraz | G | S. enteritidis |
| 33 | 1714-69 | human | Fars | Shiraz | H | S. enteritidis |
| 34 | 1714-70 | human | Fars | Shiraz | I | S. enteritidis |
| 35 | 1714-71 | human | Fars | Shiraz | J | S. enteritidis |
| 36 | 1693-58 | poultry | Markazi | Arak | K | S. enteritidis |
| 37 | 1693-57 | poultry | Markazi | Arak | S. enteritidis | |
| 38 | 1648(1) | poultry | Alborz | Karaj | L | S. enteritidis |
| 39 | 1697-10 | poultry | Qazvin | Qazvin | A | S. enteritidis |
| 40 | 1718-7 | poultry | Qazvin | Takestan | B | S. enteritidis |
| 41 | 13076 | Lab Strain | NA | NA | ATCC | S. enteritidis |
| 42 | 1693-1 | poultry | Markazi | Farahan | P | S. enteritidis |
| 43 | 1693-3 | poultry | Markazi | Farahan | S. enteritidis | |
| 44 | 1693-4 | poultry | Markazi | Farahan | S. enteritidis | |
| 45 | 1693-5 | poultry | Markazi | Farahan | S. enteritidis | |
| 46 | 1693-6 | poultry | Markazi | Farahan | S. enteritidis | |
| 47 | 1693-7 | poultry | Markazi | Farahan | S. enteritidis | |
| 48 | 1693-8 | poultry | Markazi | Farahan | S. enteritidis | |
| 49 | 1693-17 | poultry | Markazi | Farahan | S. enteritidis | |
| 50 | 1693-22 | poultry | Markazi | Govar | Q | S. enteritidis |
| 51 | 1693-26 | poultry | Markazi | Mahalat | R | S. enteritidis |
| 52 | 1693-28 | poultry | Markazi | Mahalat | S. enteritidis | |
| 53 | 1693-35 | poultry | Markazi | Farahan | S | S. enteritidis |
| 54 | 1693-36 | poultry | Markazi | Farahan | S. enteritidis | |
| 55 | 1693-38 | poultry | Markazi | Farahan | S. enteritidis | |
| 56 | 1693-40 | poultry | Markazi | Mahalat | O | S. enteritidis |
| 57 | 1693-41 | poultry | Markazi | Mahalat | S. enteritidis | |
| 58 | 1693-42 | poultry | Markazi | Mahalat | S. enteritidis | |
| 59 | 1693-43 | poultry | Markazi | Mahalat | S. enteritidis | |
| 60 | 1693-44 | poultry | Markazi | Mahalat | S. enteritidis | |
| 61 | 1693-46 | poultry | Markazi | Mahalat | S. enteritidis | |
| 62 | 1693-48 | poultry | Markazi | Tafresh | T | S. enteritidis |
| 63 | 1693-50 | poultry | Markazi | Tafresh | S. enteritidis | |
| 64 | 1693-51 | poultry | Markazi | Tafresh | S. enteritidis | |
| 65 | 1693-52 | poultry | Markazi | Tafresh | S. enteritidis | |
| 66 | 1693-53 | poultry | Markazi | Tafresh | S. enteritidis | |
| 67 | 1693-54 | poultry | Markazi | Tafresh | S. enteritidis | |
| 68 | 1714-19 | poultry | Fars | Shiraz | C | S. enteritidis |
| 69 | 1714-28 | poultry | Fars | Shiraz | S. enteritidis | |
| 70 | 1714-37 | poultry | Fars | Shiraz | S. enteritidis | |
| 71 | 1714-56 | poultry | Fars | Shiraz | S. enteritidis | |
| 72 | 1595-4 | poultry | Alborz | Karaj | N | S. enteritidis |
| 73 | 2490-1 | poultry | Khorasan | Mashhad | U | S. enteritidis |
| 74 | 2490-3 | poultry | Khorasan | Mashhad | V | S. enteritidis |
| 75 | 2490-4 | poultry | Khorasan | Mashhad | W | S. enteritidis |
| 76 | 2490-5 | poultry | Khorasan | Neishabour | X | S. enteritidis |
Serotyping.
All strains were serotyped at Razi Vaccine and Serum Research Institute, Microbiology Department according to Kauffmann-White-Le Minor scheme (3) by agglutination with “O” and “H” antigen specific sera (Mast, Bootle, England).
DNA extraction.
To perform MLVA and MLST analysis bacterial DNA was extracted by simple boiling method as described previously (24) with little modification. Briefly a loopful of overnight culture at 37 °C on nutrient agar plates was harvested and suspended in 0.5 ml of TE buffer and vortexed. Cell suspension was boiled at 98 °C for 10 min and immediately cooled on ice for 5 min. The suspension was centrifuged at 12,000 g for 5 min. Small portion (1/64 v/v) of a 10-mg/ml Proteinase K solution (Roche, Germany) was added to the supernatant and this mixture was directly used for PCRs.
VNTR.
Seven MLVA loci including SE2, SE3, SE5, SE7, SE8, SENTR4, and SENTR7 were selected (Table 2). Amplification details were described previously (25). To compare the diversity between MLVA loci, Nei’s diversity index was calculated as 1−Σ (Allele)2. Simpson’s diversity index of the seven-loci MLVA system was measured according to Hunter and Gaston (26). The BioNumerics software v4.61 (Applied Maths, Belgium) was used to cluster the MLVA types represented in dendrogram using categorical coefficient.
Table 2.
MLVA & MLST Primers and their references
| MLVA Loci (Alias) | Range of Amplicon Size | Primer Sequence (5′-3′) | References |
|---|---|---|---|
| SE2 (SENTR6, ENTR20) | 208–229 | F-CTTCGGATTATACCTGGATTG R-TGGACGGAGGCGATAG |
Cho et al. 2008 |
| SE3 | 308–320 | F-CAACAAAACAACAGCAGCAT R-GGGAAACGGTAATCAGAAAGT |
|
| SE5 (SENTR5, STTR5) | 200–224 | F-CGGGAAACCACCATCAC R-CAGGCCGAACAGCAGGAT |
|
| SE7 | 484–545 | F-CCGACCCAATAAGGAG R-CTTACCGTTGGTAGTTTGTTA T |
|
| SE8 | 469–556 | F-TTGCCGCATAGCAGCAGAAGT R-GCCTGAACACGCTTTTTAATAGGCT |
|
| SENTR4 (SE1, ENTR13) | 119–126 | F-GACCAACACTCTATGAACCAATG R-ACCAGGCAACTATTCGCTATC |
Malorny et al. 2008 |
| SENTR7 (SE9) | 126–135 | F-ACGATCACCACGGTCACTTC R-CGGATAACAACAGGACGCTTC |
| MLST Loci (Alias) | Data Base Standard Size | Primer Sequence (5′-3′) | References |
|---|---|---|---|
| thrA (aspartokinase+homoserine dehydrogenase) | 501 | F-GTCACGGTGATCGATCCGGT R-CTCCAGCAGCCCCTCTTTCAG |
Achtman-1 et al. 2015 |
| purE (phosphoribosylaminoimidazole carboxylas) | 399 | F-CGCATTATTCCGGCGCGTGT R-GAACGCAAACTTGCTTCAT |
|
| sucA (alpha ketoglutarate dehydrogenase) | 501 | F-AGCACCGAAGAGAAACGCTG R-GGTTGTTGATAACGATACGTAC |
|
| hisD (histidinol dehydrogenase) | 501 | F-GTCGGTCTGTATATTCCCGG R-GGTAATCGCATCCACCAAATC |
|
| aroC (chorismate synthase) | 501 | F-CCTGGCACCTCGCGCTATAC R-CCACACACGGATCGTGGCG |
|
| hemD (uroporphyrinogen III cosynthase) | 432 | F 5′-GTGGCCTGGAGTTTTCCACT R-GACCAATAGCCGACAGCGTAG |
|
| dnaN (DNA polymerase III beta subunit) | 501 | F-CCGATTCTCGGTAACCTGCT R-CCGCGGAATTTCTCATTCGAG |
MLST.
To Characterize Salmonella Eteritidis isolates based on MLST scheme, internal fragments of seven housekeeping genes were utilized as follows: thrA (aspartokinase+homoserine dehydrogenase), purE (phosphoribosylaminoimidazole carboxylas), sucA (alpha ketoglutarate dehydrogenase), hisD (histidinol dehydrogenase), aroC (chorismate synthase), hemD (uroporphyrinogen III cosynthase), dnaN (DNA polymerase III beta subunit)
Amplification protocols proposed by the Warwick University MLST database (27) were used in this study, including primers which have been selected from the set of primers presented under the titles of amplifications & sequences. All the primers were provided and went under optimization tests to find the best match for pair primers in final test. Next to this experimental evaluation the annealing temperatures was modified to 62.9°C for hemD and 58°C for the 6 remaining of genes. Amplifications for the hemD gene were carried out with approximately 12 μl reactions containing 6 μl PCR master mix (Ampliqon, Denmark), 1.5 μl of working solution (5 pM/μl) from each flanking primer and 1.5 μl of DNA template plus 1.5 μl of molecular-grade PCR water. In the second protocol used for thrA, purE, sucA, hisD, aroC, dnaN, reaction mixtures contained 6 μl PCR master mix (Ampliquor, Denmark), 0.3 μl of each flanking primer (5 pM/μl) and 1.5 μl of DNA template and 4 μl molecular-grade PCR water to a volume of 12 μl. PCR reactions were run on an Eppendorf PCR system (Eppendorf, Germany) were an initial 1 min denaturation at 95°C followed by 35 cycles of denaturation (94°C for 40 sec), annealing (62.5°C for 20 sec) and extension (72°C for 30 sec) with a final elongation step of 72°C for 5 min was used for the first protorol reactions. For the second group this was the same except the annealing temperature which was lowered to 58°C (Table 3).
Sequence typing data analysis.
PCR products including DNA amplicons (40pg/μl of concentration) along with primers (10ng/μl of concentration) were sent to Macrogen® (South Korea – Seoul) company to be sequenced by Sanger method. The results were received in chromatogram format, that needed to be analyzed by related bioinformatics softwares, like Chromas Lite Ver. 2.01(28) to evaluate the sequence results, Clustal X Ver.2.0.11(29) to align the segments and compare their nucleotides, and Artemis (30) as a DNA sequence viewer and annotation tool that allows visualization of sequence features and the results of analyses within the context of the sequence.
RESULTS
VNTR.
Successful amplification of all isolates recorded four alleles for SE5 and two alleles for SE7 and no allelic formation in all other five loci with monomorphic state. To validate these findings selected isolates were gone under sequencing procedures with 175 PCR products. Considering 11 various VNTR scores previously observed from gel electrophoresis (25), Nie’s diversity index was calculated, ranging from 0 (SE2, SE3, SE7, SE8, and SENTR4) up to 0.15 (SENTR7) and 0.58 (SE5). Altogether six “MLVA-type” as allele combination with Single-locus variant (SLV) was identified, in which three MLVA-type were common between human and chicken isolates.
MLST.
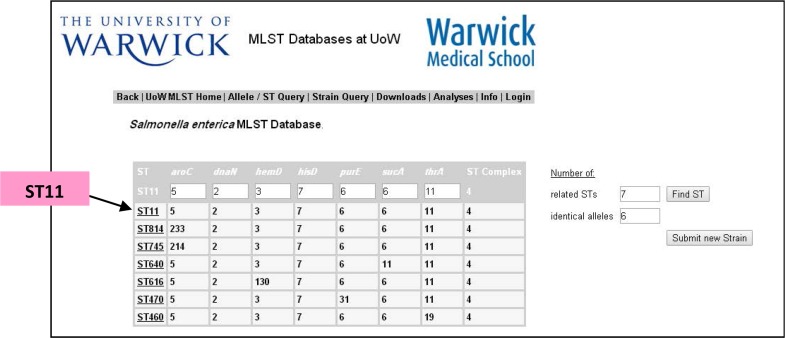
MLST analysis did not represent any nucleotide differences among Seventy-five S. Enteritidis isolates collected from various Iranian cities and also standard strain (ATCC13076). The only allelic profile which was obtained from Warwick University MLST data base (27) according to the panel of mentioned seven housekeeping genes were respectively (2,3,5, 6,7,11) which was categorized as the Sequence Type (ST)11 in MLST data base. Considering 6 out of 7 loci identical, ST814, ST745, ST640, ST616, ST470, ST460 were regarded as the closest sequence type registered on MLST data base (Fig. 1).
Fig. 1.
MLST data-base (27) result related to Sequence Type (ST) findings of this study
DISCUSSION
As the first report of Multi locus sequence typing of Salmonella Enteritidis population in Iran our study characterized all isolates as a unique ST (ST 11) which was in notable contrast with diversity observed by Multi locus VNTR analysis in the same collection. Similar report has been presented in Japan (31) that among 30 S. Enteritidis isolates collected between 1980 and 1990, all were categorized in the same sequence type 11. Referring to MLST data base, Denmark was among the countries with the most frequent report of this sequence type such as No.9968721-11 registered in 2003, along with a broad range of records from US and Europe (32). Another recent report from Brazil (33) covering a period of more than two decades showed 44 out of 46 isolates Salmonella Enter-itidis have been typed as belonging to ST 11.
Generally, molecular typing methods are intended to tackle two different levels of epidemiological problems, which reflect different insights toward solving a local or global epidemiology in different timeframes. In one hand localized outbreak of disease in a short period of time should be assessed and on the other, relation between strains causing a disease in one geographic area with those observed around the world during a longer period would be investigated. These two different conceptual views demand different appropriate scheme of molecular typing. However, they should have one capability in common, which is their inevitable discriminatory power (20), So that isolates recorded in same molecular type are likely to be descended from a younger ancestor and those belonging to more distant ancestors are expected to differ in type unless a relative higher clonal population would be under study.
Two pivotal phenomena have been revealed based on our VNTR scheme study. Firstly, fragment length polymorphism was detected in just two out of seven loci with rather slight 0.15 up to 0.58 Nie’s diversity index for SENTR7 and SE5 respectively. This means other five loci which have been recorded in literature as the most commonly used markers with the highest reported levels of diversity has failed to provide any heterogeneity. Secondly, all six allele combinations presented in this study has been in Single locus Variation state. These two key factors have shown that our Iranian Enteritidis population has a low level of diversity within its entity, which accommodate with what has been detected by MLST methodology.
Seven house keeping genes represent only a fraction of the bacteria whole genome in MLST scheme which hardly exceed 0.2% of it, but have proved a staggering level of diversity for numerous bacteria (34), to let it claim among the highest discriminatory techniques of population structure analysis (35). Considering this privilege attributed to MLST scheme, observation of the homogeneity between our population and one standard (ATCC 13076) strain, seems something of a paradox in our fundamental principles. Detecting this odd similarity, a verification trial test consists of 3 different serotypes including S. Infantis, S. Typhimurium and S. Enteritidis were arranged with an all in one PCR operation. This experimental trial repeated for 3 times to evaluate 3 out of seven housekeeping genes namely hisD, thrA, sucA. Comparing amplified segments with those acquired in our previous studies next to similar fragments in standard genome, were in accordance to what was expected to see. Three similar serotypes integrated to different clusters, based on SNP differences between them, which was also in agreement with what has been reported previously by Achtman and colleagues (2). Also S. Enteritidis standard strain (ATCC 13076) though verified to be identical with S. Enteritidis isolates were in quite contrast with other remaining two serotypes.
Back to identical entity of standard strain in compare to our population, it might be due to the fact that they all belonged to the same lineage with persistent consensus genotype. Based on data released on MLST data base, there are few records of Salmonella Enter-itidis strains belonging to Sequence type 11, regarding the homogenus character of this serotype one cautious explanation would be presumably that they have a common historical origin.
Regarding the specific clonal character of Salmonella Enteritidis ST11 population two hypotheses have been presented by Noda and colleagues (31): first existence of a niche in chicken reproductive tissues and second, higher clonal population in those geographical regions in which this sequence type is in circulation. While it’s been proved housekeeping genes might possess less effective role in chicken oviduct colonization than genes related to factors like fimbriae or flagellae or stress tolerance, the second hypothesis would more probably interpret our conserved population. Under the light of food-borne character of Salmonella Enter-itidis and existence of human cases representing the similar sequence type 11, its higher clonal population can be attributed to the potential transmission of this pathogen between human beings and chickens without substantial genetic changes like mutation. More over having three of the six MLVA types found in this study shared by chickens and human enteritis cases suggests a strong association between the two.
In conclusion, notwithstanding high discriminatory power, nucleotide changes accumulate in housekeeping genes in long period of time. That is why the allelic profile of isolates persists unchanged over a longer timeframe, which make MLST a desirable tool for global epidemiology. If it hadn’t been for unambiguous and portable character of this scheme there would have been no chance of finding closely related isolates to our population around the world. Finding traces of isolates with the same ancestral lineage would be of great importance in global circulation of different pathogens, like what has been recorded in our study about high clonal Iranian population of Salmonella Enteritidis with those in other countries and continents in terms of evolutionary, fundamental relatedness and population structural studies. Further global investigations covering more isolates and methods like whole genome sequencing would be advisable.
ACKNOWLEDGEMENT
This work was fully financed with the state funds from Razi Vaccine and Serum Research Institute, under grant no. 2-18-18-90036.
REFERENCES
- 1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 2010; 50: 882– 889. [DOI] [PubMed] [Google Scholar]
- 2. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 2012; 8: 1– 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grimont PA, Weill FX. (2007). Antignenic formulas of the Salmonella serovars 9th. ed WHO Collaboration Center for Resistance and Research on Salmonella, Institute Pasteur; Paris. [Google Scholar]
- 4. Litrup E, Torpdahl M, Malorny B, Huehn S, Christensen H, Nielsen EM. Association between phylogeny, virulence potential and serovars of Salmonella enterica. Infect Genet Evol 2010; 10: 1132– 1139. [DOI] [PubMed] [Google Scholar]
- 5. Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 2011; 8: 887– 900. [DOI] [PubMed] [Google Scholar]
- 6. Hald T, Lo Fo Wong DM, Aarestrup FM. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog Dis 2007; 4: 313– 326. [DOI] [PubMed] [Google Scholar]
- 7. Braden CR. Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin Infect Dis 2006; 43: 512– 517. [DOI] [PubMed] [Google Scholar]
- 8. Sadeyen JR, Trotereau J, Velge P, Marly J, Beaumont C, Barrow PA, et al. Salmonella carrier state in chicken: comparison of expression of immune response genes between susceptible and resistant animals. Microbes Infect 2004; 6: 1278– 1286. [DOI] [PubMed] [Google Scholar]
- 9. Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. Journal of clinical microbiology 1999; 37: 1661– 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olson AB, Andrysiak AK, Tracz DM, Guard-Bouldin J, Demczuk W, Ng LK, et al. Limited genetic diversity in Salmonella enterica serovar Enteritidis PT13. BMC Microbiol 2007; 7: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Popoff MY, Bockemuhl J, Gheesling LL. Supplement 2001 (no. 45) to the Kauffmann–White scheme. Res Microbiol 2003; 154: 173– 174. [DOI] [PubMed] [Google Scholar]
- 12. Threlfall EJ, Frost JA. The identification, typing and fingerprinting of Salmonella: laboratory aspects and epidemiological applications. J Appl Bacteriol 1990; 68: 5– 16. [DOI] [PubMed] [Google Scholar]
- 13. Fadl AA, Khan MI. Genotypic evaluation of Salmonella enteritidis isolates of known phage types by arbitrarily primed polymerase chain reaction. Avian Dis 1997; 41: 732– 737. [PubMed] [Google Scholar]
- 14. Thong KL, Ngeow YF, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol 1995; 33: 1070– 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng J, Keys CE, Zhao S, Meng J, Brown EW. Enhanced subtyping scheme for Salmonella enteritidis. Emerg Infect Dis 2007; 13: 1932– 1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kruy SL, Van Cuyck H, Koeck JL. Multilocus variable number tandem repeat analysis for Salmonella enterica subspecies. Eur J Clin Microbiol Infect Dis 2011; 30: 465– 473. [DOI] [PubMed] [Google Scholar]
- 17. Jolley KA, Maiden MC. Using MLST to study bacterial variation: prospects in the genomic era. Future Microbiol 2014; 9: 623– 630. [DOI] [PubMed] [Google Scholar]
- 18. Malorny B, Junker E, Helmuth R. Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype Enteritidis. BMC Microbiol 2008; 8: 1– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hopkins K, Peters T, de Pinna E, Wain J. Standardisation of multilocus variable-number tandem-repeat analysis (MLVA) for subtyping of Salmonella enterica serovar Enteritidis. Euro Surveill 2011; 16: 1– 29. [PubMed] [Google Scholar]
- 20. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 1998; 95: 3140– 3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Golab N, Khaki P, Noorbakhsh F. Molecular Typing of Salmonella Isolates in Poultry by Pulsed-Field Gel Electrophoresis in Iran. Int J Enteric Pathog 2014; 2: 1– 5. [Google Scholar]
- 22. Shiroodi AA, Jamshidian M, Salehi TZ, Reza G, Boroujeni N, Amini K. Genotyping of Salmonella enterica subsp. enterica serovar Entritidis, isolated from poultry, cattle and human in Iran by ERIC-PCR. International Journal of Biosciences (IJB) 2014; 5: 147– 153. [Google Scholar]
- 23. Peighambari SM, Akbarian R, Morshed R, Yazdani A. Characterization of Salmonella isolates from poultry sources in Iran. Iranian Journal of Veterinary Medicine 2013; 7: 35– 41. [Google Scholar]
- 24. Karimnasab N, Tadayon K, Khaki P, Moradi Bidhendi S, Ghaderi R, Sekhavati M, et al. An optimized affordable DNA-extraction method from Salmonella enterica Enteritidis for PCR experiments. Archives of Razi 2013; 68: 105– 109. [Google Scholar]
- 25. Ghaderi R, Tadayon K, Avagyan S, Khaki P, Moradi Bidhendi S, Forbes KJ, et al. The population structure of Salmonella enterica Enteritidis in Iran analyzed by multiple-locus variable-number tandem repeat analysis. Trop Anim Health Prod 2012; 45: 1– 6. [DOI] [PubMed] [Google Scholar]
- 26. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 1988; 26: 2465– 2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Achtman-1 M. MLST Databases at UoW. 2015. [cited 2015/05/29]; Available from: http://mlst.warwick.ac.uk/mlst/dbs/Senterica.
- 28. McCarthy C. (1996). Chromas: version 1.3. Griffith University; Brisbane, Australia. [Google Scholar]
- 29. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947– 8. [DOI] [PubMed] [Google Scholar]
- 30. Anonymous Artemis: Genome Browser and Annotation Tool. [cited 2012/12/20]; Available from: http://www.sanger.ac.uk/resources/software/artemis/. [Google Scholar]
- 31. Noda T, Murakami K, Asai T, Etoh Y, Ishihara T, Kuroki T, et al. Multi-locus sequence typing of Salmonella enterica subsp. enterica serovar Enteritidis strains in Japan between 1973 and 2004. Acta Vet Scand 2011; 53: 1– 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Achtman-2 M. MLST Databases at UoW. 2015. [cited 2015/05/29]; Available from: http://mlst.warwick.ac.uk/mlst/dbs/Senterica/handlers/getFileData/home/cbailster/mlst/zope/Extensions/gadfly/Senterica/DB/PublicStrains.txt.
- 33. Campioni F, Pitondo-Silva A, Bergamini AM, Falcão JP. Comparison of four molecular methods to type Salmonella Enteritidis strains. Apmis 2015; 123: 422– 426. [DOI] [PubMed] [Google Scholar]
- 34. Anon Comprehensive list of all MLST databases. 2015. [cited 2005/05/30]; Available from: http://pubmlst.org/databases.
- 35. Maiden MCJ. Multilocus sequence typing of bacteria. Annu Rev Microbiol 2006; 60: 561– 588. [DOI] [PubMed] [Google Scholar]