Abstract
AIM: To construct the natural immune Fab antibody phage display libraries of colorectal cancer and to select antibodies related with colorectal cancer.
METHODS: Extract total RNA from tissue of local cancer metastasis lymph nodes of patients with colorectal cancer. RT-PCR was used to amplify the heavy chain Fd and light chain κ and the amplification products were inserted successively into the vector pComb3 to construct the human libraries of Fab antibodies. They were then panned by phage display technology. By means of Dot immunoblotting and ELISA, the libraries were identified and the Fab phage antibodies binding with antigens of colorectal cancer were selected.
RESULTS: The amplified fragments of Fd and κ gained by RT-PCR were about 650 bp. Fd and κ PCR products were subsequently inserted into the vector pComb3, resulting in a recombination rate of 40% and the volume of Fab phage display library reached 1.48 × 106. The libraries were enriched about 120-fold by 3 cycles of adsorption-elution-multiplication (panning). Dot immunoblotting showed Fab expressions on the phage libraries and ELISA showed 5 clones of Fab phage anti bodies which had binding activities with antigens of colorectal cancer.
CONCLUSION: The natural immune Fab antibody phage display libraries of colorectal cancer were constructed. They could be used to select the relative antibodies of colorectal cancer.
Keywords: colorectal neoplasms, immunology, bacterio phages, antibody library
INTRODUCTION
The incidence of colorectal cancer is growing in China. It has been clear that the prognosis of colorectal cancer is related to early diagnosis and treatment[1-15]. Since the development and evolution of the colorectal cancer is one kind of complicated procedures involving multi-genes, multi-factors and multi-steps, there have been no highly peculiar molecular pathologic changes and tumor[16-39]. A major focus of cancer immunology is on the isolation of antibodies that react selectively with human tumor cells[40,41], because the antibodies could have important applications for targeting diagnostic and therapeutic agents to tumors and for identifying tumorigenic antigens[42-47]. The established approach has been to generate large panels of monoclonal antibodies (mAb) from mice immunized with human tumor cells and to screen the antibodies for reactivity against the tumor[48-50]. Despite the enormous efforts put on this approach, few antibodies that react preferentially with human tumors, and none that react specifically with one type of tumor (such as colorectal cancer), have been reported. Further attempts to isolate more specific and high affinity antibodies will require improved methods of generating and selecting antibodies against human tumors[51]. One is the introduction of method for synthesizing virtually the entire repertoire of a person’s antibody genes of variable regions by PCR technique and for expressing the encoded antibodies on the surface of a phage vector[52]. The resulting phage antibody library can be panned to select and clone rare antibodies on the basis of their binding specificities[53,54]. It can resolve the problems of generating humanization mAb by hybridoma approach. Humans can be immunized or are immune to many antigens (including tumor antigens) but only local lymph nodes, a productive source of antibody-producing cells, is readily available. Here we describe the construction of the natural immune phage display libraries expressing Fab antibodies derived from local cancer metastasis lymph nodes of co lorectal cancer patients and report the initial results of panning the libraries for anti-colorectal cancer antibodies.
MATERIALS AND METHODS
Vector, E. coli and helper phage
The vector (pComb3), about 4894 base pairs, contains ampicillin resistance (Ampr) gene, start sites of plasmid CO1E1 and f1 phage replication. The expression of inserting fragments is controlled by the same LacZ promoter on its upstream sequence. E. coli XL1-Blue contains tetracycline resistance (Tetr) gene on its gene type of Tn10. Helper phage (VCSM13) contains kanamycin resistance (Kanar) gene with valency of 1015 pfu•L¯¹. It is amplified in SOC culture medium and preserved in 4 °C.
Lymph nodes total RNA preparation
Lymph nodes in mesenterium were resected during surgical operation on patients with colorectal cancer and preserved in li quid nitrogen immediately. The nodes of patients (case number 260280, 260583 and 260476) were defined as tumor metastatic lymph nodes by pathological examination. One hundred mg of each node was used to extract total RNA by the standard method of guanidinium isothiocyanate.
Amplifying Fd and κ chain genes of antibodies by RT-PCR
Total RNA (20-50 μg) was added to 60 pmol primer of Oligo (dt) and heated at 65 °C for 10 min. The mixture was then used in a 20 μL reverse transcription reaction containing 200 μmol•L¯¹ each dNTPs and 20 U of reverse transcriptase (Promega), which was incubated at 37 °C for 1 h. The RNA-cDNA mixture (5 μL) was then used in 50 μL PCR reaction mixture containing all four dNTPs at 60 μmol•L¯¹, 5 U of Taq polymerase (Promega), and 50 pmol•L¯¹ of appropriate 5’ and 3’ primers[55,56]. VK1a and VK3a are 5’ primers for amplification of the κ chain with the Sac I site for cloning into the vector pComb3. CK1a is a 3’ primer corresponding to the 3’ end of the light chain κ, Xba I site. VH1a and VH3a are 5’ primers for the heavy chain (Fd), Xho I site. CG1z is the 3’ primer for the Fd and corresponds to part of the hinge region, Spe I site.
Vκ5’ primers: VK1a, 5’-GACATCGAGCTCACCCAGTCTCCA -3’;
Vκ3a, 5’-GAAATTGAGCTCACGCAGTCTCCA-3’;
Vκ3’ primers: CK1a, 5’-GCGCCGTCTAGAACTAACACTCTCCCCTGTTGAAGCTCTTTGTGACGGGCAAG-3’;
VH (Fd) 5’ primers: VH1a, 5’-CAGGTGCAGCTCGAGCAGTCTGGG-3’;
VH3a, 5’-GAGGTGCAGCTCGAGGAGTCTGGG-3’;
VH (Fd) 3’ primers: CG1z, 5’-GCATGTACTAGTTTTGTCACAAGATTTGGG -3’.
The reaction mixtures were then subjected to 35 rounds of amplification (PE/Cetus thermal cycler) at 94 °C for 1 min, 52 °C for 1.5 min and 72 °C for 2 min followed by a final incubation at 72 °C for 10 min. An aliquot of the reaction mixture (5 μL) was run on a 10 g•L¯¹ agarose gel.
Cloning heavy chain Fd into pComb3
The Fd fragment product of PCR (isolated by agarose gel electrophore sis) was cut with an excess of the restriction enzymes Xho I and Spe I and typically about 350 ng was ligated with 2 μg of Xho/Spe I -linearized pComb3 vector (isolated by agarose gel electrophoresis) in a total volume of 150 μL with 10 U of ligase (Promega) at 16 °C for 15 h. Following ligation, DNA was precipitated at -20 °C for 2 h by the addition of 15 μL of 3 mol•L¯¹ sodium acetate (pH5.2), and 330 μL of ethanol. DNA was pelleted by microcentrifugation at 4 °C for 15 min. The DNA pellet was resuspended in 20 μL of water and transformed into 150 μL XL1-blue (porated by calcium chloride). After transformation, XL1-blue samples (10, 1, 0.1 μL) were withdrawn for plating to determine the rate of transformation. The insertion of target Fd fragments were detected by PC R from the plasmids extracted from several random XL1-blue monoclones. The plasmids with Fd insertion were named p+Fd.
Cloning light chain κ into p+Fd and antibodies Fab libraries construction
The κ fragment product of PCR and recombined p+Fd (isolated by agarose gel electrophoresis) were cut with an excess of the restriction enzymes Sac I and Xba I. The ligation, transformation, E. coli amplification and determination of the transformation rate were same as described above. The insertion of target κ fragments was detected by digestion with Sac I/Xba I from the plasmids extracted from several random XL1-blue monoclones. The Fab fragments’ insertion was detected by digestion with Xho I and Xba I. The plasmids with Fd together with κ insertion were named Fd+κ. Helper phage VCSM13 (1012 pfu) was added to XL1-blue samples contained Fab gene libraries. Following the superinfection, the primer Fab phage display libraries of two patients were constructed, named K1 and K2, and preserved at 4 °C.
Panning of the primer Fab phage display libraries
The colorectal cancer antigens were derived from fresh cancer tissues of patients 1, 2, and 3 by sonicating the tissues (each mass 500 mg). The supernatants were named Ag1, Ag2 and Ag3. Colon cancer cell line of Lovo (109•L¯¹) was sonicated also, and its supernatants were named Lovo. The wells of a microtiter plate were coated overnight at 4 °C with the antigen supernatants above respectively (antigens were diluted with 0.1 mol•L¯¹ bicarbonate buffer, pH9.6). Cancer tissue antigens coated the microtiter plate wells in mixture of Ag1 and Ag2. Lovo cells of 200 μL (0.5 × 108•L¯¹) were attached to the wall of the microtiter plate wells, and 50 μL mixture of K1 and K2 was added to the wells coated with antigens respectively. The panning procedure is a modification[57] of that originally described by Parmley and Smith. The phage binding with antigens re-infected the XL1-Blue by eluting from the wells. A round of panning was finished after superinfection by VCSM13. Following 3 rounds of panning, the percent yield of phage was determined as (no. of phage eluted/no. of phage applied) × 100.
Dot immunoblotting analysis of Fab displaying[58]
Five μL library of each round of panning was added to nitrocellulose (NC) filter (with diameter of 0.5 cm). After being dried at room temperature, the NC filters were blocked in 50 mL•L¯¹ de-lipid milk and then react with the biotinylated anti-human IgG (1:100 dilution) and alkaline phosphatase (AP)-avidin at room temperature successively. Finally the NBT+BCIP was added and color developed. The control unit was the suspension of the phage contained vacant vector pComb3.
ELISA analysis of Fab displaying[59]
The wells of a microtiter plate were coated overnight at 4 °C with the antigen supernatants Ag1, Ag2, Ag3 and Lovo respectively (antigens were diluted in 1:10 0 with 0.1 mol•L¯¹ bicarbonate buffer, pH9.6). The suspension of K1 and K2 (50 μL each) was added to the wells respectively, and cultivated at 37 °C. Following 3 washes with TBS buffer, 25 μL of a 1:100 dilution of biotinylated goat anti-human IgG and HP-conjugated biotin were added successively and incubated at 37 °C. Finally 50 μL of TMB was added and color development was monitored at 490 nm. The A 490 values of positive clones were higher than that of negative clones at least two folds. After 3 rounds of panning, 5 clones of positive XL1-blue were superinfected with VCSM13 and the supernatants of Fab displaying were prepared. Fifty μL of the Fab display supernatants were added to the wells (coated with antigens of Ag1, Ag2, Ag3 and Lovo) respectively, and ELI SA analysis was applied by the method described above.
RESULTS
Amplifying the fragments of Fd and κ chain genes of antibodies by RT-PCR
The total RNAs of the lymph nodes defined as tumor metastasis in mesenterium of patients with colorectal cancer were isolated. The integrity of RNAs was shown by alkaline denaturing agarose gel. Their purity (A260/A280) reached 2.04. Fd and κ immunoglobulin chains were PCR-amplified after reverse transcription from this RNA. For amplifying Fd and κ, two 5’primers and one 3’primer were used respectively which contained some sites of the restriction enzymes corresponding to the vector pCom b3. The PCR amplification of Fd and κ was about 650 bp (Figure 1).
Figure 1.
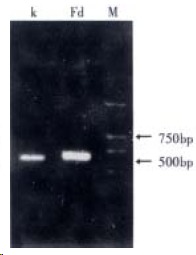
PCR products of Fd and κ.
Construction and identification of recombinant p+Fd
The recombinants of p+Fd were constructed by cloning Fd fragments into pComb3. After transforming XL1-blue, the Fd gene libraries were formed. And the quantity of the transformants reached 5.53 × 106. After extracting the plasmids from eight random XL1-blue monoclones, the insertion of target Fd fragments was detected by PCR with the primers same as that in amplifying Fd described above. Four of eight PCR could amplify the fragments of 650 bp (Figure 2). This meant that the recombination fre quency was 50%, so the practical volume of Fd library was 2.77 × 106.
Figure 2.
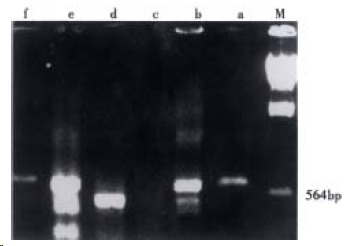
Fd insertion by PCR identification.
Construction and identification of Fab gene recombinant
The recombinants of Fd+κ were constructed by cloning κ fragments into p+Fd. After transforming XL1-blue, the Fab gene libraries were formed and the quantity of the transformants reached 7.4 × 106. Following extracting the plasmids from ten random XL1-blue monoclones, the insertion of target κ fragments was detected by digestion with Sac I and Xba I, and five of these plasmids discharged the fragments of 650 bp (Figure 3). By digesting with Xho I and Xba I, four of these plasmids discharged the fragments about 2.4 kb (Figure 4). The recombination frequency of fragments Fd together with κ insertion was 40%, so the practical volume of Fab library was 7.4 × 106 × 50% × 40% = 1.48 × 106.
Figure 3.
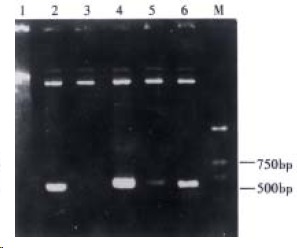
Identification of κ insertion by Sac I/Xba I digestion.
Figure 4.
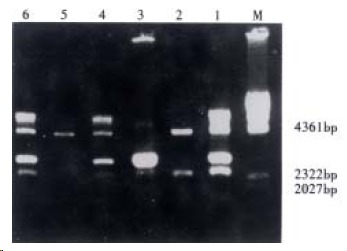
Identification of κ and Fd insertion by Xho I/Xba I digestion.
ELISA of binding activity of colorectal cancer antigens and Fab displaying on primer phage libraries
The activity of colorectal cancer antigens binding with K1 (or K2) was identified by ways of ELISA with control unit of wash buffer. The primer phage display libraries integrated with the colorectal cancer antigens of not only its immune derivation, but also other immune derivation (Table 1).
Table 1.
Binding of Fab displayed by prime phage display librar ies with colorectal cancer antigens (A490)
| Ag1 | Ag2 | Ag3 | Lovo | Wash buffer | |
| K1 | 1.285 | 1.496 | 1.332 | 0.326 | 0.105 |
| K2 | 1.324 | 1.552 | 1.352 | 0.322 | 0.122 |
| BC | 0.115 | 0.105 | 0.110 | 0.092 | 0.087 |
Panning and identification of the Fab phage display libraries
After 3 rounds of panning, the libraries were enriched about 120-fold. Dot immunoblotting applied by the system of AP-goat anti-human IgG showed that there were Fab expressions on the phage libraries of each panning round (Figure 5). There was no obvious color development in the control units (C). The supernatants of Fab displaying prepared from 5 clones of positive XL1-Blue after 3 rounds of panning showed the significant binding activity with the antigens related with colorectal cancer by ELISA analysis (Table 2). The control units were the rat colon antigens and wash buffer.
Figure 5.

Dot immunoblotting analysis.
Table 2.
Binding activity of phage antibodies with antigens (A490)
| Phage antibodies | Ag1, Ag2 and Ag3 | Lovo | Rat colon antigens | Wash buffer |
| Clone 1 | 1.236 | 1.042 | 0.121 | 0.096 |
| Clone 2 | 1.117 | 0.873 | 0.110 | 0.100 |
| Clone 3 | 1.450 | 1.329 | 0.182 | 0.121 |
| Clone 4 | 1.106 | 0.912 | 0.106 | 0.087 |
| Clone 5 | 1.345 | 1.024 | 0.090 |
DISCUSSION
Fd parts and κ immunoglobulin chains (650 bp) were PCR-amplified after reverse transcription from the RNAs, which were isolated from the lymph nodes defined as tumor metastasis in mesenterium of patients with colorectal cancer. For mimicking the diversity of the antibody genes, we used 5’ primers based on the leader exon and the N terminus of first framework (FR1) sequences according to their relatively constant regions, and 3’ primers within the constant regions during amplifying the V genes of immunoglobulin Fab. To maximize complementarity, degeneracy was incorporated into the primers, or different primers were designed for different families of antibody V genes. For cloning of the amplified DNA into expression vectors, rare restriction sites were introduced within the PCR primer s. Following mixing the 2-group of Fd PCR products, we cloned them into the vector pComb3. Then the 2-group of κPCR products were inserted into the recombinants of p+Fd. Therefore the gene libraries of humanization antibody were constructed, which contained parts of 2-subgroup of heavy chains and 2-subgroup of κ chains. Previously reports had shown that the volume of the phage display libraries was 1.0 × 107 at most constructed by this vectors and ways[57]. In review, the volume was 106~107 according to the reports abroad, and about 10 6 according to the domestic reports[60,61]. We reported the volume being 1.48 × 106.
By antibody technology, the mAbs can be prepared by-passing hybridoma approach. Construction of the natural immune antibody library is the method of by-passing unnatural immunization. The natural immune antibody library is constructed from the natural donors, such as lymphocyte B and plasma cell. In our present study, the natural immune Fab antibody phage display libraries were constructed from the lymphocytes in the lymph nodes of 2 patients with colorectal cancer. There have been several studies on the library of immunity antibody in recent years[60-62], but no reports on construction of the natural immune antibody library of human colorectal cancer. Theoretically the humanization antibody library constructed from the lymphocytes B in lymph nodes defined as tumor metastasis of patients with tumor can aim at screening the antibodies, because the genes of heavy and light chains have been rearranged and ligated specifically due to immunizing of tumor antigens. By this way the promiscuous work of preparation of antigens by immunizing rats was relieved and the problems of weak antigens, heterological antibodies, low fusion, instability and low antibodies products of hybridoma were resolved. From the natural immune antibody library, a few of phage antibodies immunized by some for eign antigens (such as bovine serum album, lysozyme, bovine thyroid globulin, etc.) were screened, and some self-antigens (such as tumor necrosis factor-α, carcinoembryonic antigen, CD4, thyroid globulin, etc.). Many of these phage antibodies showed high affinity and specificity, and the potential value for application.
It is important to assemble antibody molecules that have correctly folded heavy and light chains and thus retain antigen-recognition capabilities. Previously it had been demonstrated that Fd chains (comprising VH and CH1, the variable region and constant domain 1 of the immunoglobulin heavy chain) and κ chains targeted to the periplasm of E. coli assemble to form functional Fab molecules. If the Fd chain was anchored in the membrane and concomitantly provided with secreted κ chain, functional Fab molecules could form on the membrane surface facing the periplasm. The coexpression behind the PelB leader sequence of Fd fused to cpIII (the major coat protein of M13 phage) and κ chains lead to membrane anchoring of the Fd chain and compartmentalization of the κ chains in the periplasm. These two chains could assemble in situ to allow accumulation of functional Fab on the membrane surface, which, by virtue of the cpIII sequences, would be incorporated along the entire length of the filamentous phage particles on subsequent infection with helper phage. These phages displaying Fab could infect E. coli again. Following rounds of panning by adsorption-wash-amplification, the phages comprising specific antibody molecules could be enriched[63]. Our study showed that the phage Fab libraries were enriched about 120-fold after 3 rounds of panning. It suggested that the colorectal cancer antigens had selected the phages in the libraries displaying Fab. Dot immunoblotting showed that there were Fab expressions on the phage libraries of each panning round. ELISA analysis showed that the phage displaying Fab had the significant binding activity with the antigens related with colorectal cancer. Considering that nowadays there are few and hypo-specific antibodies related with colorectal cancer, the natural immune Fab antibody phage display libraries of colorectal can be used to select the affinity and specificity antibodies related to colorectal cancer. It is important to diagnose and treat colorectal cancer using these antibodies.
Footnotes
Edited by Xu JY
Supported by the Natural Science Foundation of Guangdong Provin ce, China, No.980120 and the Foundation of Excellent Youth Teacher, China, 2001
References
- 1.Zhou DY, Feng FC, Zhang YL, Lai ZS, Zhang WD, Li LB, Xu GL, Wan TM, Pan DS, Zhou D. Comparison of Shams' test for rectal mucus to an immunological test for fecal occult blood in large intestinal carcinoma screening. Analysis of a check-up of 6480 asymptomatic subjects. Chin Med J ( Engl) 1993;106:739–742. [PubMed] [Google Scholar]
- 2.Jia L, Chen TX, Sun JW, Na ZM, Zhang HH. Relationship between microvesselnsity and proliferating cell nuclear antigen and prognosis in colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:74–76. [Google Scholar]
- 3.Jiang XL, Quan QZ, Liu T, Dong XC. Recent advances in research of ulcerative colitis. Shijie Huaren Xiaohua Zazhi. 2000;8:216–218. [Google Scholar]
- 4.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 5.Gu HP, Ni CR, Zhan RZ. Relationship of expressions of CD15, CD44v6 and nm23 h1 mRNA with metastasis and prognosis of colon carcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:887–891. [Google Scholar]
- 6.Yang JH, Rao BQ, Wang Y, Tu XH, Zhang LY, Chen SH, Oua2Yang XN, Dai XH. Clinical significance of detecting the circulating cancer cells in peripheral blood from colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:187–189. [Google Scholar]
- 7.Zhou TX, Li JS, Xing LW, You SH. Determination of lipid peroxide and superoxide dismutase in blood and tissue of patients with gastrointestinal cancer. World J Gastroenterol. 2000;6(Suppl 3):56. [Google Scholar]
- 8.Futamura M, Takagi Y, Koumura H, Kida H, Tanemura H, Shimokawa K, Saji S. Spread of colorectal cancer micrometastases in regional lymph nodes by reverse transcriptase-polymerase chain reactions for carcinoembryonic antigen and cytokeratin 20. J Surg Oncol. 1998;68:34–40. doi: 10.1002/(sici)1096-9098(199805)68:1<34::aid-jso8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Kobaek-Larsen M, Thorup I, Diederichsen A, Fenger C, Hoitinga MR. Review of colorectal cancer and its metastases in rodent models: comparative aspects with those in humans. Comp Med. 2000;50:16–26. [PubMed] [Google Scholar]
- 10.Liu R, Wang YH, Tang Y, Cao GS. Effect of Octreotide on cell-cycle kinetics and serum CEA level in hepatic metastasesof colonic adenocarcinoma. China Natl J New Gastroenterol. 1997;3:69–71. doi: 10.3748/wjg.v3.i2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khubchandani IT, Karamchandani MC, Kleckner FS, Sheets JA, Stasik JJ, Rosen L, Riether RD. Mass screening for colorectal cancer. Dis Colon Rectum. 1989;32:754–758. doi: 10.1007/BF02562123. [DOI] [PubMed] [Google Scholar]
- 12.Xu GL, Chen ZM, Zhang WD. The clinical and pathology characteristics of young people colorectal cancer. Xin Xiaohuabingxue Zazhi. 1997;5:304. [Google Scholar]
- 13.Ren DL, Yuan HX, Luo ZB, Lin DS, Fan XH, Zhang SF. Analysis of 186 cases of children colorectal cancer in China. Xin Xiaohuabingxue Zazhi. 1997;5:626–628. [Google Scholar]
- 14.Tannapfel A, Katalinic A, Köckerling F, Wittekind C. The prediction of lymph node metastases in colorectal cancer by expression of the nucleoside diphosphate kinase/nm23-H1 and histopathological variables. Am J Gastroenterol. 1997;92:1182–1186. [PubMed] [Google Scholar]
- 15.Ratto C, Sofo L, Ippoliti M, Merico M, Doglietto GB, Crucitti F. Prognostic factors in colorectal cancer. Literature review for clinical application. Dis Colon Rectum. 1998;41:1033–1049. doi: 10.1007/BF02237397. [DOI] [PubMed] [Google Scholar]
- 16.Ji DJ, Cao Y, Zhang YL, Jiang P, Yu N, Feng FC, Zhou DY. Synchronous studies on variations of p53 gene transcriptions and expressions in colorectal ca rcinomas HT-29 and Lovo cell lines. Shijie Huaren Xiaohua Zazhi. 2000;8:77–79. [Google Scholar]
- 17.Xu SH, Feng JG, Li DC, Mou HZ, Lou RC. Relationship between CD44in the peripheral blood of patients with colorectal cancer and clinicopathological features. Shijie Huaren Xiaohua Zazhi. 2000;8:432–435. [Google Scholar]
- 18.Yamaguchi A, Urano T, Fushida S, Furukawa K, Nishimura G, Yonemura Y, Miyazaki I, Nakagawara G, Shiku H. Inverse association of nm23-H1 expression by colorectal cancer with liver metastasis. Br J Cancer. 1993;68:1020–1024. doi: 10.1038/bjc.1993.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao MF, Mao H, Zheng JX, Yuan YW. Effect of vascular endothelial growth factor on adhesion of large intestine cancercell HT-29. Shijie Huaren Xiaohua Zazhi. 2000;8:646–649. [Google Scholar]
- 20.Liu H, Wu JS, Li LH, Yao X. The expression of plateletderived growth factor and angio genesis in human colorectal carcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:661–664. [Google Scholar]
- 21.Xu QW, Li YS, Zhu HG. Relationship between expression p53 protein, PCNA and CEA in colorectal cancer and lymphnode metastasis. World J Gastroenterol. 1998;4:218. [Google Scholar]
- 22.Wu BP, Zhang YL, Zhou DY, Gao CF, Lai ZS. Microsatellite instability, MMR gene expression and proliferation kinetics in colorectal cancer with famillial predisposition. World J Gastroenterol. 2000;6:902–905. doi: 10.3748/wjg.v6.i6.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo WJ, Zhou GD, Wu HJ, Liu YQ, Wu RG, Zhang WD. Ultrastructural localization of glutathione S-transferase-pi in human colorectal cancer cells. World J Gastroenterol. 2000;6:454–455. doi: 10.3748/wjg.v6.i3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Lu GY. Analysis of ras p21 and DNA contents in colorectal cancer. Shijie Huaren Xiaohua Zazhi. 1999;7:706–707. [Google Scholar]
- 25.Hu JY, Wang S, Zhu JG, Zhou GH, Sun QB. Expression of B7 costimulation molecules by colorectal cancer cells reducestumorigenicity and induces anti-tumor immunity. World J Gastroenterol. 1999;5:147–151. doi: 10.3748/wjg.v5.i2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang DC, Luo YH, Lu R, Liu WW, Liu FX, Liang ZY. Loss of heterozygosity at APC, MCC and DCC genetic loci in colorectal cancers. China Natl J New Gastroenterol. 1995;1:21–24. [Google Scholar]
- 27.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, Izumi Y, Irimura T. Involvement of carbohydrate antigen sialyl Lewis (x) in colorectal cancer metastasis. Dis Colon Rectum. 1997;40:420–431. doi: 10.1007/BF02258386. [DOI] [PubMed] [Google Scholar]
- 28.Zheng CX, Zhan WH, Zhao JZ, Zheng D, Wang DP, He YL, Zheng ZQ. The prognostic value of preoperative serum levels of CEA, CA19-9 and CA72-4 in patients with colorectal cancer. World J Gastroenterol. 2001;7:431–434. doi: 10.3748/wjg.v7.i3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vauthey JN, Dudrick PS, Lind DS, Copeland EM. Management of recurrent colorectal cancer: another look at carcinoembryonic antigen-detected recurrence. Dig Dis. 1996;14:5–13. doi: 10.1159/000171535. [DOI] [PubMed] [Google Scholar]
- 30.Park JG. Genetic diagnosis and management of hereditary nonpolyposis colorectal cancer. World J Gastroenterol. 1998;4(Suppl 2):55. [Google Scholar]
- 31.Luo F, Kan B, Lei S, Yan LN, Mao YQ, Zou LQ, Yang YX, Wei YQ. Study on p53 protein and C-erbB2 protein expression inprimary hepatic cancer and colorectal cancer by flow cytometry. World J Gastroenterol. 1998;4(Suppl 2):87. [Google Scholar]
- 32.Wang YX, Ruan CP, Li L, Shi JH, Kong XT. Clinical significance of changes of perioperative T cell and expression of its activatedantigen in colorectal cancer patients. World J Gastroenterol. 1999;5:181–182. doi: 10.3748/wjg.v5.i2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jass JR. Future role of the pathologist in reporting colorectal cancer. World J Surg. 1997;21:688–693. doi: 10.1007/s002689900292. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Luo HS, Yu BP. Telomerase and colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:800–802. [Google Scholar]
- 35.Wang LS, Pan LJ, Chen CL, Li MS, Shun Y, Zhang YL, Zhou DY. Effect of Bifidobacterium on proliferation and apoptosis ofexperimental large bowel carci noma in situ. Shijie Huaren Xiaohua Zazhi. 2000;8:429–431. [Google Scholar]
- 36.Sheng QG. Relationship between related factors with lymph node metastasis of colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:654–657. [Google Scholar]
- 37.Ratto C, Sofo L, Ippoliti M, Merico M, Doglietto GB, Crucitti F. Prognostic factors in colorectal cancer. Literature review for clinical application. Dis Colon Rectum. 1998;41:1033–1049. doi: 10.1007/BF02237397. [DOI] [PubMed] [Google Scholar]
- 38.McLeod HL, Murray GI. Tumour markers of prognosis in colorectal cancer. Br J Cancer. 1999;79:191–203. doi: 10.1038/sj.bjc.6690033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin J, Liang YJ, Wang HZ, Li SQ. Expression of endocrine cells in colorectal cancer and its adjacent mucosa. Huaren Xiaohua Zazhi. 1998;6:315–317. [Google Scholar]
- 40.Owens RJ, Young RJ. The genetic engineering of monoclonal antibodies. J Immunol Methods. 1994;168:149–165. doi: 10.1016/0022-1759(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 41.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 42.Fan DM. The clinical application with monoclonal antibodies on digestion neoplasm. China Natl J New Gastroenterol. 1993;1:68–70. [Google Scholar]
- 43.Shi YQ, Xiao B, Miao JY, Li MF, Qiao TD, Chen BJ, Chen Z, Han JL, Zhou SJ, Fan DM. A novel cDNA fragment associated with gastric cancer drug resistance screened from a library by mAb MGr1. Huaren Xiaohua Zazhi. 1998;6:656–659. [Google Scholar]
- 44.Song ZQ, Hao F, Zhang J, Gu CH. Detection of antibodies against hypervariable region 1 in sera from patients with hepatitis C. Shijie Huaren Xiaohua Zazhi. 1999;7:666–668. [Google Scholar]
- 45.Qiu K, Wang BC, Chen ZN, Fang P, Liu CG, Wan WX, Liu YF. 99 mTc-labeled HAb18 McAb Fab fragment for radioimmunoimaging in nude mice bearing human hepatocellular carcinoma. World J Gastroenterol. 1998;4:117–120. doi: 10.3748/wjg.v4.i2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu SP, Zheng SJ, Zhou HY. Evaluation of colorectal carcinoma screening with fecal monoclonal antibody. World J Gastroenterol. 1998;4(Suppl 2):95. [Google Scholar]
- 47.Fan DM, Xiao B, Shi YQ, Ming F, Qiao TD, Chen BJ, Chen Z. A novel cDNA fragment associated with gastric cancer drug resistance was screened out from a library by monoclonal antibody MGr1. World J Gastroenterol. 1998;4(Suppl 2):110–111. [Google Scholar]
- 48.Zhang J, Liu YF, Yang SJ, Sun ZW, Qiao Q, Zhang SZ. Construction and expression of mouse/humanized scFv and their fusion to humanized mutant TNFα against hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:616–620. [Google Scholar]
- 49.Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoogenboom HR, de Bruïne AP, Hufton SE, Hoet RM, Arends JW, Roovers RC. Antibody phage display technology and its applications. Immunotechnology. 1998;4:1–20. doi: 10.1016/s1380-2933(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 51.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 52.Zheng W, Tan H, Song SB, Lu HY, Wang Y, Yu YX, Yin R. The construction and expression of a fusion protein consistinganti-HBsAg antibody fragment Fab and interferon a in E. coli. World J Gastroenterol. 2000;6(Suppl 3):83. [Google Scholar]
- 53.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Wang Y, Li QX, Wang YM, Xu JJ, Dong ZW. Cloning of 3 h11 mAb variable region gene and expression of 3 h11 human-mouse chimeric light Chain. World J Gastroenterol. 1998;4:41–44. doi: 10.3748/wjg.v4.i1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huse WD, Sastry L, Iverson SA, Kang AS, Alting-Mees M, Burton DR, Benkovic SJ, Lerner RA. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 56.Barbas CF, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbas CF, Lerner RA. Combinatorial immunoglobulin libraries on the surface of phage (phabs): rapid selection of antigen-specific Fabs. Methods: Comp Methods Enzymol. 1991;2:119–124. [Google Scholar]
- 58.Wu W, Xu DZ, Yan YP, Zhang JX, Liu Y, Li RL. Evaluation of dot immunogold filtration assay for anti-HAV IgM antibody. World J Gastroenterol. 1999;5:132–134. doi: 10.3748/wjg.v5.i2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen B, Zhou SJ, Fan DM. Establishment of a quick ConA-mAb-ELISA for detection of serum MC3-Ag, a novel colorectalcancer-associated antigen. Xin Xiaohuabingxue Zazhi. 1995;3(Sup pl 4):12–14. [Google Scholar]
- 60.Zhao YF, Wang HT, Wang QL, Du ZY, Yang JG, Sun HY, Ma LR, Yin Z. Scree ning of “1F7” idiotypic anti-gp160 antibodiesfrom phage antibody library. Chinese Journal of Biochemistry and Molecular Biology. 1998;14:20–24. [Google Scholar]
- 61.Wang Y, Hua B, Liu QY, Gao RK, Zhu YC, Chen YP. By passing immunization: Construction of a phage antibody librarywith randomized CDR3. Chin J Microbiol Immunol. 1997;17:449–452. [Google Scholar]
- 62.Lan FH, Gao H, Liu YF, Chen SM. Construction of a naive IgG phage display library from a normal human individual. J Fourth Mil Med Univ. 1999;20:464–467. [Google Scholar]
- 63.Pini A, Viti F, Santucci A, Carnemolla B, Zardi L, Neri P, Neri D. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. 1998;273:21769–21776. doi: 10.1074/jbc.273.34.21769. [DOI] [PubMed] [Google Scholar]


