Abstract
OBJECTIVES
To assess a novel application of the Prostate Health Index (phi) and biopsy tissue DNA content in benign-adjacent and cancer areas to predict which patients would eventually require treatment of prostate cancer in the Proactive Surveillance cohort.
METHODS
We identified 71 men who had had serum and biopsy tissue from their diagnosis banked and available for the present study. Of the 71 patients, 39 had developed unfavorable biopsy findings and 32 had maintained favorable biopsy status during surveillance. The serum total prostate-specific antigen (tPSA), free PSA (fPSA) and [−2]proPSA were measured using the Beckman Coulter immunoassay. The DNA content measurements of Feulgen-stained biopsy sections were performed using the AutoCyte imaging system.
RESULTS
The ratio of phi was significantly greater (37.23 ± 15.76 vs 30.60 ± 12.28; P = .03) in men who ultimately had unfavorable biopsy findings. The serum phi ratio (P = .003), [−2]proPSA/%fPSA (P = .004), biopsy tissue DNA content (ie, benign-adjacent excess of optical density, P = .019; and cancer area standard deviation of optical density, P = .002) were significant predictors of unfavorable biopsy conversion on Cox regression analysis. However, phi and [−2]proPSA/%fPSA showed a highly significant correlation (rho = 0.927, P < .0001) and no difference in accuracy (c-index, 0.6247 vs 0.6158; P = .704) for unfavorable biopsy conversion prediction. Furthermore, phi and [−2]proPSA/%fPSA remained significant (P = .047 and P = .036, respectively) in the multivariate models and, combined with the biopsy tissue DNA content, showed improvement in the predictive accuracy (c-index, 0.6908 and 0.6884, respectively) for unfavorable biopsy conversion.
CONCLUSIONS
The Prostate Health Index to proPSA/%fPSA, combined with biopsy tissue DNA content, improved the accuracy to about 70% to predict unfavorable biopsy conversion at the annual surveillance biopsy examination among men enrolled in an Active Surveillance program.
Prostate cancer (PCa) is the second leading cause of cancer death among men in the United States, with approximately 192 280 newly diagnosed cases and 27 360 deaths in 2009.1 With prostate-specific anti gen (PSA) screening, a dramatic increase has occurred in the PCa incidence since its introduction, with a shift of the disease at diagnosis to much earlier and potentially more curable stage.2 The European randomized study of screening for PCa showed a 20% reduction in PCa death using PSA screening.3 However, 1410 men need to be screened and 48 additional PCa cases need to be treated to prevent 1 death from PCa.3 The Prostate, Lung, Colon, and Ovary cancer screening trial of the National Cancer Institute found no difference in the rate of PCa deaths between the screened and control groups after 7–10 years of follow-up.4 The results of the Prostate, Lung, Colon, and Ovary cancer trial should be reviewed with caution owing to prescreening of the study population with PSA determination and the high percentage of men in the control group who had undergone PSA screening. Although the benefits of PCa screening are uncertain, the burdens associated with the early detection and treatment of PCa are known. It has been estimated that 23%–42% of screen-detected PCa cases would never have been diagnosed in the absence of screening.2
Active surveillance (AS) as an alternative to immediate surgery has been proposed for low-grade, low-stage tumors in an effort to reduce the unnecessary treatment of PCa.5 Epstein et al6 proposed a PSA density of <0.15 ng/mL/cm3 and favorable diagnostic needle biopsy characteristics (ie, Gleason score of ≤6, ≤2 cores involved with cancer, and ≤50% of any core involved with cancer) as criteria to identify low-grade, low-stage tumors that could be followed up with serial measurements of PSA and repeated biopsies, without immediate intervention. The recent National Comprehensive Cancer Network clinical practice guidelines in oncology7 have described tumors that meet the Epstein criteria6 as “very low risk PCa for recurrence” and recommended AS as a treatment option for men with very low risk PCa who have a life expectancy of <20 years.
Our research group earlier reported that the DNA content of diagnostic biopsy benign-adjacent (BA) and cancer (CA) tissue areas8 and serum [−2]proPSA/percentage of free PSA (%fPSA)9 can be used to predict unfavorable biopsy conversion among men enrolled in the AS program. proPSA is the precursor form of PSA and contains a 7-amino acid pro-leader peptide. Additional truncated forms of proPSA exist in serum, primarily those with leader sequences of 5, 4, and 2 amino acids.10 Cleavage activity of the leader sequences by human kallikrein-2 and trypsin to activate PSA decreases with the decreasing size of the pro-peptide leader sequence, with [−2]proPSA resistant to activation. Recently, Beckman Coulter (Brea, CA) developed a mathematical formula combining total PSA (tPSA), fPSA, and [−2]proPSA and called the Prostate Health Index {phi = ([−2]proPSA/fPSA) × (square root of tPSA)}, which has been approved for use in European countries and is currently pending U.S. Food and Drug Administration approval. The purpose of the present study was to assess phi, phi versus [−2]proPSA/%fPSA, and their combination with diagnostic biopsy tissue DNA content to predict unfavorable biopsy conversion at the annual surveillance examination in our AS cohort.
MATERIAL AND METHODS
Patient Sample
Patients who provided written informed consent were enrolled in our institutional review board-approved AS program if they had met the following inclusion criteria: nonpalpable tumor found on digital rectal examination (ie, Stage T1c tumors), PSA density of ≤0.15 ng/mL/cm3 (PSA level before diagnosis divided by the prostate volume determined using transrectal ultrasound measurement), and favorable diagnostic needle biopsy characteristics (Gleason score of ≤6, ≤2 cores involved with cancer, and ≤50% of any core involved with cancer). The patients were followed up semiannually with serum tPSA and fPSA determination and digital rectal examination and annually with a ≥12-core surveillance biopsy examination. Curative intervention was triggered by unfavorable biopsy conversion (Gleason score of ≥7 or any Gleason pattern 4/5 or ≥3 cores involved with cancer, or >50% of any core involved with cancer) found on the annual surveillance biopsy examination.
Serum PSA Isoform Measurement
Serum was obtained before biopsy and stored at −80°C until testing. The serum specimens were analyzed using the Beckman Coulter Access immunoassay system for tPSA, fPSA, and [−2]proPSA. The assays for tPSA and fPSA were commercially available, and the assay for [−2]proPSA was for research use only.11 The [−2]proPSA assay was calibrated using [−2]proPSA purified from the AVA12-PSA mammalian cell line. The assay has linear ranges of <1–5000 pg/mL for [−2]proPSA, with an interassay precision of 4.8%–12.5% (range 20–279 pg/mL) for [−2]proPSA. The cross-reactivity with other PSA isoforms in the assay was minimal.
DNA Content Analysis
Feulgen DNA staining was performed according to the manufacturer’s instructions (TriPath Imaging, Burlington, NC) on ~5-μm tissue sections from the prostate biopsy cores. Feulgen specifically and quantitatively stains DNA in cellular material by uncovering the free aldehyde groups in the DNA during the acid hydrolysis process, which then reacts with the Feulgen reagent to form a stable, colored compound (blue) that absorbs light at 560 nm. To measure the DNA content, the imaging system was first calibrated using a control slide that contained Feulgen-stained rat hepatocytes. Next, a minimum of 125 intact, Feulgen-stained nuclei were captured in the CA and BA (from the same core as the CA) tissue areas marked by a pathologist (J.I.E.) for each patient using an AutoCyte Pathology Workstation (TriPath Imaging) and the QUIC-DNA software (TriPath Imaging). We used the variance of the DNA content measurements using the nuclei captured for each case as our input variables.
Statistical Analysis
All data were analyzed using Stata, version 10.0, statistical analysis software (StataCorp, College Station, TX). The Mann-Whitney U test was used to test the distribution differences across the favorable and unfavorable biopsy groups. Univariate Cox proportional hazard regression analysis was used to identify the significant predictors for unfavorable biopsy conversion on annual surveillance examination. Ties were handled by the Breslow method. The proportional hazard assumption was verified by examination of the residual plots and Schoenfeld residuals. We determined the optimal cutoffs to dichotomize the continuous variables using the classification and regression tree method. Kaplan-Meier plots and the log-rank test were used to test the equality of the survivor functions across the 2 groups. Harrell’s concordance indexes (c-indexes) were calculated for the significant predictors of unfavorable biopsy conversion. The c-index is the probability that, given 2 randomly selected patients, the patient with the worse outcome would, in fact, have been predicted to have a worse outcome.12 This measure, similar to the area under the receiver operating characteristic curve, ranges from 0.5 (ie, chance or a coin flip) to 1.0 (perfect ability to rank patients). Statistical significance in the present study was set at P ≤ .05.
RESULTS
Of the 71 patients with banked serum and biopsy tissue from their diagnosis available, 39 had developed unfavorable biopsy features and 32 had maintained favorable biopsy features at the annual surveillance biopsy examination (median follow-up 3.72 years). The demographic, clinical, and pathologic information for the favorable and unfavorable biopsy groups is listed in Table 1.
Table 1.
Patient characteristics at diagnosis
| Variable | Favorable (n = 32) | Unfavorable (n = 39) | P Value |
|---|---|---|---|
| Age | 65.42 ± 4.37 (65.03) | 64.82 ± 4.70 (64.97) | .991 |
| tPSA (ng/mL) | 4.61 ± 2.75 (4.36) | 5.35 ± 2.02 (5.53) | .056 |
| fPSA (ng/mL) | 0.88 ± 0.59 (0.74) | 0.97 ± 0.52 (0.89) | .298 |
| %fPSA | 19.15 ± 6.36 (18.98) | 18.40 ± 6.44 (18.21) | .587 |
| [−2]proPSA (pg/mL) | 12.21 ± 7.43 (11.36) | 15.00 ± 6.97 (14.24) | .051 |
| [−2]proPSA/%fPSA | 0.65 ± 0.36 (0.55) | 0.87 ± 0.44 (0.88) | .018 |
| Prostate Health Index | 30.60 ± 12.28 (28.86) | 37.23 ± 15.76 (37.78) | .030 |
| PSA density (ng/mL/cm3) | 0.094 ± 0.058 (0.087) | 0.100 ± 0.0369 (0.100) | .173 |
| Prostate volume (cm3) | 51.86 ± 17.50 (50.00) | 57.29 ± 29.27 (50.00) | .737 |
| Positive cores (n) | 1.19 ± 0.40 (1.00) | 1.18 ± 0.39 (1.00) | .931 |
| Maximal percentage of core involved with cancer | 7.39 ± 10.13 (1.00) | 8.29 ± 11.60 (1.00) | .741 |
tPSA, total prostate-specific antigen; fPSA, free PSA; %fPSA, percentage of fPSA.
Data presented as mean ± standard deviation, with median in parentheses.
On univariate Cox regression analysis, age (P = .860), tPSA (P = .441), fPSA (P = .948), %fPSA (P = .433), PSA density (P = .816), number of cores involved with cancer (P = .780), and the maximal percentage of core involved with cancer (P = .469) were not significant predictors for unfavorable biopsy conversion. The serum phi (hazard ratio [HR] 1.030, 95% confidence interval [CI] 1.007–1.0527; P = .011), [−2]proPSA/%fPSA (HR 2.530, 95% CI 1.181–5.414; P = .017), DNA content in BA tissue area (ie, excess of optical density [OD]; HR 1.011, 95% CI 1.002–1.021; P = .016) and CA tissue area (ie, standard deviation of OD; HR 1.135, 95% CI 1.005–1.281; P = .042) were significant predictors as a continuous variable for unfavorable biopsy conversion.
Dichotomized cutoffs for phi, [−2]proPSA/%fPSA, BA excess of OD, and CA standard deviation of OD were then defined using the classification and regression tree method because they were significant as continuous variables for unfavorable biopsy conversion prediction, and Kaplan-Meier graphs were generated (Fig. 1). Dichotomized phi, [−2]proPSA/%fPSA, BA excess of OD, and CA standard deviation of OD were significant predictors of unfavorable biopsy conversion at the annual surveillance biopsy examination (Table 2). The Harrell c-index for phi, [−2]proPSA/%fPSA, BA excess of OD, and CA standard deviation of OD was 0.6247, 0.6158, 0.5756, and 0.6216, respectively. However, no statistically significant difference was found between the accuracy of phi and [−2]proPSA/%fPSA for predicting unfavorable biopsy conversion (0.6247 vs 0.6158, respectively; P = .704). Also, a highly significant correlation was found between phi and [−2]proPSA/%fPSA (rho 0.927, P < .0001). Thus, phi and [−2]proPSA/%fPSA were considered separately for multivariate analysis with BA excess of OD and CA standard deviation of OD (Table 2). Both phi and [−2]proPSA/%fPSA remained significant in the multivariate models (Table 2) and, combined with the biopsy tissue DNA content, showed an improvement in predictive accuracy (model 1, c-index 0.6908; model 2, c-index 0.6884) for unfavorable biopsy conversion.
Figure 1.
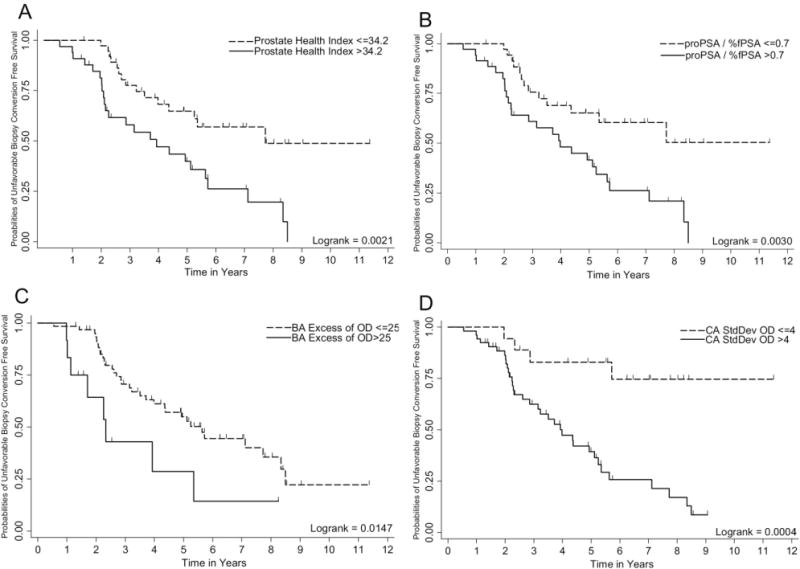
Kaplan-Meier plot showing ability of Prostate Health Index, [−2]proPSA/%fPSA, BA Excess of OD and CAStdDev of OD to predict unfavorable biopsy conversion on annual surveillance biopsy in an active surveillance cohort. Logrank test was used to test the equality of survivor functions across two groups.
Table 2.
Cox regression analysis for predicting unfavorable biopsy conversion at annual surveillance biopsy examination in an active surveillance cohort
| Variable | Patients (n) | Univariate
|
Multivariate Model 1
|
Multivariate Model 2
|
|||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | HR (95% CI) | P Value | ||
| Serum | |||||||
| Prostate Health Index | .003 | .047 | |||||
| ≤34.2 | 38 | 1.00 | 1.00 | ||||
| >34.2 | 33 | 2.65 (1.39–5.07) | 2.00 (1.008–3.97) | ||||
| [−2]proPSA/%fPSA | .004 | .036 | |||||
| ≤0.7 | 36 | 1.00 | 1.00 | ||||
| >0.7 | 35 | 2.65 (1.36–5.16) | 2.15 (1.05–4.38) | ||||
| DNA content | |||||||
| BA excess of OD | .019 | .023 | .018 | ||||
| ≤25 | 59 | 1.00 | 1.00 | 1.00 | |||
| >25 | 12 | 2.58 (1.17–5.68) | 2.69 (1.15–6.32) | 2.80 (1.19–6.57) | |||
| CA SD of OD | .002 | .004 | .003 | ||||
| ≤4 | 18 | 1.00 | 1.00 | 1.00 | |||
| >4 | 53 | 5.36 (1.89–15.24) | 4.87 (1.66–14.24) | 4.97 (1.70–14.50) | |||
HR, hazard ratio; CI, confidence interval; PSA, BA, benign-adjacent; OD, optical density; CA, cancer; SD, standard deviation.
COMMENT
The overdetection and overtreatment of PCa resulting from PSA screening is an important disease management issue among older men. The absolute decrease in PCa-specific mortality in men ≥65 years old randomized to radical prostatectomy versus watchful waiting was only 0.1% (HR 13.1, 95% CI 8.8–19.5 vs HR 13.2, 95% CI 8.9–19.6) at 12 years in the Scandinavian Prostate Cancer Group Trial.13 Furthermore, men ≥65 years old in the radical prostatectomy group had a 2.7% greater overall mortality rate than the watchful waiting group (HR 42, 95% CI 35–50.5, vs HR 39.3, 95% CI 32.5–47.7) at 12 years.13 Thus, most older men diagnosed with screen-detected PCa do not gain years of life with curative intervention. Since 1995, the Johns Hopkins Hospital (JHH) started the AS program for men with very-low-risk PCa, with curative intervention triggered by unfavorable pathology conversion at the annual surveillance biopsy examination.5
In the present study, we have shown that phi can predict unfavorable biopsy conversion at the annual surveillance biopsy examination in the AS cohort (Table 2). However, no difference was found in the accuracy between phi and [−2]proPSA/%fPSA (0.6247 vs 0.6158; P = .704) for unfavorable biopsy conversion prediction. Because phi {([−2]proPSA/fPSA) × (square root of tPSA)} and [−2]proPSA/%fPSA {([−2]proPSA × tPSA)/(fPSA × 100)} use information from the same 3 biomarkers, although in a different manner, a highly significant correlation was found between phi and [−2]proPSA/%fPSA (rho 0.927, P < .0001). Furthermore, phi and [−2]proPSA/%fPSA remained significant in the multivariate models (Table 2) and, combined with biopsy tissue DNA content, showed improvement in predictive accuracy (c-index 0.6908 and 0.6884, respectively) for unfavorable biopsy conversion. Hence, the identification of novel biomarkers and their combination can help us to predict unfavorable biopsy conversion (ie, a recommendation for curative intervention) during surveillance in a AS program.
PSA kinetics,14 p53 nuclear staining,15 Ki-67,16 neu-roendocrine differentiation,17 TMPRSS2–ERG fusions,18 and the apparent diffusion coefficient on diffusion-weighted magnetic resonance imaging19 have shown prognostic value in patients undergoing watchful waiting. Notably, the JHH PAS program enrollment criteria (Stage T1c, Gleason score of ≤6, ≤2 cores involved with cancer, and ≤50% of any core involved with cancer) is more conservative than the enrollment criteria used by other investigators,13–19 who have even enrolled men with Stage T2 lesions and Gleason score 7 tumors in PCa surveillance studies. Because patients with only very-low-risk PCa disease were enrolled and effectively matched for known clinicopathologic parameters at entry, biomarkers such as PCA3 and PSA kinetics were not predictive of unfavorable biopsy conversion during surveillance in the JHH AS cohort.20,21
Recently, a great deal of attention has been paid to the role of proPSA in the early detection and prognosis of PCa. Sokoll et al22 demonstrated that the percentage of proPSA {([−2]proPSA/fPSA) × 100} could reduce unnecessary biopsies in men with a tPSA of 2.5–4.0 ng/mL. In a National Cancer Institute Early Detection Research Network validation study, investigators showed that the percentage of proPSA was the best predictor of PCa detection, particularly in the 2–10-ng/mL tPSA range.23 However, phi was not evaluated in the National Cancer Institute Early Detection Research Network validation study. Recently, Le et al,24 in a prospective study, showed that phi provides the best discrimination between PCa and benign disease in men with a tPSA level of 2.5–10 ng/mL and negative digital rectal examination findings.
DNA alteration in both BA and CA tissue areas of the prostate is a significant event representing the upregulation of proliferation-related genes, including transcription factors, signal transducers, and growth regulators compared with prostatic tissue from healthy donors.25,26 Tumors with normal DNA content are slow growing, less likely to disseminate, and have a better prognosis than nondiploid tumors.27 Furthermore, the DNA content can be used as a surrogate for the biopsy Gleason score for PCa pathologic stage prediction.28 Tribukait29 showed that DNA ploidy has independent prognostic value for survival, particularly in low-grade, low-stage tumors, for which other known variables did not provide any prognostic value. Adolfsson et al30 showed that nondiploid tumors result in a significantly increased risk of PCa-specific death in a cohort of 119 patients with clinically localized PCa (Stage T1-T2) diagnosed during the pre-PSA era and undergoing watchful waiting (median follow-up 24 years).
Although the JHH AS program was started in 1995, serum and urine samples were routinely collected and stored at −80°C for biomarker studies starting from 2007. Thus, one limitation of our study was the lack of a large sample size to assess the phi alone and combined with DNA content in matched diagnostic biopsy tissue and serum samples to predict for unfavorable biopsy conversion. Additional research is needed to validate these results, preferably using multi-institutional cohorts and to expand the use of PSA isoforms and biopsy tissue DNA content combined with other molecular biomarkers to assess the prognosis of patients with very-low-risk PCa, which could eventually change the treatment of patients with PCa.
CONCLUSIONS
Prostate Health Index or proPSA/%fPSA ratio combined with biopsy tissue DNA content improved the accuracy to about 70% to predict unfavorable biopsy conversion on annual surveillance biopsy examinations (ie, a recommendation for curative intervention) among men with very-low-risk PCa enrolled in the AS program.
Acknowledgments
To Beckman Coulter for providing the [−2] proPSA serum assay under a materials transfer agreement.
This project was supported by a Johns Hopkins University Prostate Cancer SPORE grant (P50CA58236), the Early Detection Research Network National Cancer Institute/National Institutes of Health (grants CA086323-06 and CA115102-04), and the Prostate Cancer Foundation Patana Fund.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Draisma G, Boer R, Otto SJ, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for prostate cancer. J Natl Cancer Inst. 2003;95:868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 3.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Andriole GL, Crawford ED, Grubb RL, III, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter HB, Kettermann A, Warlick C, et al. Expectant management of prostate cancer with curative intent: an update of the Johns Hopkins experience. J Urol. 2007;178:2359–2364. doi: 10.1016/j.juro.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein JI, Walsh PC, Carmichael M, et al. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA. 1994;271:368–374. [PubMed] [Google Scholar]
- 7.Kawachi MH, Bahnson RR, Barry M, et al. NCCN clinical practice guidelines in oncology: prostate cancer early detection. J Natl Compr Cancer Netw. 2010;8:240–262. doi: 10.6004/jnccn.2010.0016. [DOI] [PubMed] [Google Scholar]
- 8.Isharwal S, Makarov DV, Carter HB, et al. DNA content in the diagnostic biopsy for benign-adjacent and cancer-tissue areas predicts the need for treatment in men with T1c prostate cancer undergoing surveillance in an expectant management programme. BJU Int. 2010;105:329–333. doi: 10.1111/j.1464-410X.2009.08791.x. [DOI] [PubMed] [Google Scholar]
- 9.Makarov DV, Isharwal S, Sokoll LJ, et al. Pro-prostate-specific antigen measurements in serum and tissue are associated with treatment necessity among men enrolled in expectant management for prostate cancer. Clin Cancer Res. 2009;15:7316–7321. doi: 10.1158/1078-0432.CCR-09-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peter J, Unverzagt C, Krogh TN, et al. Identification of precursor forms of free prostate-specific antigen in serum of prostate cancer patients by immunosorption and mass spectrometry. Cancer Res. 2001;61:957–962. [PubMed] [Google Scholar]
- 11.Weinzierl CF, Su SX, Pierson TB, et al. Measuring [−2]proPSA in serum: analytical performance of the Access p2PSA assay from Beckman Coulter. Presented at the Annual Meeting of the American Association for Clinical Chemistry. 2007 abstract C-38. [Google Scholar]
- 12.Harrell FE, Jr, Califf RM, Pryor DB, et al. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 13.Bill-Axelson A, Holmberg L, Filen F, et al. Radical prostatectomy versus watchful waiting in localized prostate cancer: the Scandinavian prostate cancer group 4 randomized trial. J Natl Cancer Inst. 2008;100:1144–1154. doi: 10.1093/jnci/djn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khatami A, Aus G, Damber JE, et al. PSA doubling time predicts the outcome after active surveillance in screening-detected prostate cancer: results from the European randomized study of screening for prostate cancer, Sweden section. Int J Cancer. 2007;120:170–174. doi: 10.1002/ijc.22161. [DOI] [PubMed] [Google Scholar]
- 15.Borre M, Stausbol-Gron B, Overgaard J. p53 accumulation associated with Bcl-2, the proliferation marker MIB-1 and survival in patients with prostate cancer subjected to watchful waiting. J Urol. 2000;164:716–721. doi: 10.1097/00005392-200009010-00023. [DOI] [PubMed] [Google Scholar]
- 16.Jhavar S, Bartlett J, Kovacs G, et al. Biopsy tissue microarray study of Ki-67 expression in untreated, localized prostate cancer managed by active surveillance. Prostate Cancer Prostatic Dis. 2009;12:143–147. doi: 10.1038/pcan.2008.47. [DOI] [PubMed] [Google Scholar]
- 17.Borre M, Nerstrom B, Overgaard J. Association between immunohistochemical expression of vascular endothelial growth factor (VEGF), VEGF-expressing neuroendocrine-differentiated tumor cells, and outcome in prostate cancer patients subjected to watchful waiting. Clin Cancer Res. 2000;6:1882–1890. [PubMed] [Google Scholar]
- 18.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 19.van As NJ, de Souza NM, Riches SF, et al. A study of diffusion-weighted magnetic resonance imaging in men with untreated localised prostate cancer on active surveillance. Eur Urol. 2009;56:981–988. doi: 10.1016/j.eururo.2008.11.051. [DOI] [PubMed] [Google Scholar]
- 20.Tosoian JJ, Loeb S, Kettermann A, et al. Accuracy of PCA3 measurement in predicting short-term biopsy progression in an active surveillance program. J Urol. 2010;183:534–538. doi: 10.1016/j.juro.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Ross AE, Loeb S, Landis P, et al. Prostate-specific antigen kinetics during follow-up are an unreliable trigger for intervention in a Prostate Cancer Surveillance Program. J Clin Oncol. 2010;28:2810–2816. doi: 10.1200/JCO.2009.25.7311. [DOI] [PubMed] [Google Scholar]
- 22.Sokoll LJ, Chan DW, Mikolajczyk SD, et al. Proenzyme PSA for the early detection of prostate cancer in the 2.5–4.0 ng/mL total PSA range: preliminary analysis. Urology. 2003;61:274–276. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 23.Sokoll L, Sanda MG, Feng Z, et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [−2]proPSA: improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol Biomarkers Prev. 2010;19:1193–1200. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le BV, Griffin CR, Loeb S, et al. [−2]Proenzyme prostate specific antigen is more accurate than total and free prostate specific antigen in differentiating prostate cancer from benign disease in a prospective prostate cancer screening study. J Urol. 2010;183:1355–1359. doi: 10.1016/j.juro.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deitch AD, Miller GJ, deVere WRW. Significance of abnormal diploid DNA histograms in localized prostate cancer and adjacent benign prostatic tissue. Cancer. 1993;72:1692–1700. doi: 10.1002/1097-0142(19930901)72:5<1692::aid-cncr2820720533>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Chandran UR, Dhir R, Ma C, et al. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isharwal S, Miller MC, Epstein JI, et al. Prognostic value of Her-2/neu and DNA index for progression, metastasis and prostate cancer-specific death in men with long-term follow-up after radical prostatectomy. Int J Cancer. 2008;123:2636–2643. doi: 10.1002/ijc.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isharwal S, Miller MC, Epstein JI, et al. DNA ploidy as surrogate for biopsy Gleason score for preoperative organ versus nonorgan-confined prostate cancer prediction. Urology. 2009;73:1092–1097. doi: 10.1016/j.urology.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tribukait B. Nuclear deoxyribonucleic acid determination in patients with prostate carcinomas: clinical research and application. Eur Urol. 1993;23(suppl 2):64–76. doi: 10.1159/000474709. [DOI] [PubMed] [Google Scholar]
- 30.Adolfsson J, Tribukait B, Levitt S. The 20-yr outcome in patients with well- or moderately differentiated clinically localized prostate cancer diagnosed in the pre-PSA era: the prognostic value of tumour ploidy and comorbidity. Eur Urol. 2007;52:1028–1035. doi: 10.1016/j.eururo.2007.04.002. [DOI] [PubMed] [Google Scholar]


