Abstract
A highly consistent trait of tumor stromal fibroblasts is the induction of the membrane-bound serine protease fibroblast activation protein-α (FAP), which is overexpressed on the surface of reactive stromal fibroblasts present within the stroma of the majority of human epithelial tumors. In contrast, FAP is not expressed by tumor epithelial cells or by fibroblasts or other cell types in normal tissues. The proteolytic activity of FAP, therefore, represents a potential pan-tumor target that can be exploited for the release of potent cytotoxins from inactive prodrugs consisting of an FAP peptide substrate coupled to a cytotoxin. To identify FAP peptide substrates, we used liquid chromatography tandem mass spectroscopy based sequencing to generate a complete map of the FAP cleavage sites within human collagen I derived gelatin. Positional analysis of the frequency of each amino acid at each position within the cleavage sites revealed FAP consensus sequences PPGP and (D/E)-(R/K)-G-(E/D)-(T/S)-G-P. These studies further demonstrated that ranking cleavage sites based on the magnitude of the LC/MS/MS extracted ion current predicted FAP substrates that were cleaved with highest efficiency. Fluorescence-quenched peptides were synthesized on the basis of the cleavage sites with the highest ion current rankings, and kinetic parameters for FAP hydrolysis were determined. The substrate DRGETGP, which corresponded to the consensus sequence, had the lowest Km of 21 μM. Overall the Km values were relatively similar for both high and low ranked substrates, whereas the kcat values differed by up to 100-fold. On the basis of these results, the FAP consensus sequences are currently being evaluated as FAP-selective peptide carriers for incorporation into FAP-activated prodrugs.
The growth of epithelial neoplasms requires the formation of a supporting tumor stroma to supply nutrients and growth factors for tumor cell survival and continued growth. This invasive growth is associated with characteristic changes in the supporting stroma that include induction of tumor blood vessel formation, the recruitment of reactive stromal myofibroblasts, lymphocytes, and macrophages, the release of peptide signaling molecules and proteases, and the production of an altered extracellular matrix (1–5). The tumor stroma compartment represents a major component of the mass of most carcinomas, with 20–50% commonly seen in breast, lung, and colorectal cancers and reaching >90% in carcinomas that have desmoplastic reactions such as breast and pancreatic cancers (5, 6).
Unlike malignant epithelial cells, activated tumor stromal fibroblasts are not transformed genetically and do not demonstrate the genetic and phenotypic heterogeneity seen in malignant cells. Reactive tumor stromal fibroblasts differ from fibroblasts of normal adult tissues in regard to morphology, gene expression profiles, and production of important biological mediators such as growth factors and proteases (1, 6, 7). For example, a highly consistent trait of tumor stromal fibroblasts is the induction of fibroblast activation protein-α (FAP).1 FAP was originally identified as an inducible antigen expressed on reactive stroma and given the name “fibroblast activation protein”. FAP was independently identified by a second group as a gelatinase expressed by aggressive melanoma cell lines and was given the name “seprase” for surface-expressed protease (8). Subsequent cloning of FAP and seprase revealed that they are the same cell-surface serine protease.
FAP was originally reported to be a cell-surface antigen recognized on human astrocytes and sarcoma cell lines in vitro by the F19 monoclonal antibody (MAb) (9). In one series using human tissues, FAP was detected in the stroma of over 90% of malignant breast, colorectal, skin, and pancreatic tumors (7, 10). In contrast, most normal adult tissues have demonstrated no detectable FAP protein expression (7). FAP expression has been most characterized in breast tissue. Garin-Chesa et al. used the F19 MAb to demonstrate strong (12/14) and moderate (2/14) expression of FAP in the stroma of human breast carcinomas but observed no expression by breast cancer epithelial cells and no expression in adjacent normal breast tissue. Additionally, little to no expression was observed in the stroma or epithelial cells of 10/10 samples of fibrocystic disease and 2/2 samples of fibroadenomas (7).
FAP is a member of the enzyme class known as post-prolyl peptidases that are uniquely capable of cleaving the Pro–X amino acid bond. These enzymes have been demonstrated to play a role in cancer biology and are capable of modifying bioactive peptides (11). This group of proteases includes the well-characterized dipeptidyl peptidase IV (DPPIV) as well as DPPII, DPP6, DPP7, DPP8, DPP9, prolyl carboxypeptidase, and prolyl endopeptidase (11). The substrate preferences for many of these prolyl peptidases are not entirely known, but like DPPIV, they all have dipeptidase functionality. FAP is highly homologous to DPPIV (11). Like DPPIV, FAP is a type II integral membrane protein that is able to cleave peptides with proline as the penultimate amino acid (12). However, FAP differs from DPPIV in that it also has gelatinase and, possibly, collagenase activity (8, 11). Using zymography, FAP was demonstrated to cleave gelatin and human collagen I but was unable to cleave human fibronectin, laminin, or collagen IV (12). These results suggest that FAP’s physiologic function may be primarily that of an endopeptidase that can degrade proteins rather than a dipeptidase like DPPIV. This additional gelatinase/collagenase activity may be unique to FAP among the family of prolyl proteases. Unlike DPPIV, FAP is also not widely expressed in most normal tissues (11).
Our laboratory has been engaged in the development of prodrugs that can be selectively activated by tissue-specific proteases (13–17). These prodrugs are produced by coupling a cytotoxic agent to a peptide carrier to produce an inactive compound that can only become activated upon release of the cytotoxin from the peptide by proteolysis. Our initial efforts have focused on the development of prodrugs activated by the prostate cancer serine proteases prostate-specific antigen (PSA) and human glandular kallikrein 2 (hK2) that are both members of the kallikrein family (13–16). In contrast to this tissue-specific approach, FAP represents a potential pan-tumor target. To develop FAP-activated prodrugs requires the identification of a peptide substrate that is selectively hydrolyzed by FAP. In the current study, we present results using liquid chromatography/tandem mass spectrometry (LC/MS/MS) to generate a complete map of FAP cleavage sites within recombinant forms of human collagen I derived gelatin. We have synthesized selected peptides on the basis of these cleavage maps and analyzed them for hydrolysis by FAP to identify peptides that could be used to target cytotoxins to FAP-expressing tumor tissue.
EXPERIMENTAL PROCEDURES
Materials
The Drosophila Expression System (DES) was from Invitrogen (Rockville, MD). Peptide Ala-Pro-AFC [AFC = 7-amino-4-(trifluoromethyl)coumarin] was from Bachem (Heidelberg, Germany). Gly-Pro-AMC, the MMP substrate sampler kit, and all other peptide synthesis reagents were from Anaspec (San Jose, CA). Novatag Dnp resin, N-(7-methoxycoumarin-4-acetyloxy)succinimide (Mca-Osu), 1-hydroxybenzotriazole (HOBt), and N-methyl-2-pyrrolidone (NMP) were from Novabiochem, San Diego, CA. Unless otherwise indicated all the other reagents were from Sigma-Aldrich (St. Louis, MO).
FAP Cloning and Expression
A PCR approach was used to amplify and attach a His6 tag to the amino terminus of the extracellular domain of FAP (Genbank accession number NM_004460). The primers used were (forward BglII) 5′-GGAAGATCTCATCATCACCATCACCATCGCCCT-TCAAG-3′ and (reverse XhoI) 5′-GGCCTCGAGTCATT-AGTCTGACAAAGAGAAACACTGC-3′. Template amplification was performed using Pfu polymerase (Promega, Madison) as per the suggested protocol. A PCR reaction began with an initial denaturation step (94 °C for 2 min) followed by three cycles of amplification (94 °C for 30 s, 40 °C for 1 min, 72 °C for 2 min), followed by 30 cycles of amplification (94 °C for 30 s, 58 °C for 1 min, 72 °C for 2 min), and ended with a final extension step (72 °C for 10 min). A 2 kb PCR fragment was purified by gel electrophoresis, digested with BglII/XhoI, and cloned into pMT/BiP/V5–HisA (Invitrogen, California) previously digested with the same set of enzymes. The final construct was designated as pMT–His–FAP.
Transfection of Insect Cells and Stable Cell Line Generation
Schneider’s S2 cells (Invitrogen) were maintained in DES medium (Gibco, Rockville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at room temperature. Before transfection, the cells were seeded in a 35 mm dish and grown until they reached a density of (2–4) × 106 cells/mL. The cells were cotransfected with 19 μg of pMT–His–FAP and 1 μg of a pCoHYGRO selection vector using a kit for calcium phosphate-mediated transfection (Invitrogen). The calcium phosphate solution was removed 16 h post-transfection, and fresh DES medium supplemented with 10% FBS was added (a complete medium). The cells were grown for an additional 2 days, and then the medium was replaced with the complete medium containing 400 μg/mL hygromycin B (Invitrogen). The selection medium was changed every 3–4 days. Extensive cell death of nontransfected cells was evident after about 1 week, and cells resistant to hygromycin B started to grow out 2–3 weeks post-transfection.
His-Tagged FAP Large-Scale Expression and Purification
The hygromycin-resistant cells were seeded in 10 T-150’s at a density of 1 million cells/mL. When the cells reached a density of 2–3 million cells/mL, 500 μM CuSO4 was added to induce FAP expression. The cells were grown until they reached a density of 10–15 million cells/mL (8–9 days). A 2 mL portion of 200 mm L-glutamine was added to the cell suspension on days 2 and 6. Conditioned medium containing secreted FAP was collected after 12–14 days. The medium was concentrated, and excess CuSO4 was removed by three rounds of ultrafiltration using an Amicon 8480 membrane (Millipore) with a 30 000 kDa cutoff. After each round of ultrafiltration, the volume was made up using sterile water. Final purification was obtained by incubating the concentrate with Ni–NTA resin (Qiagen, California) in manufacturer-recommended salt and imidazole concentrations. FAP was eluted from the resin using 250 mM imidazole. The final 30 mL of eluate was diluted with water to 300 mL, and imidazole was removed by two rounds of ultrafiltration. The purity was checked by SDS–PAGE and Coomassie staining. Western blots were probed with anti-His tag [penta-His–horse radish peroxidase (HRP) conjugate from Qiagen). Overall, a yield of 1–2 mg was obtained from a 700 mL culture. Final purified aliquots were stored in reaction buffer at −20 °C.
FAP Enzyme Activity
FAP dipeptidyl peptidase activity was determined by digesting 500 μM Ala-Pro-AFC (Calbiochem) with rhFAP as described by Park et al. (12). Assays were performed at 23 °C in 100 mM Tris, 100 mM NaCl at pH 7.8 in 10% DMSO and 0.3% Brij-35. The fluorescence output was monitored every 30 s using a DTX 880 multimode detector (Beckman Dickinson). Standard curves of AFC (Calbiochem) fluorescence vs concentration were run with each assay to convert relative fluorescence units to moles of product generated. Excitation/emission wavelengths of 370 and 535 nm, respectively, were used to monitor liberation of the AFC fluorophore. The rate of hydrolysis (mol/min) was ascertained by determining the slope during the first minute of the reaction. This rate was then used to calculate the enzyme units per unit volume with 1 unit of enzyme activity being defined as the cleavage of 60 μmol of substrate/min.
FAP Gelatinase Assay
Quenched gelatin and collagen conjugates were used to detect and confirm FAP’s gelatinase and collagenase activity. DQ gelatin from pig skin and DQ collagen type IV from human placenta fluorescence-quenched conjugates (Invitrogen, Rockville, MD) were digested with FAP, and digestion was monitored on a fluorescence plate reader. Protein substrates were dissolved in reaction buffer (100 mM NaCl, 100 mM Tris, pH 7.8) to a final concentration of 100 μg/mL. Trypsin digestion of each protein was used as a positive control. As a negative control the His-tagged extracellular domain of prostate-specific membrane antigen (PSMA), which was similarly purified from S2 cells under the same conditions as FAP, was also incubated with the quenched proteins. Fluorescence-quenched DQ bovine serum albumin (BSA) was used as a negative control for FAP protease activity.
Digestion of Recombinant Gelatin with FAP for Cleavage Mapping
Recombinant human gelatins of 100 and 8.5 kDa (Fibrogen, San Francisco, CA) were dissolved in reaction buffer, and 1 μg of FAP was added per 100 μg of protein substrate. Digestion was done for 4–6 h at 37 °C. As a positive control, trypsin digestion was performed. As a negative control, protein solutions were incubated with either BSA/buffer or buffer alone. Peptide fragments of size <30 kDa were purified using a 30 kDa Microcon spin filter (Millipore, Billerica, MA). The fragments were further purified with C18 spin tubes (Agilent, Palo Alto, CA) as per the suggested protocol with the substitution of 0.5% acetonitrile in place of 5% for binding and washing of the C18 columns. Samples were prepared for matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) analysis on an Applied Biosystems Voyager-DE STR (mass accuracy ≥100 ppm) by 100:1 dilution with 2,5-dihydroxybenzoic acid (DHB) as the matrix. Analysis was performed in the linear mode over a mass acquisition range of 500–5000 Da with 50 laser shots performed per spectrum. Instrument calibration was performed routinely by the Mass Spectrometry Core Facility and was not performed prior to individual analyses.
Nanoflow HPLC and Mass Spectrometry
Peptides obtained from the FAP/gelatin digests were dried using a SpeedVac (Eppendorf), resuspended in LC/MS loading buffer (3% ACN, 0.1% formic acid), and analyzed using nanoflow LC/MS/MS on an Agilent 1100 series nano-LC system (Agilent) coupled to an LCQ DUO ion trap mass spectrometer (ThermoFinnigan). The LCQ was calibrated (m/z 50–2000) using the automatic calibration procedure outlined by the manufacturer. A fused silica capillary was loaded with a solution containing the Met-Arg-Phe-Ala (MRFA) peptide, caffeine, and Ultramark 1621 in 50% acetonitrile/0.1% formic acid. The capillary was connected to the nanoflow HPLC instrument and sprayed into the LCQ using the same flow rate and spray voltage used in all subsequent experiments. The calibration was tested using a standard 100 fmol tryptic digest of BSA and demonstrated a mass accuracy of ≤0.5 Da.
Peptides were preconcentrated on a 5 mm Zorbax C18 trap column (Agilent) and then eluted onto a 100 × 0.075 mm custom-packed Biobasic C18 (ThermoElectron) reversed-phase capillary column connected to a laser-pulled electrospray ionization emitter tip (New Objective) at a flow rate of 300 nL/min. Peptides were eluted from the nanospray source of the LCQ (Proxeon, Denmark) using the following gradient: 0% B at 0 min, 5% B at 8 min, 45% B at 50 min, 90% B at 55 min, 90% B at 60 min (solvent B is 0.1% formic acid in acetonitrile) at a spray voltage of 2.5 kV. The LCQ was operated in data-dependent mode using the Xcalibur software (ThermoFinnigan) in which every MS scan (400–1800 m/z) was followed by MS/MS scans (400–1800 m/z) on the three most intense ions using an isolation window of ±1.5 Da. Ions selected for MS/MS fragmentation were dynamically excluded for 30 s.
MS/MS data were searched against a collagen FASTA database using the SEQUEST search algorithm built into the Bioworks Browser (ThermoFinnigan), allowing for the variable modification of methionine oxidation. The peptides were initially filtered in a charge-dependent manner using an XCorr filter of 1.5, 2, and 2.5 for singly, doubly, and triply charged peptides. All MS/MS spectra used to identify peptides were manually inspected for validation of the y and b ion series. To quantify the relative abundance of each identified peptide, we compared the ion current for each of the observed peptide parent ions from the MS spectra. The contribution of each parent ion to the total ion current was extracted and integrated over the peptide elution peak.
Synthesis of Substrates Based on Determined Cleavage Sites
Quenched peptide substrates were prepared by using the 7-methoxycoumarin-4-acetic acid (Mca)/Dnp fluorophore/quencher pair. Synthesis of peptides was done using standard Fmoc solid-phase coupling on NovaTag Dnp resin with a substitution level of 0.4 mmol/g (Novabiochem, San Diego, CA). N-terminal capping was done twice overnight with Mca-Osu and HOBt in NMP. The peptides were cleaved with 95% TFA, 2.5% TIS, and 2.5% water. The purity and mass of each quenched peptide was confirmed by reversed-phase HPLC and MALDI-TOF analysis.
Protease Assays
The substrates were dissolved in DMSO at a stock concentration of 10 mM. Working concentrations of each substrate were made by making 2-fold dilutions ranging from 500 to 7.9 μM. rhFAP (10E–4 units, ~210–230 nM depending on the specific activity of the enzyme) was added to each assay where enzyme was included. The fluorescence of the cleavage product resulting from protease activity was monitored every 30 s using a DTX 880 multimode detector (Beckman Dickinson). Standard curves of Mca (Novabiochem) fluorescence vs concentration were run with each assay to convert relative fluorescence units to moles of product generated. Excitation and emission wavelengths of 340 and 435 nm, respectively, were used to monitor liberation of the Mca fluorophore peptide cleavage fragment. Assays were performed at 23 °C in 100 mM Tris, 100 mM NaCl at pH 7.8 in 10% DMSO and 0.3% Brij-35. Kinetic constants (Km and kcat) were calculated on the basis of the rate of hydrolysis (v) during the first minute of the reaction. Kinetic parameters were calculated from Michaelis–Menten plots (v vs [S]) with nonlinear regression analysis using SigmaPlot software.
RESULTS
Purification of Enzymatically Active FAP
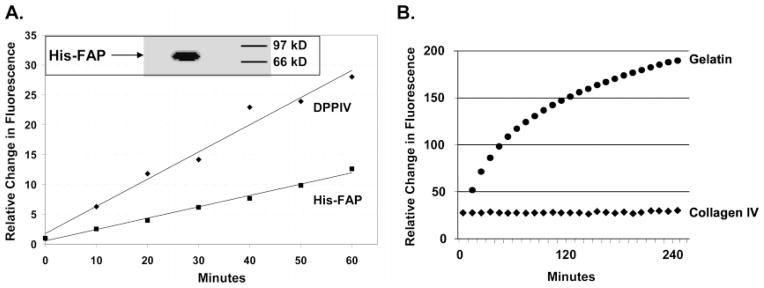
The characterization of protease substrate specificity requires that the protease be of maximum purity and correctly folded to maintain enzymatic activity. Previously it had been shown that full-length FAP, cloned and expressed in Drosophila S2 cells, yielded highly pure protein that was enzymatically similar to the human form (18). Therefore, the extracellular domain of FAP was cloned with a His6 tag at its N-terminus to generate a stable FAP-producing Drosophila S2 cell line. On induction with CuSO4, FAP was secreted into the medium, which was then concentrated by ultrafiltration and purified using Ni–NTA beads. Purified FAP was demonstrated to be enzymatically active via its ability to cleave the dipeptide substrate Ala-Pro-AFC (18) (Figure 1A). Western blot analysis with an anti-His tag MAb documented the correct protein size of ~80 kDa (Figure 1A, inset).
Figure 1.
Characterization of recombinant His-tagged FAP. (A) Hydrolysis of the dipeptide substrate Ala-Pro-AFC (500 μM) by FAP (5 μg/mL) compared to DPPIV. The inset shows Western blot analysis of purified His–FAP following denaturing SDS–PAGE. (B) Digestion of indicated FITC-labeled proteins (100 μg/mL) by FAP (5 μg/mL).
Recombinant FAP Retains Gelatinase Activity
Quenched forms of gelatin and collagen were used to confirm the gelatinase and collagenase activity of recombinant FAP. Quenching is achieved by heavily labeling these proteins with the fluorophore FITC such that the fluorescence signal from the intact protein is minimal due to self-quenching by the fluorophore. Protein digestion releases FITC-labeled fragments that result in a measurable increase in overall fluorescence signal from the reaction mixture. Previously, it had been demonstrated using gel zymography that FAP can cleave gelatin and collagen I but could not cleave collagen IV (12). It remains unclear whether FAP can digest collagen I directly as recent studies suggest that the observed digestion may be due to an artifactual conversion of collagen I to gelatin during preparation (12, 19, 20). To confirm that our recombinant FAP maintained the ability to cleave gelatin, we used the FITC-quenched protein DQ gelatin from pig skin, with DQ collagen IV from human placenta serving as a negative control. In this assay, gelatin was readily hydrolyzed by FAP while collagen IV was not hydrolyzed (Figure 1B). MALDI-TOF analysis of the digested fragments was performed to confirm FAP hydrolysis. As a negative control, we demonstrated no digestion of any of the proteins using His-tagged human carboxypeptidase PSMA purified from Drosophila S2 cells under the same conditions (data not shown). These results confirm that gelatin hydrolysis was due to FAP and not due to the presence of some other protease contaminating our purification system.
MALDI for FAP Digest of Denatured Human Collagen I
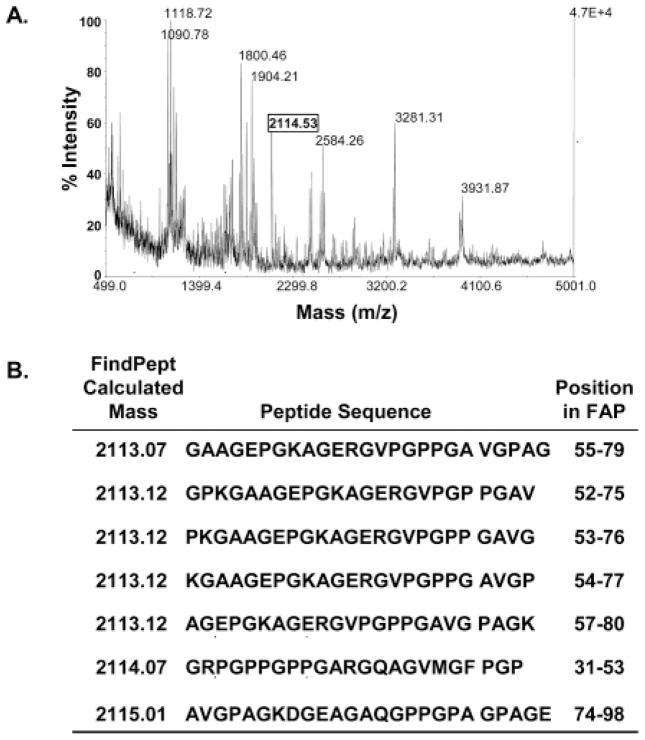
To elucidate the substrate specificity of FAP, we examined FAP digests of unlabeled denatured human collagen I using MALDI-TOF mass spectrometry (Figure 2A). Digestion reactions were performed at a substrate to protease mass ratio of 200:1 using recombinant FAP or modified trypsin as a control. Two negative controls of collagen alone and FAP alone were also included to identify any peptides due to autolysis/degradation of these proteins. SDS–PAGE analysis of FAP-digested denatured collagen I (not shown) produced a smear of continuous size fragments, suggesting the presence of many cleavage sites. To simplify cleavage mapping by MALDI-TOF, small fragments (<5 kDa) were isolated by ultrafiltration and further purified using reversed-phase chromatography, Figure 2A. MALDI-TOF was subsequently performed using serial dilutions of the isolated peptides.
Figure 2.
(A) MALDI-TOF mass spectra demonstrating FAP digestion of human collagen I. (B) Example of cleavage site sequence(s) determination using the FindPept tool for a selected mass of 2114.5 from MALDI-TOF mass spectra. Multiple potential cleavage site sequences within ±1 mass unit are identified by this analysis.
The masses of singly charged ions [M + H]+ obtained from MALDI spectra were entered into the FindPept search tool at the ExPASy proteomics server (http://www.expasy-.org/tools/findpept.html) and used to perform a peptide mass fingerprint (PMF) search against the known collagen sequence. MALDI spectra suggest that denatured human collagen I is cleaved by FAP at numerous specific sites (Figure 2A); however, we were unable to unambiguously identify the cleavage sequences using the FindPept tool (Figure 2B). In most cases, multiple peptide sequences were matched for the same mass, and in some instances, more than 30 sequences were obtained for one particular mass. This result was most likely due to the fact that human collagen I is a heterotrimeric polymer made up of repeating sequences containing the (GXY)n motif (where X = Pro and Y = hydroxy-Pro). Human collagen I is also known to be glycosylated and cross-linked randomly throughout its sequence (21). These post-translational modifications in human collagen have not been well characterized and, therefore, make it more difficult to determine the exact cleavage sites using MALDI-TOF coupled with other proteomics tools. These results, while demonstrating that FAP cleaved denatured human collagen I, demonstrated the difficulties in obtaining correct cleavage sequences by mass spectrometry due to the polymeric nature and poorly defined qualities of human collagen I.
LC and Tandem MS/MS Analysis of FAP Digestion of Recombinant Gelatin
As collagen I derived gelatin is currently one of the only known protein substrates for FAP, we needed to develop an alternative method to identify FAP-selective cleavage sites within these proteins. To solve the problem of post-translational modification, we identified a source of recombinant human gelatin and collagen from FibroGen (South San Francisco, CA) that was prepared by cloning the human collagen I sequence in a strain of Pichia pastoris which lacks the enzyme prolyl hydroxylase (22). This human collagen I based gelatin is well characterized and has no post-translational modifications. This gelatin is used to produce drug capsules and vaccine adjuvants. Therefore, this recombinant gelatin was used in subsequent FAP cleavage mapping studies.
We next used MALDI-TOF to analyze FAP digestion of the 100 kDa recombinant form of human gelatin. SDS–PAGE separation and MALDI-TOF spectra showed that FAP readily digests the recombinant gelatin (data not shown). The masses of fragments <3 kDa were purified for MALDI-TOF spectra and again analyzed using the FindPept tool. However, once again multiple peptide sequences were obtained for each cleavage fragment.
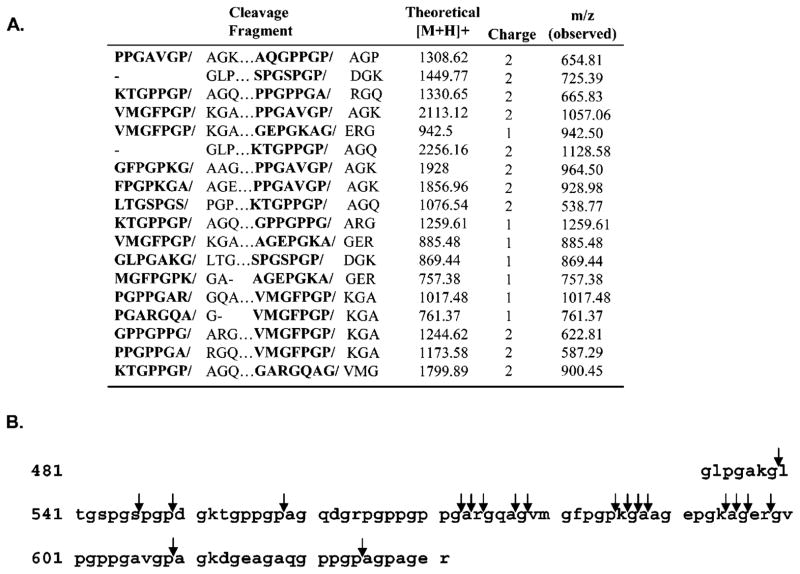
Therefore, to resolve each particular mass fragment, an LC/MS/MS method was developed. To initially work out the methodological issues, peptide fragments from the FAP digest of the 8.5 kDa recombinant gelatin were time-resolved by nano-reversed-phase LC and then sequenced using an ion trap mass spectrometer operating in MS/MS mode (Figure 3A). MS/MS spectra were searched against the 8.5 kDa gelatin sequences using the SEQUEST algorithm with no cleavage specificity. In this analysis most of the identified cleavage sites occurred after proline. However, FAP cleavage sites in these gelatins were not restricted to proline alone. FAP was also found to cleave after Ala, Arg, Gly, Lys, and Ser. FAP was also able to cleave after multiple adjacent amino acids within the 8.5 kDa gelatin fragment (Figure 3B).
Figure 3.
(A) FAP cleavage sites within the 8.5 kDa fragment of recombinant human collagen I derived gelatin. Cleavage sites are bracketed by “/”, and [M + H]+ denotes the mass of the cleavage fragment. Amino acids on the N-terminal side of the cleavage site (i.e., P7–P1) are in bold font. (B) Amino acid sequence of the 8.5 kDa fragment with FAP cleavage sites indicated by arrows.
FAP Cleavage Map for Full-Length 100 kDa Gelatin
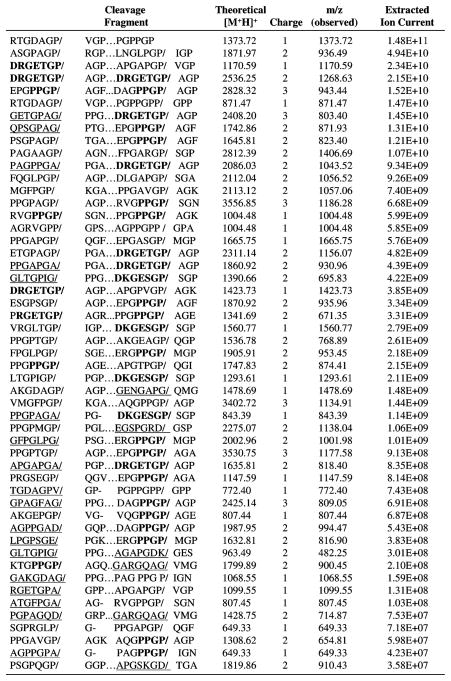
On the basis of the results with the 8.5 kDa gelatin fragment, we proceeded to use this LC/MS method to identify all of the FAP cleavage sites within the full-length 100 kDa recombinant gelatin. In an effort to identify the most preferentially cleaved sites, we also quantified the relative abundance of each of the identified peptides by integrating the ion current generated by each peptide throughout the chromatogram (Figure 4). After identification of a peptide from an MS/MS spectrum, we extracted the ion current for the parent mass of the ion from the total MS ion chromatogram using a 1.5 Da tolerance window and then integrated under the peak. In the case of multiple peaks, we chose the peak nearest to the retention time of the MS/MS spectrum matched to the peptide sequence of interest.
Figure 4.
FAP cleavage sites within the 100 kDa recombinant human collagen I derived gelatin. Cleavage sites are bracketed by “/”. The mass of the cleavage fragment, [M + H]+, and extracted ion current are included for each cleavage site. Underlined sequences indicate cleavage sites which do not contain Pro (P) in the P1 position. The consensus sequences PPGP and D-(R/K)-G-E-(T/S)-G-P are indicated by bold text.
This analysis revealed a total of 51 FAP cleavage fragments within the 100 kDa recombinant gelatin (Figure 4). The corresponding fragment was mapped within the recombinant gelatin to determine the sequence of the amino acids upstream of the FAP cleavage site toward the N-terminus. Since each fragment was produced by two cleavage events, 101 peptide sequences corresponding to the amino acids from P7 to P′1 could be mapped. Cleavage fragments were then ranked on the basis of the extracted ion current generated by each peptide (Figure 4). This ranking again revealed that FAP preferred to cleave after the G-P dipeptide. FAP was also able to cleave after other amino acids (i.e., Ala, Asp, Gly, Glu, Lys, and Val), but the majority of these cleavage fragments had very low extracted ion currents and were ranked in the lower half of the list (Figure 4). Often these non-proline cleavage sites were adjacent to a proline cleavage site. This result was also observed with the 8.5 kDa digest and suggested that the gelatin protein may misalign within the FAP catalytic site, resulting in low-level cleavage after non-proline amino acids.
On the basis of these cleavage sites, the frequency of individual amino acids within the P7–P′1 positions of each cleavage site was next determined. This analysis was somewhat limited by the repetitive nature of the collagen I sequence, and it was, therefore, not necessarily unexpected that the most frequent amino acids at each position were either proline or glycine, producing a consensus peptide with the sequence PPGPPGP. This exact sequence is found in 3 of the sequences within the map, while the peptide PPGP is found in 21 of the 101 sequences.
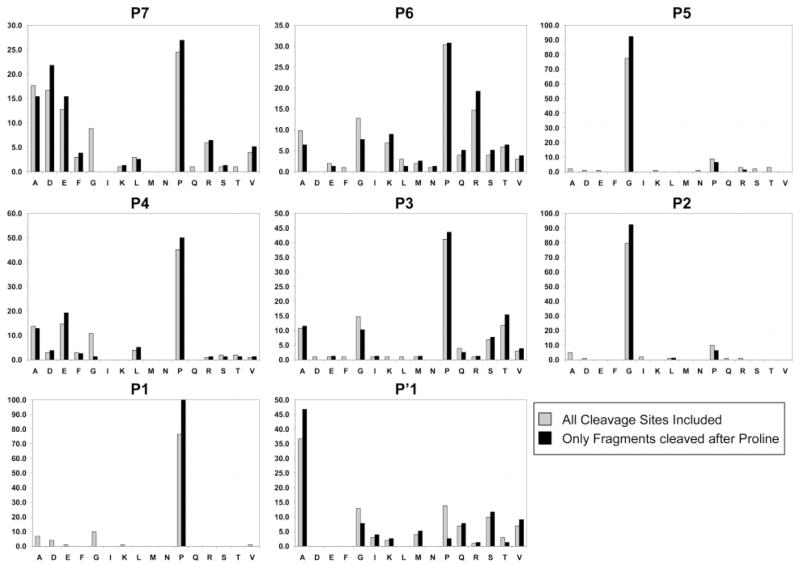
The frequency analysis, however, did reveal some additional trends that could bear on the identification of consensus sequences to be used in the development of FAP-activated peptide prodrugs (Figure 5). After proline in P7, for instance, the acidic amino acids Asp (23%) and Glu (15%) were found in ~38% of the sequences. In P6, 30% of the sequences contained one of the basic amino acids Lys (10%) and Arg (20%). In P4, Glu was observed in 20% of the sequences and Asp in 4%. In P3, the small polar amino acids Ser and Thr, which both contain a -OH functional group, were found in 24% of the sequences and Ala and Gly with a small or no side chain were found in 26%. Finally, in the P′1 position, Ala was observed in almost half of the sequences (Figure 5). On the basis of this analysis, a second consensus sequence emerged [i.e., (D/E)-(R/K)-G-(E/D)-(A/G/T/S)-G-P-A or (acidic AA)-(basic AA)-G-(acidic AA)-(small/polar OH AA)-G-P-A)]. This consensus sequence is found in 15 of the 101 peptide sequences within the cleavage map (Figure 4).
Figure 5.
Frequency (%) of each amino acid in positions P7–P′1 within the FAP cleavage sites within the 100 kDa recombinant human collagen I derived gelatin. Gray bars indicate the frequency for all 101 cleavage sites. Black bars indicate the frequency in only those cleavage sites containing Pro (P) at the P1 position. The amino acids C, H, W, and Y are not present in human collagen I and, therefore, are not included.
To determine whether these consensus sequence motifs represented FAP-preferred cleavage sites or instead were merely observed due to a relative abundance of these particular sequences within the overall gelatin amino acid sequence, we evaluated the frequency of occurrence of these sites within the gelatin protein (Table 1). Overall there were 66 FAP cleavage sites within the 100 kDa recombinant gelatin. A total of 116 G-P sites are found within the protein, and 40 (34%) of these were cleaved by FAP, Table 1. To emphasize the importance of the Ala in the P′1 position, 28 sites with the tripeptide G-P-A exist in the gelatin sequence (24% of total G-P-containing sites). However, when present, 71% of these sites were cleaved by FAP. In addition, on the basis of the cleavage site analysis, we evaluated the frequency of the consensus sequence (D/E)-(K/R)-(D/E)-G-(A/G/S/T)-G-P, the expanded consensus sequence (A/D/E/P)-(K/R)-(D/E/P)-G-(A/G/S/T/P)-G-P, in which Pro is also included in the analysis and the P6 position only included the basic amino acids K/R, and the expanded consensus sequence (D/E)-(K/R/P)-(D/E/P)-G-(A/G/S/T/P)-G-P, in which the P7 position included only the acidic amino acids D/E and not Pro or Ala. The (D/E)-(K/R)-(D/E)-G-(A/G/S/T)-G-P sequence is cleaved by FAP three out of the four times it appears in the gelatin sequence, Table 1. Overall unique consensus sequences were found in only 13% of the total G-P cleavage sites in gelatin. However, when present, 60% of these sequences were cleaved by FAP, Table 1. The presence of this sequence, however, was not sufficient to produce FAP cleavage in the absence of proline in the P1 amino acid, thus further confirming the importance of proline in this position. Seven such sequences containing (A/D/E/P)-(K/R/P)-(D/E/P)-G-(A/G/S/T/P)-G-(non-proline) were identified in the gelatin sequence, and none of these were cleaved by FAP (Table 1). These results demonstrate that the frequent appearance of the (D/E)-(K/R)-(D/E)-G-(A/G/S/T)-G-P sequence in the cleavage fragments is not due to the frequent occurrence of this motif in the overall gelatin sequence, but is rather due to FAP preference for cleavage after this consensus site.
Table 1.
Frequency Analysis of Consensus Sequences within the 100 kDa Gelatin Amino Acid Sequence
| amino acids
|
total no. of G-P cleavage sitesa (%) | no. of FAP-cleaved sitesb (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | ||
| G | P | 116 (100) | 40 (34) | ||||||
| G | P | A | 28 (24) | 20 (71) | |||||
| D/E | K/R | G | D/E | A/G/S/T | G | P | 4 (3) | 3 (75) | |
| A/D/E/P | K/R | G | D/E/P | A/G/S/T/P | G | P | 14 (12) | 8 (57) | |
| D/E | K/R/P | G | D/E/P | A/G/S/T/P | G | P | 8 (7) | 6 (75) | |
| combinedc | 15 (13) | 9 (60) | |||||||
| D/E/P | K/R/P | G | D/E/P | A/G/S/T/P | G | non-Pro | 7 (6) | 0 (0) | |
Occurrence of the G-P dipeptide sequence in the 100 kDa gelatin sequence.
Number of times FAP cleaves after G-P sites.
Total unique sites from the consensus sequence indicated in rows 3–5.
FAP Hydrolysis of Fluorescence-Quenched Peptides Based on the Gelatin Cleavage Map
While previous studies had already determined that FAP preferred to cleave after the dipeptide GP, the normalized ion current analysis also suggested a strong preference for cleavage after GP as well as a strong preference for cleavage of the peptide with GP in P2–P1 and Ala in P′1. To determine whether this type of cleavage site ranking based on the abundance of each extracted ion in the MS spectra would help predict which sequences represented good FAP substrates, we proceeded to synthesize a series of fluorescently quenched peptide substrates based on the cleavage sequences. For this analysis, we generated peptide substrate from sequences with high (i.e., ASGPAGPA, DRGETGPA), intermediate (i.e., EKGES-GP, VGPAGK), and low (i.e., GARGQAG, APGSKGDA) extracted ion currents, Figure 4. Michaelis–Menten kinetic parameters (i.e., Km, Vmax, kcat) of FAP hydrolysis of these fluorescence-quenched peptides were determined. kcat/Km ratios were calculated to rank these peptides as FAP substrates (Table 2).
Table 2.
Enzyme Kinetics of FAP Substrates Generated from Cleavage Sites within 100 kDa Gelatin
| no. | substrate sequence | Km (μM) | Vm (mol/s) | kcat (s−1) | kcat/Km (M−1 s−1) | normalized ion current rank |
|---|---|---|---|---|---|---|
| 1 | Mca-ASGPAGPA-Dnp | 36.1 ± 2.7 | 2.26E–12 ± 8.14E–14 | 0.196 ± 0.007 | 5441 ± 478 | 2 |
| 2 | Mca-EPGPPGPA-Dnp | 26.9 ± 0.7 | 1.32E–12 ± 9.53E–14 | 0.115 ± 0.006 | 4270 ± 90 | 5 |
| 3 | Mca-DRGETGPA-Dnp | 21.0 ± 2.2 | 8.50E–13 ± 1.89E–14 | 0.081 ± 0.002 | 3851 ± 366 | 3, 4, 7 |
| 4 | Mca-VGPAGK-Dnp | 52.0 ± 4.4 | 1.09E–12 ± 4.41E–14 | 0.095 ± 0.004 | 1822 ± 100 | 13, 21 |
| 5 | Mca-DKGESGPA-Dnp | 46.0 ± 3.5 | 7.69E–13 ± 2.11E–14 | 0.067 ± 0.001 | 1453 ± 99 | 20, 24 |
| 6 | Mca-APGSKGDA-Dnp | 51.0 ± 11.8 | 1.68E–13 ± 1.95E–13 | 0.015 ± 0.017 | 286 ± 391 | 51 |
The Mca-ASGPAGPA-Dnp substrate (1) had the highest kcat/Km ratio (Table 2). This peptide sequence was also based on a cleavage fragment with the second highest extracted ion current. Substrate 2, based on the fragment with the fifth ranked extracted ion current, had the second highest kcat/Km ratio. In addition, the Mca-DRGETGPA-Dnp substrate (3) had the third highest ratio. This substrate is based on a consensus sequence that is the second most common cleavage site in the map and which is found in cleavage fragments with the third, fourth, and seventh highest extracted ion currents. In contrast, the Mca-APGSKGDA-Dnp substrate (6), which was least efficiently cleaved by FAP, was based on the cleavage fragment with the lowest overall extracted ion current (Figure 4). Formal kinetic studies were not performed on a second substrate, Mca-GARGQAG-Dnp, based on the fragment with the 47th ranked extracted ion current, but this substrate demonstrated a low hydrolysis rate compared to rates from an equimolar concentration (i.e., 200 μM) of substrates 1–5. Two other substrates with intermediate Kcat/Km ratios [i.e., Mca-VGPAGK-Dnp (4) and Mca-DKGESGPA-Dnp (5)] were found in cleavage fragments with intermediate extracted ion currents (Table 2).
In a final set of studies, we synthesized additional fluorescence-quenched peptides to explore the importance of amino acids in the P′ positions on the carboxy terminal side of the cleavage site (Table 3). A comparison of the Kcat/Km ratios of substrate 7 with no P′1 amino acid to 8 and substrate 12 to 13 demonstrated that FAP prefers an amino acid in the P′1 position. Both substrates lacking the P′1 alanine were poorly hydrolyzed by FAP. The addition of a P′2 Gly as in 10 or a P′3 Gly as in substrate 11 did not substantially improve the kinetic ratio. Other comparisons demonstrated a slight preference for Asp in the P7 position as in substrate 8 compared to Glu as in substrate 9. In the P6 position, substitution of Ser as in substrate 13 for Arg as in substrate 8 resulted in some decrease in the kinetic ratio. Finally, in the P′1 position, while Ser was only observed in 12% of the sequences compared to 47% for Ala, the substitution of Ser in the P′1 position as in substrate 14 resulted in a substantial increase in the kcat/Km ratio compared to that of the comparable substrate 10 containing Ala in this position (Table 3). The substitution of Ser in the P′1 position generated a peptide substrate with a kcat that was 2–5 fold higher than that of other active substrates.
Table 3.
Enzyme Kinetics of FAP Substrates Derived from the Consensus Sequence DRGETGPA
| no. | substrate sequence | Km (μM) | Vm (mol/s) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| 7 | Mca-DRGETGP-Dnp | 23.8 ± 6.8 | 3.49E–14 ± 8.02E–15 | 0.003 ± 0.001 | 127 ± 31.9 |
| 8 | Mca-DRGETGPA-Dnp | 21.0 ± 2.2 | 8.50E–13 ± 1.89E–14 | 0.081 ± 0.002 | 3851 ± 366 |
| 9 | Mca-ERGETGPA-Dnp | 56.7 ± 0.7 | 1.62E–12 ± 5.20E–14 | 0.141 ± 0.004 | 2483 ± 58 |
| 10 | Mca-ERGETGPAG-Dnp | 43.0 ± 3.8 | 1.39E–12 ± 8.41E–14 | 0.121 ± 0.007 | 2810 ± 149 |
| 11 | Mca-ERGETGPAGG -Dnp | 37.1 ± 1.1 | 9.05E–13 ± 2.69E–12 | 0.079 ± 0.004 | 2120 ± 35 |
| 12 | Mca-DSGETGP-Dnp | 39.6 ± 29.7 | 3.88E–14 ± 8.11E–15 | 0.003 ± 0.001 | 85 ± 60 |
| 13 | Mca-DSGETGPA-Dnp | 20.2 ± 0.5 | 6.74E–13 ± 4.65E–14 | 0.059 ± 0.004 | 2900 ± 221 |
| 14 | Mca-ERGETGPSG-Dnp | 51.3 ± 1.9 | 3.08E–12 ± 1.77E–13 | 0.268 ± 0.015 | 5218 ± 316 |
DISCUSSION
The overall goal of this study was to develop a method to identify substrates on the basis of FAP’s collagenase or gelatinase activity. This knowledge of substrate specificity can be used to elucidate FAP’s biological role as well as for development of targeted therapeutic prodrugs. Substrate specificity can be defined by using either by high-throughput methods such as the positional scanning synthetic combinatorial library (15) and one-bead one-peptide library (23) or phage display (24, 25). However, these methods have the disadvantage of an artificial scaffold which can alter the physiological substrate specificity of a protease. Here we have described an approach to take a known protein substrate for a protease and map its cleavage site using proteomics. In the case of FAP, the problem was more complicated because of the polymeric nature of the collagen I protein coupled with multiple types of post-translational modifications.
A large number of collagenases and gelatinases have been previously reported in the literature. However, for most of them substrate characterization was done using combinatorial libraries or synthetic model substrates (24, 26–29). To our knowledge there is only one prior reported study in which the collagen protein was digested with a new type of collagenase and cleavage sites determined by Edman sequencing of specific bands isolated on SDS–PAGE (30). However, in this previous study only 10–12 cleavage fragments were obtained. Thus, it was possible to easily separate them by SDS–PAGE (30). In contrast, in our study the FAP digest of collagen or gelatin produced more than 100 fragments, which produced a smear on SDS–PAGE, rather than discrete bands. This result suggested that FAP cleaved collagen at many sites, thus making traditional Edman sequencing impractical. On this basis, we proceeded to develop the LC/MS/MS-based approach to map these FAP cleavage sites.
Denatured human collagen I was digested with FAP, and small size fragments (<3 kDa) were purified for MALDI-TOF analysis. Previously, MADLI-TOF has been used to identify proteins by analysis of tryptic digests (31). Mass spectra revealed that FAP was cleaving collagen I at specific sites. However, sites could not be identified by MALDI-TOF because each mass peak frequently matched more than 10 individual sequences within the collagen I protein. An attempt was also made to perform an LC/tandem MS/MS analysis on these fragments. However, the mass spectroscopic data could not be solved because of collagen’s several post-translational modifications (PTMs) that include proline hydroxylation, glycosylation, and cross-linking. Many of these PTMs have not been characterized within collagen I, and therefore, we determined that nonrecombinant, purified human collagen I could not be easily used for such an analysis.
To solve these problems, we identified a source of recombinant gelatin derived from human collagen I that had no PTMs and which could be used for FAP cleavage mapping (22). After validating the LC/MS based method using the 8.5 kDa gelatin fragment, we proceeded to generate the map of all FAP cleavage sites within full-length 100 kDa human collagen I derived gelatin. Sequence analysis of the 51 peptide fragments from this digest confirmed earlier reported studies documenting that FAP has a preference for the dipeptide GP in the P2–P1 position. However, these earlier studies, based on a positional scanning method, reported a strict requirement for G-P for FAP hydrolysis. In contrast to these results, we documented that, while FAP prefers cleavage after the G-P dipeptide, FAP can also cleave peptides containing other amino acids in the P1 position (i.e., Ala, Arg, Asp, Gly, Glu, Lys, Ser, and Val).
One disadvantage of the positional scanning synthetic approach is that it often does not yield much specific sequence information beyond the P2–P3 position of the peptide. For the case of FAP, Edosada et al., using positional scanning, documented a preference for Ser or Ala in the P3 position (32). This finding also emerged from our cleavage map analysis, in which Ser/Thr was observed in 24% of the sequences in P3 and Ala was observed in 11%. In the study of Edosada et al., no preference could be observed in P4 (32). Using a map of a known protein substrate, in this case, allowed us to obtain additional sequence information beyond P3, with sequence preferences emerging for P4, P6, and P7 from the analysis of the cleavage map.
An added advantage of this LC/MS/MS sequencing approach is that it allowed us to quantify the abundance of each identified peptide by comparing the extracted ion current for each of the observed peptide parent ions from the MS spectra. The contribution of each parent ion to the total ion current could then be extracted and integrated over the peptide elution peak. This approach allowed us to rank the peptide fragments on the basis of the extracted ion current. Traditional Edman sequencing does not provide the potential for such fragment stratification. While it was already known before this study that FAP preferred cleavage after proline, this information could also have been learned initially from analysis of the cleavage fragments ranked by the extracted ion current.
Since the abundance of each individual fragment depends on cleavage from two sites, the extracted ion current for each fragment represents an “average” based on the efficiency of FAP cleavage at both sites. Therefore, initially, we thought the ranking would provide no additional information beyond the prediction of a preferred amino acid in the P1 position. To evaluate the utility of this ranking based on ion abundance, we synthesized seven fluorescently quenched peptide substrates based on peptide sequences from fragments with high, intermediate, and low extracted ion currents and determined FAP hydrolysis kinetics for each. The extracted ion current rankings corresponded well with the kcat/Km ratios of these peptide substrates, with the highest ranked having the highest ratio, etc. Although based on a limited sample of peptide substrates, these results suggest that this method of ranking cleavage sites could be applied to the analysis of cleavage maps of other proteases with unknown substrate specificity to help identify the best substrates for further synthesis and evaluation.
In conclusion, we have used an LC/MS/MS-based method to generate a map of all FAP cleavage sites within recombinant human collagen I derived gelatin. From this map, we identified potential consensus peptide sequences that could be used to generate FAP-selective peptide–cytotoxin prodrugs. These FAP-activated prodrugs would selectively target FAP-producing myofibroblasts within the stroma of epithelial tumors and, therefore, would represent a potential “pan-tumor” therapeutic agent. Further studies are under way in our laboratory to assess the efficiency and specificity of these consensus peptides for FAP hydrolysis compared to those of other proteases. In addition, we are further evaluating the role of proline hydroxylation of some of these substrates to determine whether such modification enhances FAP hydrolysis.
Acknowledgments
We acknowledge the Mass Spectrometry/Proteomics Facility at the Johns Hopkins University School of Medicine (www.hopkinsmedicine.org/msf/) for assistance with MALDI-TOF analyses and Dr. John Isaacs for helpful comments related to preparation of the manuscript.
Footnotes
This work supported by funding from the Susan G. Komen Breast Cancer Foundation (to S.R.D.), the Department of Defense Breast Cancer Research Program (Grant DAMD17-03-1-0304) (to S.R.D.), and a Department of Defense Prostate Cancer Research Program Pre-Doctoral Award (to W.N.B.). The Mass Spectrometry/Proteomics Facility at the Johns Hopkins University School of Medicine is supported by NCRR Grant 1S10-RR14702, the Johns Hopkins Fund for Medical Discovery, and the Institute for Cell.
Abbreviations: FAP, fibroblast activation protein; LC/MS/MS, liquid chromatography/tandem mass spectrometry; MALDI-TOF, matrix-assisted laser desorption/ionization time of flight; Mca, 7-methoxy-coumarin-4-acetic acid; Dnp, dinitrophenyl.
References
- 1.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 2.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 3.Basset P, Bellocq JP, Wolf C, Stoll I, Hutin P, Limacher JM, Podhajcer OL, Chenard MP, Rio MC, Chambon P. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 1990;348:699–704. doi: 10.1038/348699a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown LF, Guidi AJ, Schnitt SJ, Van De Water L, Iruela-Arispe ML, Yeo TK, Tognazzi K, Dvorak HF. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin Cancer Res. 1999;5:1041–1056. [PubMed] [Google Scholar]
- 5.Haslam SZ, Woodward TL. Host micro-environment in breast cancer development: epithelial-cell-stromal-cell interactions and steroid hormone action in normal and cancerous mammary gland. Breast Cancer Res. 2003;5:208–215. doi: 10.1186/bcr615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iozzo RV. Tumor stroma as a regulator of neoplastic behavior. Agonistic and antagonistic elements embedded in the same connective tissue. Lab Invest. 1995;73:157–160. [PubMed] [Google Scholar]
- 7.Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci USA. 1990;87:7235–7239. doi: 10.1073/pnas.87.18.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoyama A, Chen WT. A 170-kDa membrane-bound protease is associated with the expression of invasiveness by human malignant melanoma cells. Proc Natl Acad Sci USA. 1990;87:8296–8300. doi: 10.1073/pnas.87.21.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rettig WJ, Garin-Chesa P, Beresford HR, Oettgen HF, Melamed MR, Old LJ. Cell-surface glycoproteins of human sarcomas: differential expression in normal and malignant tissues and cultured cells. Proc Natl Acad Sci USA. 1988;85:3110–3114. doi: 10.1073/pnas.85.9.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scanlan MJ, Raj BK, Calvo B, Garin-Chesa P, Sanz-Moncasi MP, Healey JH, Old LJ, Rettig WJ. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA. 1994;91:5657–61. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly T. Fibroblast activation protein-alpha and dipeptidyl peptidase IV (CD26): cell-surface proteases that activate cell signaling and are potential targets for cancer therapy. Drug Resist Updates. 2005;8:51–8. doi: 10.1016/j.drup.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Park JE, Lenter MC, Zimmermann RN, Garin-Chesa P, Old LJ, Rettig WJ. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 13.Denmeade SR, Nagy A, Gao J, Lilja H, Schally A, Isaacs J. Enzymatic activation of a doxorubicin-peptide prodrug by prostate-specific antigen. Cancer Res. 1998;58:2537–2540. [PubMed] [Google Scholar]
- 14.Denmeade SR, Jakobsen CM, Janssen S, Khan SR, Garrett ES, Lilja H, Christensen SB, Isaacs JT. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. J Natl Cancer Inst. 2003;95:990–1000. doi: 10.1093/jnci/95.13.990. [DOI] [PubMed] [Google Scholar]
- 15.Janssen S, Jakobsen CM, Rosen DM, Ricklis RM, Reineke U, Christensen SB, Lilja H, Denmeade SR. Screening a combinatorial peptide library to develop a human glandular kallikrein 2-activated prodrug as targeted therapy for prostate cancer. Mol Cancer Ther. 2004;3:1439–50. [PubMed] [Google Scholar]
- 16.Janssen S, Rosen DM, Ricklis RM, Dionne CA, Lilja H, Christensen SB, Isaacs JT, Denmeade SR. Pharmacokinetics, biodistribution, and antitumor efficacy of a human glandular kallikrein 2 (hK2)-activated thapsigargin prodrug. Prostate. 2006;66:358–68. doi: 10.1002/pros.20348. [DOI] [PubMed] [Google Scholar]
- 17.Mhaka A, Gady AM, Rosen DM, Lo KM, Gillies SD, Denmeade SR. Use of methotrexate-based peptide substrates to characterize the substrate specificity of prostate-specific membrane antigen (PSMA) Cancer Biol Ther. 2004;3:551–8. doi: 10.4161/cbt.3.6.846. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, Albright CF, Fish BH, George HJ, Selling BH, Hollis GF, Wynn R. Expression, purification, and kinetic characterization of full-length human fibroblast activation protein. Protein Expr Purif. 2002;24:274–281. doi: 10.1006/prep.2001.1572. [DOI] [PubMed] [Google Scholar]
- 19.Piñeiro-Sánchez ML, Goldstein LA, Dodt J, Howard L, Yeh Y, Tran H, Argraves WS, Chen WT. Identification of the 170-kDa melanoma membrane-bound gelatinase (seprase) as a serine integral membrane protease. J Biol Chem. 1997;272:7595–601. doi: 10.1074/jbc.272.12.7595. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen VJ, Jackson KW, Lee KN, McKee PA. Effect of fibroblast activation protein and alpha2-antiplasmin cleaving enzyme on collagen types I, III, and IV. Arch Biochem Biophys. 2007;457:177–86. doi: 10.1016/j.abb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulleid NJ, John DC, Kadler KE. Recombinant expression systems for the production of collagen. Biochem Soc Trans. 2000;28:350–353. [PubMed] [Google Scholar]
- 22.Olsen D, Jiang J, Chang R, Duffy R, Sakaguchi M, Leigh S, Lundgard R, Ju J, Buschman F, Truong-Le V, Pham B, Polarek JW. Expression and characterization of a low molecular weight recombinant human gelatin: development of a substitute for animal-derived gelatin with superior features. Protein Expr Purif. 2005;40:346–357. doi: 10.1016/j.pep.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Backes BJ, Harris JL, Leonetti F, Craik CS, Ellman JA. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat Biotechnol. 2000;18:187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- 24.Deng SJ, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, Carter HL, Neugebauer J, Pahel G, Weiner MP, Moss ML. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J Biol Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 25.Matthews DJ, Wells JA. Substrate phage: selection of protease substrates by monovalent phage display. Science. 1993;260:1113–1117. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- 26.Knauper V, Lopez-Otin C, Smith B, Knight G, Murphy G. Biochemical characterization of human collagenase-3. J Biol Chem. 1996;271:1544–1550. doi: 10.1074/jbc.271.3.1544. [DOI] [PubMed] [Google Scholar]
- 27.McGeehan GM, Bickett DM, Green M, Kassel D, Wiseman JS, Berman J. Characterization of the peptide substrate specificities of interstitial collagenase and 92-kDa gelatinase. Implications for substrate optimization. J Biol Chem. 1994;269:32814–32820. [PubMed] [Google Scholar]
- 28.Matsushita O, Yoshihara K, Katayama S, Minami J, Okabe A. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J Bacteriol. 1994;176:149–156. doi: 10.1128/jb.176.1.149-156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hori H, Nagai Y. Purification of tadpole collagenase and characterization using collagen and synthetic substrates. Biochim Biophys Acta. 1979;566:211–221. doi: 10.1016/0005-2744(79)90263-8. [DOI] [PubMed] [Google Scholar]
- 30.Tsu CA, Perona JJ, Schellenberger V, Turck CW, Craik CS. The substrate specificity of Uca pugilator collagenolytic serine protease 1 correlates with the bovine type I collagen cleavage sites. J Biol Chem. 1994;269:19565–19572. [PubMed] [Google Scholar]
- 31.Patterson SD, Aebersold R. Mass spectrometric approaches for the identification of gel-separated proteins. Electrophoresis. 1995;16:1791–1814. doi: 10.1002/elps.11501601299. [DOI] [PubMed] [Google Scholar]
- 32.Edosada CY, Quan C, Wiesmann C, Tran T, Sutherlin D, Reynolds M, Elliott JM, Raab H, Fairbrother W, Wolf BB. Selective inhibition of fibroblast activation protein protease based on dipeptide substrate specificity. J Biol Chem. 2006;281:7437–7444. doi: 10.1074/jbc.M511112200. [DOI] [PubMed] [Google Scholar]