Summary
The Cancer Genome Atlas has reported that 96% of ovarian high-grade serous carcinomas (HGSCs) have TP53 somatic mutations suggesting that mutation of this gene is a defining feature of this neoplasm. In the current study, 5 gynecologic pathologists independently evaluated hematoxylin and eosin slides of 14 available cases from The Cancer Genome Atlas classified as HGSC that lacked a TP53 mutation. The histologic diagnoses rendered by these pathologists and the accompanying molecular genetic data are the subject of this report. Only 1 case (Case 5), which contained a homozygous deletion of TP53, had unanimous interobserver agreement for a diagnosis of pure HGSC. In 1 case (Case 3), all 5 observers (100%) rendered a diagnosis of HGSC; however, 3 observers (60%) noted that the histologic features were not classic for HGSC and suggested this case may have arisen from a low-grade serous carcinoma (arisen from an alternate pathway compared with the usual HGSC). In 2 cases (Cases 4 and 12), only 3 observers (60%) in each case, respectively, interpreted it as having a component of HGSC. In the remaining 10 (71%) of tumors (Cases 1, 2, 6–11, 13, and 14), the consensus diagnosis was not HGSC, with individual diagnoses including low-grade serous carcinoma, high-grade endometrioid carcinoma, HGSC, metastatic carcinoma, clear cell carcinoma, atypical proliferative (borderline) serous tumor, and adenocarcinoma, not otherwise specified. Therefore, 13 (93%) of the tumors (Cases 1–4 and 6–14) were either not a pure HGSC or represented a diagnosis other than HGSC, all with molecular results not characteristic of HGSC. Accordingly, our review of the TP53 wild-type HGSCs reported in The Cancer Genome Atlas suggests that 100% of de novo HGSCs contain TP53 somatic mutations or deletions, with the exception of the rare HGSCs that develop from a low-grade serous tumor precursor. We, therefore, propose that lack of molecular alterations of TP53 are essentially inconsistent with the diagnosis of ovarian HGSC and that tumors diagnosed as such should be rigorously reassessed to achieve correct classification.
Keywords: The Cancer Genome Atlas, TCGA, TP53, High-grade serous carcinoma
Ovarian cancer is the most lethal gynecologic malignancy accounting for approximately 21,980 new cases in 2014 and resulting in 14,270 deaths annually in the United States (1). High-grade serous carcinoma (HGSC) accounts for approximately 75% of all ovarian carcinomas and is responsible for nearly 90% of the fatalities (2). The recent recognition of a microscopic precursor lesion in the fallopian tube, serous tubal intraepithelial carcinoma, which is morphologically similar to ovarian HGSC and contains a TP53 mutation in >90% of cases, (3,4) suggests that mutation of TP53 is an early and important molecular event in the pathogenesis of HGSC. A genome-wide analysis of HGSC by The Cancer Genome Atlas (TCGA) Research Network reported TP53 mutations in 96% of specimens (5), supporting this view. In that study, only 15 HGSCs analyzed lacked a TP53 mutation raising the question as to what distinguished this small group from the remainder. The aim of the present study was to evaluate the morphologic features and molecular genetic data of this particular group of tumors to determine whether the lack of TP53 mutations characterized a rare subset of HGSCs or whether the tumors had been misclassified.
MATERIALS AND METHODS
All samples were part of the previously reported TCGA study on ovarian cancer that was IRB approved at all participating sites (5). In the TCGA study, cases were included based on the original pathology report. Specimens were reviewed by the Biospecimen Core Resource (a centralized laboratory that reviews and processes specimens and their associated data for all of the TCGA Research Network). However, whether specific histologic criteria were used is unknown. Immunohistochemistry was not employed as inclusion/exclusion criteria in the TCGA, and the original pathology reports for the cases in the current study do not indicate that immunohistochemistry was performed at the time of the initial diagnosis.
All tumor-bearing slides from the 15 TCGA cases with wild-type TP53 sequences were retrieved from tissue source sites. One case with insufficient tissue for review was excluded. All hematoxylin and eosin slides from the remaining 14 cases were reviewed by 1 author (R.J.K.), and representative slides were selected for this study. Those representative slides were reviewed independently by 5 gynecologic pathologists (R.V., I.-M.S., R.A.S., C.Z., R.J.K.) who were blinded to all clinical and molecular information with the exception that all cases lacked a TP53 mutation, and a diagnosis was rendered based on criteria used in routine practice. Molecular data were obtained from the cBioPortal for Cancer Genomics website (6), and those results were then correlated with the rendered rereview diagnoses.
RESULTS
The 5 pathologists’ diagnoses in this study and reported molecular data for each tumor are shown in Table 1. All 5 pathologists agreed in 8 (57%) of the 14 cases, and at least 3 pathologists agreed in 11 (79%) of the cases. Of the 8 cases with a unanimous diagnosis, 4 were classified as low-grade serous carcinoma (LGSC) (Cases 6, 11, 13, and 14), 1 as an atypical proliferative serous tumor (typical serous borderline tumor) (Case 9), 1 as a high-grade endometrioid carcinoma (Case 8), 1 as an unusual HGSC with features suggesting evolution from LGSC (Case 3), and 1 as a pure HGSC (Case 5). Therefore, the panel of observers uniformly agreed that only 1 (7%) of 14 TCGA TP53 wild-type cases originally diagnosed as HGSC was unequivocally an HGSC (Case 5) (Fig. 1). This tumor had a BRCA1 germline mutation, substantial level of somatic copy number alterations, high number of mutations, and homozygous TP53 deletion.
TABLE 1.
Rereview diagnoses and molecular data for TP53 wild-type high-grade serous carcinomas from the TCGA study
| Rereview pathologist | Molecular data from the TCGA(6) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | TCGA* | A | B | C | D | E | TP53 HD† | BRCA‡ | No. Muts§ | CNA (%)‖ |
| 1 | 09-2056 | LGSC | HG Endo | LGSC | HGSC | HGSC | − | − | 68 | 46 |
| 2 | 10-0933 | Met | HG Endo | HG Endo | Met | Met | − | − | 33 | 47 |
| 3 | 13-0727 | HGSC¶ | HGSC¶ | HGSC¶ | HGSC | HGSC | − | − | 21 | 22 |
| 4 | 13-0755 | CCC | LG/HGSC# | CCC | HGSC | HGSC | − | − | 73 | 18 |
| 5 | 13-1408 | HGSC | HGSC | HGSC | HGSC | HGSC | + | + | 78 | 31 |
| 6 | 13-1477 | LGSC | LGSC | LGSC | LGSC | LGSC | − | − | 42 | 9 |
| 7 | 24-1544 | Met | Met | Met | HGSC | HGSC | − | − | 17 | 31 |
| 8 | 24-1565 | HG Endo | HG Endo | HG Endo | HG Endo | HG Endo | − | − | 25 | 9 |
| 9 | 24-2038 | APST | APST | APST | APST | APST | − | − | 10 | 11 |
| 10 | 25-1316 | LGSC | HG Endo | LGSC | HGSC | HGSC | − | − | 13 | 30 |
| 11 | 25-1328 | LGSC | LGSC | LGSC | LGSC | LGSC | − | − | 10 | NA |
| 12 | 25-2042 | CCC | Adeno, NOS | HGSC | HGSC | HGSC | − | − | 56 | 19 |
| 13 | 25-2408 | LGSC | LGSC | LGSC | LGSC | LGSC | − | − | 13 | 9 |
| 14 | 61-2095 | LGSC | LGSC | LGSC | LGSC | LGSC | − | − | 109 | 1 |
TCGA study number.
Homozygous deletion of TP53.
Mutation of BRCA1 or BRCA2.
No. mutations.
Fraction of copy number altered genome.
HGSC with LGSC architecture.
Serous carcinoma with features of both LGSC and HGSC.
+ indicates Present; −, Absent; Adeno, adenocarcinoma; APST, atypical proliferative serous tumor (typical serous borderline tumor); CCC, clear cell carcinoma; HG Endo, high-grade endometrioid carcinoma; HGSC, high-grade serous carcinoma; LGSC, invasive low-grade serous carcinoma; Met, metastatic carcinoma; NA, not available; NOS, not otherwise specified; TCGA, The Cancer Genome Atlas
FIG. 1.
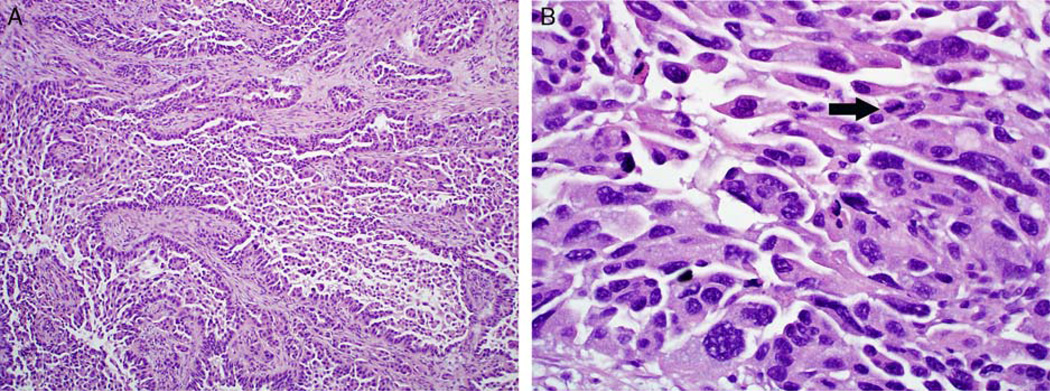
Case 5: all 5 observers classified this case as high-grade serous carcinoma. (A) The architectural features are notable for large papillae lined by stratified epithelium with irregular slit-like spaces. Numerous detached and small epithelial clusters are present. (B) The cytologic features consist of large nuclei, nuclear pleomorphism, hyperchromasia, bizarre nuclear atypia, and mitotic activity (arrow).
The other tumor diagnosed by all panel members as an HGSC, but for which 3 of 5 observers noted features suggesting evolution from LGSC (Case 3) (Fig. 2) had a substantially lower number of mutations and relatively lower level of somatic copy number alterations. Morphologically, this tumor had a micropapillary-rich architecture but exhibited greater cytologic atypia and more mitotic figures than the usual LGSC, warranting a diagnosis of HGSC. It lacked the molecular features characteristic of HGSC, specifically a TP53 mutation, large number of mutations, and high level of somatic copy number alterations (6–9). In contrast, it lacked KRAS and BRAF mutations, which are found in approximately half of LGSCs (10,11). Thus, neither the morphologic nor molecular findings allowed for the definitive classification of this neoplasm.
FIG. 2.
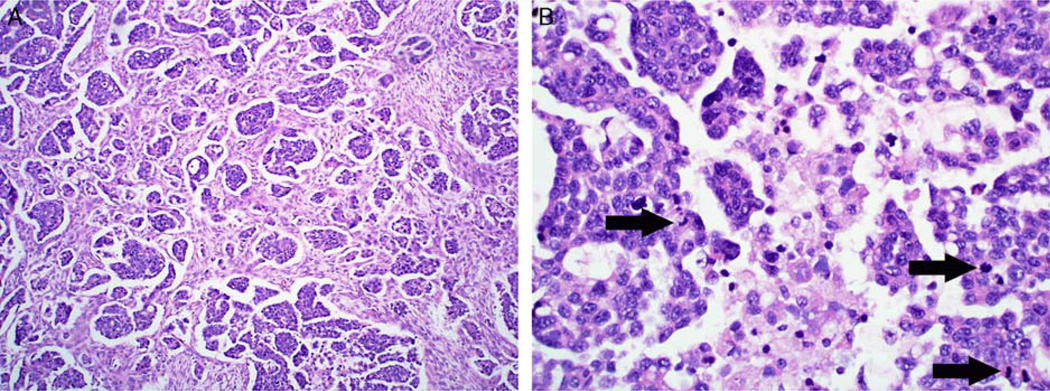
Case 3: all 5 observers classified this case as high-grade serous carcinoma; however, 3 noted features suggestive of evolution from lowgrade serous carcinoma. (A) The tumor is predominantly composed of a micropapillary-rich pattern, commonly seen in invasive low-grade serous carcinoma, in which small micropapillary nests within clear lacunar spaces haphazardly infiltrate through stroma. (B) The nuclei are not as large as frequently seen in high-grade serous carcinoma and more uniform than in most such cases. However, the combined presence of some degree of both chromatin irregularity and variation in nuclear shape and mitotic activity (arrows) favor classification as high-grade serous carcinoma.
In each of 2 cases (Cases 4 and 12), 3 of 5 observers, respectively, diagnosed a component of HGSC. In Case 4 (Fig. 3), 1 of the 3 observers that diagnosed a component of HGSC also noted a mixed component of LGSC. However, 2 observers in this case interpreted the tumor as clear cell carcinoma. In Case 12 (Fig. 4), 2 of the observers diagnosed the tumor as something other than HGSC: 1 as clear cell carcinoma and the other as adenocarcinoma, not otherwise specified. On the basis of morphology and the relatively low level of somatic copy number alterations, both cases were not unequivocally HGSC.
FIG. 3.
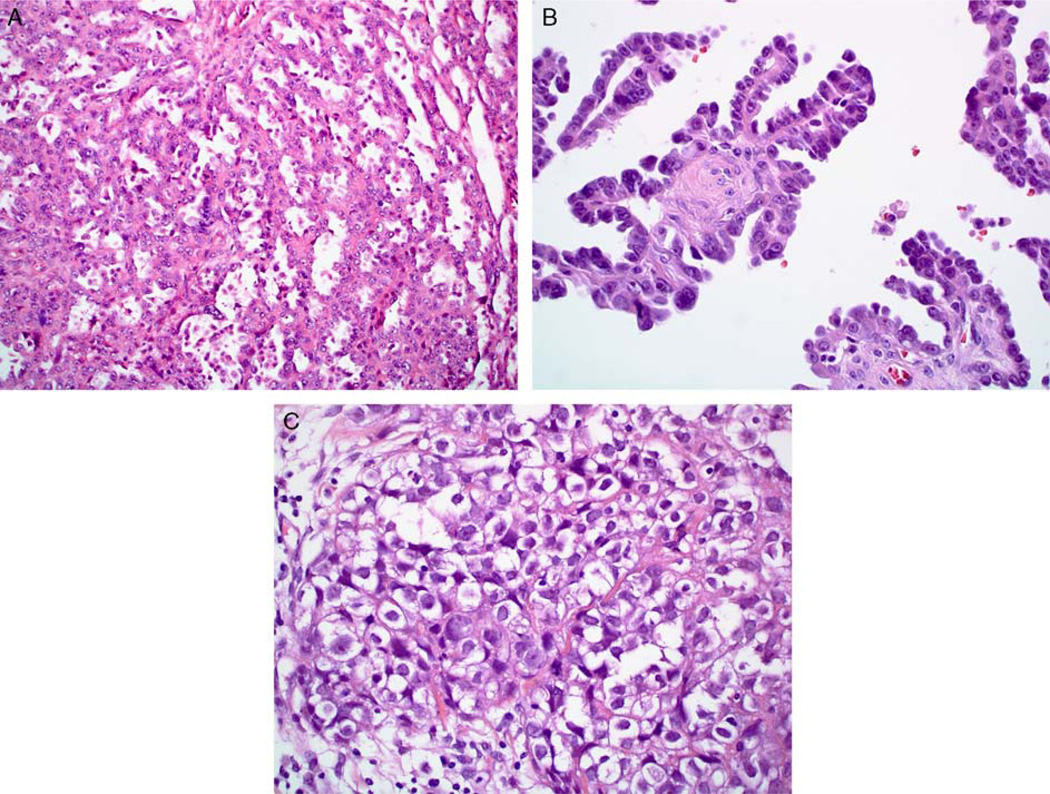
Case 4: this case was classified as high-grade serous carcinoma by 2 observers, serous carcinoma with a mixture of low-grade and high-grade features by 1, and clear cell carcinoma by 2. (A) This tumor featured areas with tubulocystic-like architecture and a vague suggestion of hobnail cell shapes. (B) The papillae in other foci were lined by a single layer of low-cuboidal cells with a slight degree of monotony and without the degree of stratification seen in many high-grade serous carcinomas. A vague suggestion of hobnail cell shapes is also present. (C) Some areas had solid architecture consisting of polygonal cells with abundant clear cytoplasm.
FIG. 4.
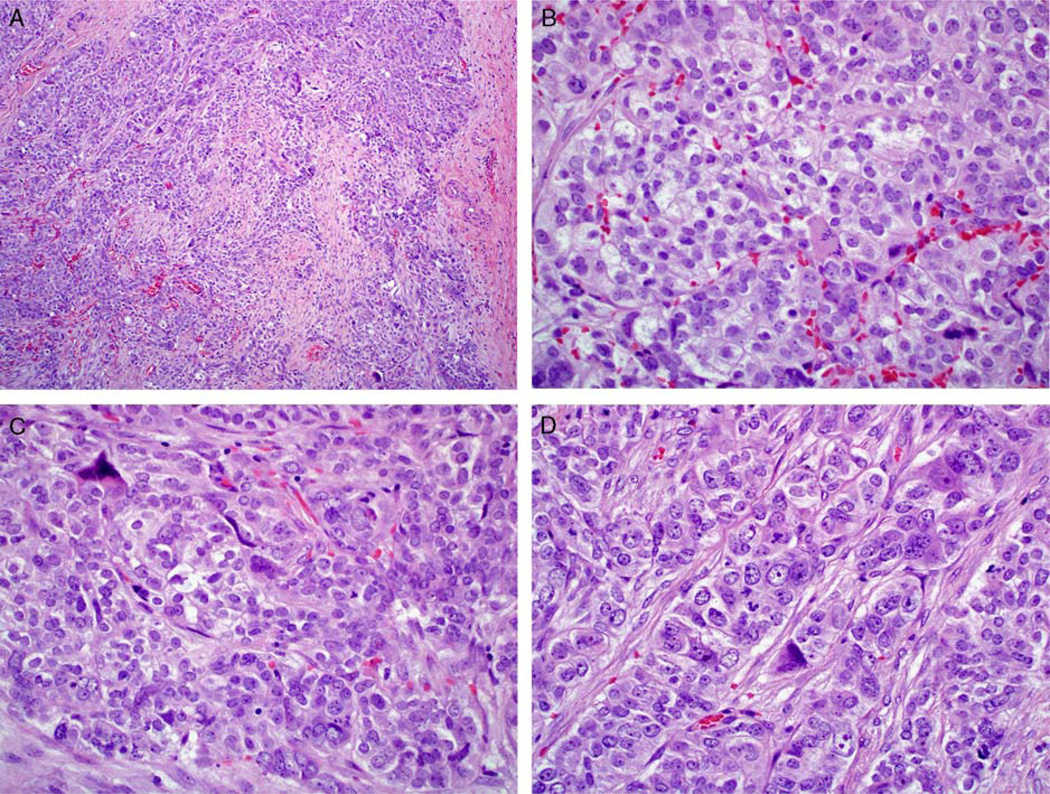
Case 12: this case was classified as high-grade serous carcinoma by 3 observers, clear cell carcinoma by 1, and adenocarcinoma, not otherwise specified by 1. (A) The tumor shows solid architecture with variably sized nests haphazardly arranged within stroma. (B) Many areas had polygonal cells with round and relatively uniform nuclei and abundant clear cytoplasm. (C) A bizarre atypical cell (upper left) is present. (D) Some foci exhibited cord-like architecture. Note the presence of high-grade nuclei.
In the remaining 10 cases (Cases 1, 2, 6–11, 13, and 14), between 0 and 2 observers per case rendered a diagnosis of HGSC, with non-HGSC diagnoses including LGSC, high-grade endometrioid carcinoma, metastatic carcinoma, and atypical proliferative (borderline) serous tumor (Table 1). Therefore, these cases were morphologically misclassified or unclassifiable, and some had numbers of mutations and/or levels of somatic copy number alterations that were not characteristic of HGSC (Table 1).
DISCUSSION
An accurate diagnosis of HGSC is important for proper clinical management and clinical trial stratification. HGSCs are exquisitely sensitive to platinum- based agents and respond to poly-(ADP-ribose) polymerase (PARP) inhibitors, unlike other histologic subtypes (12). In addition, HGSC is the most common subtype associated with BRCA germline mutations and warrants genetic testing, particularly in women with a strong family history of ovarian and/or breast carcinoma. Serous tubal intraepithelial carcinoma, the proposed precursor of ovarian/peritoneal HGSC, also exhibits TP53 mutations in 93% of cases, (3,4,13–16) implicating this molecular alteration as an early event in the pathogenesis of HGSC. Current screening tests for ovarian cancer utilizing serum measurement of CA125 and transvaginal ultrasound have failed to demonstrate a survival benefit (17–21). However, the recognition that nearly 100% of ovarian HGSCs contain a TP53 mutation suggests that detection of this molecular alteration may be a promising screening test. Accordingly, the likely success of such an approach is borne out by a recently reported screening test (PapGene test) that used a panel of genes commonly mutated in ovarian carcinomas, including TP53, and successfully detected 41% of ovarian cancers (22).
To properly classify a tumor as HGSC, reproducible histologic features must be utilized. Per the 2014 WHO Classification for gynecologic tumors, ovarian HGSC is characterized by solid, papillary, glandular, and cribriform architecture with slit-like spaces, necrosis, high-grade nuclei with bizarre forms, prominent nucleoli, abundant mitotic figures, and psammoma bodies (23). In a study of primary ovarian carcinomas published before the 2014 WHO Classification, Köbel et al. (24) demonstrated that, using histologic criteria for HGSC similar to those above, excellent interobserver reproducibility can be achieved for all histologic types combined. Particularly in the validation set in that study, 94% of cases originally diagnosed as HGSC were concordant with the central review diagnosis. For the discordant cases, 4% and 2% of those originally diagnosed as HGSC were interpreted as LGSC and endometrioid carcinoma, respectively, upon central review. Their overall frequency of misclassification of HGSC is similar to the readjusted data for the TCGA although only the HGSCs lacking a TP53 mutation were rereviewed in our study. Nevertheless, the findings in our study show the value of rigorous histologic assessment for properly defining HGSC.
Although it was not the goal of this study to immunohistochemically evaluate cases without TP53 mutations or those having nonconsensus, the use of immunohistochemical stains for the assessment of carcinomas involving the ovary can be useful to improve diagnostic accuracy and interobserver reproducibility of primary ovarian carcinomas in general, as well as distinguish LGSC from HGSC and exclude some metastatic adenocarcinomas (25–35). In particular, ovarian HGSC is usually positive for PAX8, ER, PR, and WT-1, exhibits an abnormal pattern of p53 expression (either diffuse strong or complete absence of staining), and often but not always shows diffuse expression of p16. Thus, careful attention to histologic features and selected use of immunohistochemical stains can help avoid misclassification as HGSC.
Even though consensus guidelines have not yet been established as to whether immunohistochemistry is necessary for diagnosing all cases of HGSC, the use of immunohistochemical stains should be performed at the discretion of the pathologist, particularly based on his/her level of experience with ovarian tumors. For cases in which histologic features are considered classic, morphology alone should suffice. However, in cases where there is diagnostic uncertainty or the distinction between HGSC and other primary ovarian carcinomas is unclear, immunohistochemical staining for WT-1, p53, and p16, as described above, should be performed. Nonetheless, it must be emphasized that establishing/excluding a diagnosis of HGSC should not be based solely on one immunohistochemical stain. This is because some HGSCs can be negative for WT-1 or lack aberrant expression patterns of p53, occasional endometrioid carcinomas can be WT-1-positive, and some nonserous carcinomas can show aberrant expression patterns of p53 (23). Thus, those immunohistochemical stains must be interpreted together as a panel of markers in the context of the morphologic appearance.
In addition to histology, it is also important to understand what defines HGSC at the molecular level. Prior work, including that of the TCGA, has established that molecular aberrations of HGSC consist of TP53 mutations in >96% of cases, BRCA mutations (germline or somatic) in 20%, recurrent mutations in other genes in 2% to 6%, 20 to 60 mutations in the majority of cases, a high fraction of copy number altered genome in most cases with particular chromosomal gains and losses, and various patterns of promoter methylation, mRNA expression, and miRNA expression (5,6,8,9,36). Given the significant pathogenic role of TP53 mutations and their high frequency in HGSC, their absence in select HGSCs in the TCGA was puzzling. The goal of the current study was to evaluate the histologic features of these TP53 wild-type tumors to determine whether they represented a specific subset of HGSCs or if these were misclassified.
Rereview of hematoxylin and eosin slides in our study showed that the panel of pathologists uniformly agreed that only 1 of 14 TCGA TP53 wild-type originally diagnosed as HGSC was unequivocally a pure HGSC (Case 5). This tumor had a BRCA1 germline mutation, extensive somatic copy number alterations, and a homozygous TP53 deletion instead of the more common somatic point mutation, underscoring the importance of loss of p53 function. Hence, our findings further support that unequivocal HGSCs have molecular alterations involving TP53. The other case diagnosed by all panel members as HGSC (Case 3) had histologic features suggestive of evolution from LGSC. The tumor lacked the molecular findings characteristic of HGSC, specifically a TP53 mutation and high level of DNA copy number alterations (7). It has been reported that, on rare occasion, HGSCs may develop along a pathway evolving from a low-grade serous tumor (borderline tumor or LGSC) independent of the more common pathway for the usual HGSC, which involves TP53 mutations and, presumably origin from serous tubal intraepithelial carcinoma (37,38). On the basis of interobserver histologic assessment and lack of molecular findings typical of HGSC (Table 1), the remaining cases in this study were either not a pure HGSC or represented a diagnosis other than HGSC.
The results of another study also support our findings. In an analysis of 126 extrauterine HGSCs, 94% were originally found to harbor TP53 mutations (36). Seven cases were mutation-negative, and rereview of those showed that 3 were not pure HGSC (1 LGSC, 1 high-grade carcinoma of uncertain primary site, and 1 mixed LGSC-HGSC). The remaining 4 cases were interpreted as HGSC after rereview. After adjusting for the 3 mutation-negative non-HGSC cases, the frequency of TP53 mutation in HGSC was 97%. Although alternative mechanisms of p53 dysfunction, such as amplification of MDM2 and MDM4, were suggested for TP53 mutation-negative HGSCs, it is unknown if these 4 TP53 mutation-negative HGSCs were subjected to rigorous interobsever testing as was done in our study, which potentially may have increased the reclassification frequency of those cases and yielded an even higher proportion of TP53 mutations in HGSC.
Of note, a recent study analyzing the clinical data from the TP53 mutation-negative cases in the ovarian TCGA reported that women with TP53 mutation-negative tumors had a significantly shorter survival and their tumors had higher chemoresistance compared with the TP53-mutated cases (39). These findings should be interpreted in the light of our study showing that nearly all of the “TP53 mutation-negative”cases were misclassified highlighting the importance of rigorous pathology rereview.
A potential limitation of the current study is that our pathology review panel was aware of the TP53 status of these cases before slide review. However, we do not believe that this introduces any significant bias as this was not a traditional interobserver reproducibility study. The goal of the study was actually to simply determine what were the real diagnoses of the TP53 wild-type cases in the TCGA. Moreover, the possibility that the panel members would not diagnose HGSC because they knew that the case lacked a TP53 mutation was limited as Table 1 shows that that a number of HGSC diagnoses were rendered by individual pathologists for various cases. Thus, the importance of our study is that it demonstrates that the vast majority of TP53 wildtype cases diagnosed as HGSC in the TCGA were not HGSC.
In conclusion, the findings in this study complement and extend those reported by the TCGA by showing that all de novo ovarian HGSCs contain alterations of TP53, most frequently through somatic mutation. Although certain morphologic features are characteristic of HGSC, we propose that the lack of a TP53 mutation is inconsistent with that diagnosis. These findings, in conjunction with genome-wide analysis of other gynecologic neoplasms, including ovarian clear cell carcinoma, (40,41) ovarian LGSC, (10) uterine endometrioid carcinoma, (42,43) and uterine serous carcinoma, (7,42,44) set the stage for the potential development of a morphologic-molecular classification of gynecologic neoplasms, similar to what has been achieved for breast carcinoma. This study demonstrates the need for establishing a rigorous definition of HGSC for enrollment in research studies and clinical trials. It also underscores the importance of careful pathology review in comprehensive cancer genome studies of cases with unusual molecular findings to determine if they represent variants of the particular tumor type or misclassifications.
Acknowledgments
Supported in part by Laura Mercier Ovarian Cancer Fund; The Honorable Tina Brozman Foundation; DOD Consortium Award W81XWH-11-2-0230.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Seidman JD, Cho K, Ronnett BM, et al. Surface epithelial tumors of the ovary. In: Kurman RJ, Ronnett BM, Ellenson LH, editors. Blaustein’s Pathology of the Female Genital Tract. New York, NY: Springer; 2011. pp. 679–784. [Google Scholar]
- 3.Kindelberger DW, Lee Y, Miron A, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: evidence for a causal relationship. Am J Surg Pathol. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn E, Kurman RJ, Vang R, et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic highgrade serous carcinoma—evidence supporting the clonal relationship of the two lesions. J Pathol. 2012;226:421–426. doi: 10.1002/path.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.cBioPortal for Cancer Genomics. [Accessed December 15, 2014];2014 Available at: http://www.cbioportal.org/public-portal/study.do?cancer_study_id=ov_tcga_pub. [Google Scholar]
- 7.Kuhn E, Wu RC, Guan B, et al. Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses. J Natl Cancer Inst. 2012;104:1503–1513. doi: 10.1093/jnci/djs345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo KT, Guan B, Feng Y, et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Wang TL, Kurman RJ, et al. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer G, Oldt R, III, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee S, Kaye SB. New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin Cancer Res. 2013;19:961–968. doi: 10.1158/1078-0432.CCR-12-2243. [DOI] [PubMed] [Google Scholar]
- 13.Crum CP, McKeon FD, Xian W. The oviduct and ovarian cancer: causality, clinical implications, and “targeted prevention.”. Clin Obstet Gynecol. 2012;55:24–35. doi: 10.1097/GRF.0b013e31824b1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przybycin CG, Kurman RJ, Ronnett BM, et al. Are all pelvic (nonuterine) serous carcinomas of tubal origin? Am J Surg Pathol. 2010;34:1407–1416. doi: 10.1097/PAS.0b013e3181ef7b16. [DOI] [PubMed] [Google Scholar]
- 15.Vang R, Wheeler JE. Diseases of the fallopian tube and paratubal region. In: Kurman RJ, Ronnett BM, Ellenson LH, editors. Blaustein’s Pathology of the Female Genital Tract. New York, NY: Springer; 2011. pp. 529–578. [Google Scholar]
- 16.Vang R, Shih I, Kurman RJ. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology. 2013;62:44–58. doi: 10.1111/his.12046. [DOI] [PubMed] [Google Scholar]
- 17.Buys SS, Partridge E, Greene MH, et al. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: findings from the initial screen of a randomized trial. Am J Obstet Gynecol. 2005;193:1630–1639. doi: 10.1016/j.ajog.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert L, Basso O, Sampalis J, et al. Assessment of symptomatic women for early diagnosis of ovarian cancer: results from the prospective DOvE pilot project. Lancet Oncol. 2012;13:285–291. doi: 10.1016/S1470-2045(11)70333-3. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi H, Yamada Y, Sado T, et al. A randomized study of screening for ovarian cancer: a multicenter study in Japan. Int J Gynecol Cancer. 2008;18:414–420. doi: 10.1111/j.1525-1438.2007.01035.x. [DOI] [PubMed] [Google Scholar]
- 20.Menon U, Gentry-Maharaj A, Hallett R, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 21.Partridge E, Kreimer AR, Greenlee RT, et al. Results from four rounds of ovarian cancer screening in a randomized trial. Obstet Gynecol. 2009;113:775–782. doi: 10.1097/AOG.0b013e31819cda77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5:1–10. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidman JD, Bell DA, Crum CP, et al. Tumours of the ovary: epithelial tumours-serous tumours. In: Kurman RJ, Carcangiu ML, Herrington CS, et al., editors. WHO Classification of Tumours of Female Reproductive Organs. 4th ed. Lyon, France: IARC Press; 2014. pp. 17–24. [Google Scholar]
- 24.Köbel M, Kalloger SE, Baker PM, et al. Diagnosis of ovarian carcinoma cell type is highly reproducible: a trans- Canadian study. Am J Surg Pathol. 2010;34:984–993. doi: 10.1097/PAS.0b013e3181e1a3bb. [DOI] [PubMed] [Google Scholar]
- 25.Kobel M, Bak J, Bertelsen BI, et al. Ovarian carcinoma histotype determination is highly reproducible, and is improved through the use of immunohistochemistry. Histopathology. 2014;64:1004–1013. doi: 10.1111/his.12349. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill CJ, Deavers MT, Malpica A, et al. An immunohistochemical comparison between low-grade and high-grade ovarian serous carcinomas: significantly higher expression of p53, MIB1, BCL2, HER-2/neu, and C-KIT in high-grade neoplasms. Am J Surg Pathol. 2005;29:1034–1041. [PubMed] [Google Scholar]
- 27.O’Neill CJ, McBride HA, Connolly LE, et al. High-grade ovarian serous carcinoma exhibits significantly higher p16 expression than low-grade serous carcinoma and serous borderline tumour. Histopathology. 2007;50:773–779. doi: 10.1111/j.1365-2559.2007.02682.x. [DOI] [PubMed] [Google Scholar]
- 28.Vang R, Ronnett BM. A practical approach to mucinous tumors involving the ovary: distinction of primary from metastatic tumors and prediction of site of origin for metastases of uncertain origin. Pathol Case Rev. 2006;11:18–30. [Google Scholar]
- 29.Vang R, Ronnett BM. Metastatic and miscellaneous primary tumors of the ovary. In: Nucci MR, Oliva E, editors. Gynecologic Pathology. Philadelphia, PA: Elsevier; 2009. pp. 539–613. [Google Scholar]
- 30.Vang R, Cheung ANY, Kommoss F, et al. Tumours of the ovary: secondary tumours. In: Kurman RJ, Carcangiu ML, Herrington CS, et al., editors. WHO Classification of Tumours of Female Reproductive Organs. 4th ed. Lyon, France: IARC Press; 2014. pp. 83–86. [Google Scholar]
- 31.Vang R, Gown AM, Barry TS, et al. Cytokeratins 7 and 20 in primary and secondary mucinous tumors of the ovary: analysis of coordinate immunohistochemical expression profiles and staining distribution in 179 cases. Am J Surg Pathol. 2006;30:1130–1139. doi: 10.1097/01.pas.0000213281.43036.bb. [DOI] [PubMed] [Google Scholar]
- 32.Vang R, Gown AM, Wu LS, et al. Immunohistochemical expression of CDX2 in primary ovarian mucinous tumors and metastatic mucinous carcinomas involving the ovary: comparison with CK20 and correlation with coordinate expression of CK7. Mod Pathol. 2006;19:1421–1428. doi: 10.1038/modpathol.3800698. [DOI] [PubMed] [Google Scholar]
- 33.Vang R, Gown AM, Farinola M, et al. p16 expression in primary ovarian mucinous and endometrioid tumors and metastatic adenocarcinomas in the ovary: utility for identification of metastatic HPV-related endocervical adenocarcinomas. Am J Surg Pathol. 2007;31:653–663. doi: 10.1097/01.pas.0000213369.71676.25. [DOI] [PubMed] [Google Scholar]
- 34.Vang R, Shih I, Kurman RJ. Ovarian low-grade and highgrade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. 2009;16:267–282. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yemelyanova A, Vang R, Kshirsagar M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24:1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehari R, Kurman RJ, Logani S, et al. The development of high-grade serous carcinoma from atypical proliferative (borderline) serous tumors and low-grade micropapillary serous carcinoma: a morphologic and molecular genetic analysis. Am J Surg Pathol. 2007;31:1007–1012. doi: 10.1097/PAS.0b013e31802cbbe9. [DOI] [PubMed] [Google Scholar]
- 38.Garg K, Park KJ, Soslow RA. Low-grade serous neoplasms of the ovary with transformation to high-grade carcinomas: a report of 3 cases. Int J Gynecol Pathol. 2012;31:423–428. doi: 10.1097/PGP.0b013e31824ae6f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong KK, Izaguirre DI, Kwan SY, et al. Poor survival with wild-type TP53 ovarian cancer? Gynecol Oncol. 2013;130:565–569. doi: 10.1016/j.ygyno.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones S, Wang TL, Shih I, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang H, Cheung LW, Li J, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao S, Choi M, Overton JD, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA. 2013;110:2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]