Abstract
IMPORTANCE
Currently, one of the most commonly available biomarkers in the treatment of patients with colorectal liver metastases (CRLM) is the Kirsten rat sarcoma viral oncogene homolog (KRAS); however, the prognostic implications of specific mutations of the KRAS gene are still not well defined.
OBJECTIVE
To investigate the prognostic impact of specific KRAS mutations on patients undergoing liver resection for CRLM.
DESIGN, SETTING, AND PARTICIPANTS
This retrospective single-center study was conducted from January 1, 2003, to December 31, 2013. Data about specific KRAS mutations for 331 patients who underwent hepatic resection for CRLM at Johns Hopkins Hospital between 2003 and 2013 were analyzed. Clinicopathological characteristics, perioperative details, and outcomes were stratified by specific KRAS mutation at codons 12 and 13.
INTERVENTION
Resection of CRLM.
MAIN OUTCOMES AND MEASURES
Overall survival (OS) and recurrence-free survival.
RESULTS
A mutated KRAS (mtKRAS) was identified in 91 patients (27.5%). At a median follow-up of 27.4 months, recurrence was observed in 48 patients (52.7%) with mtKRAS and 130 patients (54.2%) with wild-type KRAS (wtKRAS) (P = .82). Median and 5-year survival among patients with mtKRAS was 32.4 months and 32.7%, respectively, vs 58.5 months and 46.9%, respectively, for patients with wtKRAS (P = .02). Patients with KRAS codon 12 mutations had worse OS (hazard ratio [HR], 1.54; 95% CI, 1.05–2.27; P = .03) vs those with wtKRAS, whereas a KRAS codon 13 mutation was not associated with prognosis (HR, 1.47; 95% CI, 0.83–2.62; P = .19). Among the 6 most common mutations in codons 12 and 13, only G12V (HR, 1.78; 95% CI, 1.00–3.17; P = .05) and G12S (HR, 3.33; 95% CI, 1.22–9.10; P = .02) were associated with worse OS compared with patients with wtKRAS (both P < .05). Among patients who recurred, G12V (HR, 2.96; 95% CI, 1.32–6.61; P = .01), G12C (HR, 6.74; 95% CI, 2.05–22.2; P = .002), and G12S mutations (HR, 4.91; 95% CI, 1.52–15.8; P = .01) were associated with worse OS (both P < .05).
CONCLUSIONS AND RELEVANCE
G12V and G12S mutations of codon 12 were independent prognostic factors of worse OS. Among patients who recurred after resection of CRLM, G12V, G12C, and G12S mutations were associated with worse OS. Information on specific KRAS mutations may help individualize therapeutic and surveillance strategies for patients with resected CRLM.
Surgical therapy, often combined with adjuvant systemic chemotherapy, is the best therapeutic option to treat patients with colorectal liver metastasis (CRLM), However, while overall survival (OS) has improved, many patients with CRLM will recur and ultimately die of their disease.1,3 The factors used to predict outcome following surgical resection of CRLM largely focus on clinicopathological prognostic factors such as preoperative carcinoembryonic antigen (CEA) level, presentation of disease (ie, synchronous vs metachronous disease), disease-free interval between primary tumor and hepatic metastasis, and metastatic tumor number and size.4 There has been increasing interest in the use of biologic and molecular markers in the prognostic assessment of patients with metastatic colorectal cancer undergoing liver resection.4 Among patients with colorectal adeno-carcinoma, mutated Kirsten rat sarcoma viral oncogene homolog (KRAS) is the most common oncogene of the RAS family, reported in up to 30% to 40% of patients.5–9 While the frequency and prognostic impact of KRAS mutation status have been described for both primary and metastatic colorectal cancer, to our knowledge, the role of specific mutations on KRAS codons remains undefined.10–14
Most KRAS mutations are detected in codons 12 and 13, while mutations in codons 61 and 146 are less common.15–17 KRAS mutations in codons 12 and 13 include different point mutations; the most common are codon 12 Gly→Asp (G12D), codon 12 Gly→Val (G12V), and codon 13 Gly→Asp (G13D) substitutions.18 Previous evidence has suggested that different biologic characteristics of specific KRAS mutations can lead to variations in epidermal growth factor receptor resistance.19–21 In addition, some investigators have suggested that specific KRAS mutations may also be associated with a more aggressive tumor phenotype in patients with unresectable stage IV metastatic colorectal cancer.5,22 However, the prognostic implication of different point mutations on survival of patients following curative intent liver resection for CLRM has not been previously investigated. As such, the purpose of the present study was to define the incidence of different specific KRAS mutations among patients with resected CRLM. Specifically, we sought to characterize the prognostic impact of different KRAS point mutations on recurrence and the survival of patients undergoing curative intent liver resection for CRLM.
Methods
Study Design
Patients who underwent curative intent liver resection for CRLM between January 2003 and August 2013 at Johns Hopkins Hospital with available KRAS data were identified from our institutional review board–approved institutional database. Patients reported to have tumors with KRAS mutations but with unknown specific mutations were excluded from the study. Patients who underwent only an ablative procedure without concurrent hepatic resection and patients who underwent incomplete palliative surgery (R2 resection) were excluded from analysis. Patients who received anti–epidermal growth factor receptor agents in the perioperative period were also excluded. The Johns Hopkins University institutional review board approved the study. No additional patient informed consent that was specific to this study was required given its retrospective nature.
Standard demographic and clinicopathologic data were collected on each patient including sex, age, disease status, tumor characteristics, operative details, perioperative status, type and time of chemotherapy, date of last follow-up, date and type of recurrence, and date of death. Primary tumor characteristics, including tumor location (colon vs rectum), American Joint Committee on Cancer T stage, tumor site (right vs left), and nodal status, were recorded. Number, size, distribution of the hepatic metastases, and presentation (synchronous vs meta-chronous) were also recorded. Tumor number and size were defined by the resection specimen. The largest lesion was used as the index lesion in the case of patients with multiple tumors. Information on treatment-related variables, such as type of procedure (resection vs resection plus ablation), type of hepatic resection, and margin status were also obtained. A major hepatectomy was defined as a resection of at least 3 Couinaud liver segments.23 Data on KRAS mutational status were also recorded. Patients with a KRAS mutation were classified according to the specific KRAS mutation (G12A, G12C, G12D, G12V, G12S, and G13D).
Perioperative mortality was calculated based on the number of patients who died within 90 days of the operation.24 Long-term clinical outcomes were obtained including data on recurrence and OS at last follow-up. Recurrence was defined as the presence of a biopsy-proven tumor showing colorectal adenocarcinoma cells or a lesion deemed suspicious on follow-up computed tomographic imaging in the setting of an elevated CEA level.
KRAS Mutation Analysis
As previously described,10 the extracted DNA was evaluated for the presence of the most common mutations of the KRAS (codons 12 and 13) genes. These regions of interest were amplified using polymerase chain reaction and the reaction product underwent agarose gel electrophoresis against known positive and negative controls to assess the presence and size of the amplified product. The polymerase chain reaction protocol settings used were initial denaturation at 95°C for 5 minutes, followed by 40 cycles of amplification at 95°C for 40 seconds, 57°C for 40 seconds, and 72°C for 40 seconds and a final elongation at 72°C for 10 minutes. For amplification of the codon 12/13 region of the KRAS gene, the oligonucleotide primers used were 5′-TCATTATTTTTATTATAAGGCCTGCTG-′3 (sense) and 5′-TTGGATCATATTCGTCCACAA-′3 (antisense). The amplified products were subsequently column-purified using a GeneJET Polymerase Chain Reaction Purification Kit (Fermentas ThermoScientific) and then sequenced.
Statistical Analysis
Demographic, clinicopathologic, and perioperative features of the study population were stratified according to the specific KRAS mutation status. Summary statistics for the population were presented as totals and frequencies for categorical variables or as median values with interquartile ranges (IQRs) for continuous variables. The differences between wild-type and mutant KRAS or between mutant KRAS groups were assessed by the χ2, t, and Mann-Whitney U tests, as appropriate. Overall survival for the study population and recurrence-free survival (RFS) was estimated using the Kaplan-Meier method calculated from the date of surgery, and the differences in OS and RFS were assessed with the log-rank test. The Cox proportional hazards regression model was used to evaluate the association of relevant clinicopathologic variables with prognosis. Clinicopathologic variables of known prognostic importance, such as age, sex, T stage, location of primary tumor (colon or rectum), disease-free interval, regional lymph node status, size of largest liver metastasis, number of lesions, bilateral lesions, preoperative CEA levels, preoperative or adjuvant chemotherapy, use of ablation, and final resection margin status, were tested and a backward stepwise elimination with a threshold of P = .20 was used to select variables in the final multivariable analysis model. All analyses were carried out with Stata version 12.0 (StataCorp) and a P value of less than .05 (2-tailed) was considered statistically significant.
Results
Demographic, Clinicopathologic, and Perioperative Characteristics
A total of 331 patients who underwent curative intent liver resection for CRLM at Johns Hopkins Hospital and who met the inclusion criteria were identified. Baseline characteristics of the population, stratified for presence of KRAS mutation and type of mutation, are summarized in Table 1. Overall, the median patient age was 50 years (IQR, 42–61 years) and most patients were men (n = 206; 62.2%). Most patients had a primary colonic tumor (n = 241; 72.8%), while about one-fourth of patients (n = 90; 27.2%) had a primary rectal tumor. Most patients had T3-T4 colorectal tumors (n = 239; 72.2%) and nodal metastasis (N 1–2: n = 203; 61.3%). The median preoperative CEA level was 7.6 ng/mL (IQR, 3.1–27.0 ng/mL; to convert to micrograms per liter, multiply by 1.0). Most patients had a synchronous CRLM presentation (n = 184; 55.6%). Regarding the extent of CRLM, most patients (n = 241; 72.8%) had unilateral disease with an average tumor burden of 2 metastases (IQR, 1–3). The median size of the largest metastatic liver lesion was 2.5 cm (IQR, 1.8–3.8 cm). At the time of surgery, liver resection involved either a minor (n = 189; 57.1%) or major (n = 142; 42.9%) hepatectomy; 62 (18.7%) underwent a concurrent ablative procedure. Most patients (n = 256; 77.3%) received an R0 resection.
Table 1.
Characteristics of Patients With CRLM According to KRAS Mutation Status
| No. (%)
|
|||||||
|---|---|---|---|---|---|---|---|
|
KRAS
|
Mutant KRAS
|
||||||
| Characteristic | All Patients | Wild Type | Mutant | P Value | Codon 12 | Codon 13 | P Value |
| Total No. of patients | 331 | 240 | 91 | 67 | 24 | ||
|
| |||||||
| Patient characteristics | |||||||
|
| |||||||
| Male | 206 (61.7) | 150 (62.5) | 56 (61.5) | .87 | 37 (55.2) | 19 (79.2) | .04 |
|
| |||||||
| Age, median (IQR), y | 50.0 (41.5–60.6) | 51.1 (42.3–61.2) | 48.5 (38.6–56.1) | .07 | 48.5 (38.6–56.0) | 47.6 (39.6–57.6) | .93 |
|
| |||||||
| Primary tumor characteristics | |||||||
|
| |||||||
| Rectal primary tumor | 90 (27.2) | 71 (29.6) | 19 (20.9) | .11 | 10 (14.9) | 9 (37.5) | .02 |
|
| |||||||
| T stage (n = 283) | |||||||
|
| |||||||
| T1 or T2 | 44 (15.5) | 33 (16.2) | 11 (13.9) | 8 (13.6) | 3 (15.0) | ||
|
| |||||||
| T3 or T4 | 239 (84.5) | 171 (83.8) | 68 (86.1) | 51 (86.4) | 17 (85.0) | ||
|
| |||||||
| Node-positive primary tumor | 203 (61.3) | 141 (58.8) | 62 (68.1) | .12 | 46 (68.7) | 16 (66.7) | .86 |
|
| |||||||
| Preoperative factors | |||||||
|
| |||||||
| CEA, median (IQR) | 7.6 (3.1–27.0) | 8.7 (3.8–29.7) | 5.1 (2.7–24.9) | .06 | 5.1 (2.7–24.0) | 4.9 (2.7–38.8) | .93 |
|
| |||||||
| Chemotherapy | 224 (67.7) | 163 (67.9) | 61 (67.0) | .88 | 43 (64.2) | 18 (75.0) | .33 |
|
| |||||||
| Disease-free interval <12 mo | 219 (66.2) | 156 (65.0) | 63 (69.2) | .47 | 46 (68.7) | 17 (70.8) | .84 |
|
| |||||||
| CRLM characteristics | |||||||
|
| |||||||
| Synchronous CRLM | 184 (55.6) | 129 (53.8) | 55 (60.4) | .27 | 38 (56.7) | 17 (70.8) | .23 |
|
| |||||||
| No. of CRLM, median (IQR) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | .48 | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | .73 |
|
| |||||||
| Size of largest CRLM, median (IQR) | 2.5 (1.8–3.8) | 2.5 (1.9–4.0) | 2.5 (1.5–3.8) | .23 | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | .86 |
|
| |||||||
| Bilateral disease | 90 (27.2) | 68 (28.3) | 22 (24.2) | .45 | 16 (23.9) | 6 (25.0) | .91 |
|
| |||||||
| Details of surgical procedure | |||||||
|
| |||||||
| Resection only | 269 (81.3) | 195 (81.2) | 74 (81.3) | .99 | 56 (83.6) | 18 (75.0) | .36 |
|
| |||||||
| Resection plus ablation | 62 (18.7) | 45 (18.8) | 17 (18.7) | .99 | 11 (16.4) | 6 (25.0) | .36 |
|
| |||||||
| Major hepatectomy | 142 (42.9) | 101 (42.1) | 41 (45.1) | .63 | 27 (40.3) | 14 (58.3) | .13 |
|
| |||||||
| R0 margin status | 256 (77.3) | 183 (76.2) | 73 (80.2) | .44 | 51 (76.1) | 22 (91.7) | .10 |
|
| |||||||
| Postoperative chemotherapy | 210 (63.4) | 152 (63.3) | 58 (63.7) | .95 | 44 (65.7) | 14 (58.3) | .52 |
|
| |||||||
| Recurrence | 178 (53.8) | 130 (54.2) | 48 (52.7) | .82 | 33 (49.3) | 15 (62.5) | .27 |
|
| |||||||
| Death | 156 (47.1) | 109 (45.4) | 47 (51.7) | .31 | 34 (50.8) | 13 (54.2) | .77 |
Abbreviations: CEA, carcinoembryonic antigen; CRLM, colorectal liver metastases; HR, hazard ratio; IQR, interquartile range. SI conversion factor: To convert CEA to micrograms per liter, multiply by 1.0.
KRAS Mutations
Among the 331 patients evaluated, the overall incidence of any KRAS mutations was 27.5% (n = 91). No correlation was found between the presence of a KRAS mutation and any specific clinicopathologic characteristic (Table 1). Among all patients with CRLM (n = 331), KRAS codon 12 and 13 mutations were detected in 91 (27.5%); KRAS codon 12 mutations were detected in 67 patients (20.2%) and KRAS codon 13 mutations were detected in 24 patients (7.3%). Among patients with KRAS codon 12 mutations (n = 67), G12V was observed in 22 cases (32.8%), G12D was observed in 25 cases (37.3%), G12C was observed in 6 cases (9.0%), G12S in 7 cases (10.4%), and G12A in 4 cases (6.0%). Among patients with KRAS codon 13 mutations (n = 24), G13D was observed in 23 cases (95.8%). Various clinicopathologic characteristics were assessed and compared according to KRAS status (Table 1). The presence of KRAS codon 13 mutations was more common among men and patients with rectal cancer compared with patients who had KRAS codon 12 mutations (both P < .05). No other differences were observed between patients with KRAS codon 12 mutations vs KRAS codon 13 mutations.
Recurrence-Free Survival
At a median follow-up of 27.4 months, most patients (n = 178; 53.8%) developed a recurrence. The median RFS for the entire cohort was 20.9 months; 1-, 3-, and 5-year RFS were 65.9%, 37.1%, and 32.6%, respectively. Median and 5-year RFS among patients with mutated KRAS were 18.9 months and 30.6%, respectively, compared with 21.3 months and 33.2%, respectively, for patients with wild-type KRAS (wtKRAS) (P = .57). Median and 5-year RFS among patients with mutations in codon 12 were 22.0 months and 33.9%, respectively, compared with 18.9 months and 21.1%, respectively, for patients with codon 13 mutations (P = .45). Among the 6 most common KRAS codon 12 and 13 mutations, no mutation was associated with worse RFS compared with wtKRAS cases.
Overall Survival
Median OS for the entire cohort was 51.8 months; 1-, 3-, and 5-year OS were 93.3%, 61.4%, and 43.1%, respectively. Median and 5-year OS among patients with mtKRAS were 32.4 months and 32.7%, respectively, compared with 58.5 months and 46.9%, respectively, for patients with wtKRAS (P = .02) (eFigure in the Supplement).
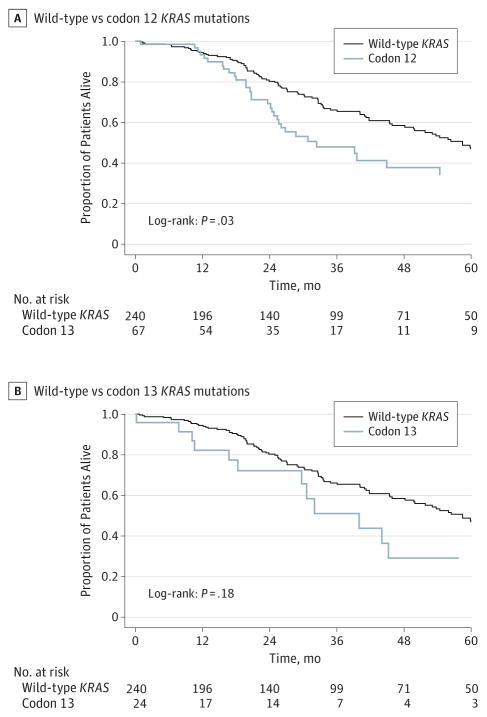
Patients with KRAS mutations had worse OS relative to patients with wtKRAS (32.4 months vs 58.5 months, respectively; P = .02). Of note, 5-year OS of patients with KRAS codon 12 and 13 mutations were 34.4% and 29.2%, respectively, compared with 46.9% for patients who had wtKRAS (P < .05 for codon 12) (Figure 1). On both Kaplan-Meier analysis (log-rank P = .03) and Cox regression analysis (univariate hazard ratio [HR], 1.54; 95% CI, 1.05–2.27; P = .03 and multivariate HR, 1.70; 95% CI, 1.13–2.55; P = .01; Table 2) patients with KRAS codon 12 mutations had a worse OS compared with patients with wt-KRAS. In contrast, KRAS codon 13 mutations were not associated with a worse prognosis compared with wtKRAS (HR, 1.47; 95% CI, 0.83–2.62) (Table 2). On univariable analysis, in addition to tumor and operative factors, such as primary tumor nodal metastasis (HR, 1.80; 95% CI, 1.24–2.62; P < .001), concurrent use of ablation (HR, 1.54; 95% CI, 1.08–2.20; P = .02), and R1 surgical margin status (HR, 1.55; 95% CI, 1.09–2.21; P = .02), codon 12 KRAS mutation (HR, 1.54; 95% CI, 1.05–2.27; P = .03) was associated with a worse long-term survival (all P < .05) (Table 2). On multivariable analysis, after controlling for other competing risk factors, codon 12 KRAS mutations (HR, 1.7; 95% CI, 1.13–2.55; P = .01) remained independently associated with a worse OS (Table 2).
Figure 1.
Overall Survival of the Study Group Stratified by Codon Location
Table 2.
Univariable and Multivariable Cox Proportional Hazard Analysis for Overall Survival
| Prognostic Factor | Hazard Ratio (95% CI) | |||
|---|---|---|---|---|
| Univariable | P Value | Multivariable | P Value | |
| KRAS | ||||
| Wild type | 1 [Reference] | 1 [Reference] | ||
| All codon 12 mutants | 1.54 (1.05–2.27) | .03 | 1.7 (1.13–2.55) | .01 |
| All codon 13 mutants | 1.47 (0.83–2.62) | .19 | 1.61 (0.87–2.97) | .13 |
| Age | 1.01 (0.99–1.02) | .38 | … | … |
| Female | 1.11 (0.80–1.54) | .52 | … | … |
| AJCC 7th edition T stage | ||||
| T1/T2 | 1 [Reference] | |||
| T3/T4 | 1 (0.62–1.60) | .99 | … | … |
| Location of tumor | ||||
| Colon | 1 [Reference] | |||
| Rectal | 1.08 (0.76–1.52) | .67 | … | … |
| Disease-free interval <12 mo | 1.06 (0.76–1.48) | .74 | … | … |
| Regional lymph node status | ||||
| Negative | 1 [Reference] | 1 [Reference] | ||
| Positive | 1.8 (1.24–2.62) | <.001 | 2.06 (1.39–3.04) | <.001 |
| Tumor size | 1.06 (0.97–1.15) | .20 | ||
| No. of lesions | 1.07 (0.99–1.15) | .09 | … | … |
| Bilobar lesions | 1.06 (0.75–1.50) | .75 | … | … |
| CEA ≥ 30 ng/mL | 1.00 (1.00–1.00) | NA | … | … |
| Chemotherapy | ||||
| Preoperative | 1.19 (0.82–1.74) | .36 | … | … |
| Adjuvant | 0.84 (0.61–1.17) | .30 | … | … |
| Ablation | 1.54 (1.08–2.20) | .02 | 1.81 (1.25–2.62) | .002 |
| Margin | ||||
| R0 | 1 [Reference] | 1 [Reference] | ||
| R1 | 1.55 (1.09–2.21) | .02 | 1.73 (1.17–2.57) | .01 |
Abbreviations: AJCC, American Joint Committee on Cancer; CEA, carcinoembryonic antigen; NA, not applicable; ellipses, not included in the multivariable analysis.
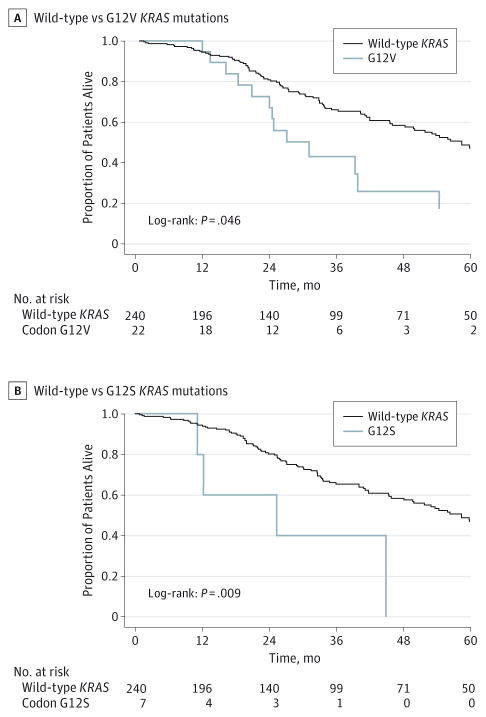
On further analysis of the 6 most common KRAS codon 12 and 13 mutations, the G12V and G12S mutations were the point mutations most associated with worse long-term prognosis. Specifically, G12V (n = 22) (HR, 1.78; 95% CI, 1.00–3.17; P = .05) and G12S (n = 7) (HR, 3.33; 95% CI, 1.22–9.10; P = .02) mutations were associated with a roughly 2- to 3-fold increased risk for long-term death compared with wtKRAS (Figure 2; Table 3). In addition, in the subgroup of patients who recurred after the curative intent liver resection, the presence of the KRAS mutations G12V (HR, 2.96; 95% CI, 1.32–6.61; P = .01), G12S (HR, 4.91; 95% CI, 1.52–15.8; P = .01), and G12C (HR, 6.74; 95% CI, 2.05–22.2; P = .002) were associated with an increased risk for death after recurrence compared with patients with wtKRAS who recurred (Table 3).
Figure 2.
Overall Survival of the Study Group Stratified by Mutation
Table 3.
Univariable Cox Proportional Hazard Analysis for Overall Survival According to KRAS Mutation in the Whole Cohort and Recurred Group
| KRAS | Univariate Analysis, Hazard Ratio (95% CI) | |||
|---|---|---|---|---|
| Whole Cohort | P Value | Recurred Group | P Value | |
| Wild type | 1 [Reference] | 1 [Reference] | ||
| G12D | 1.14 (0.62–2.13) | .67 | 1.78 (0.85–3.73) | .13 |
| G12V | 1.78 (1.00–3.17) | .05 | 2.96 (1.32–6.61) | .01 |
| G12A | 0.86 (0.12–6.20) | .88 | NA | NA |
| G12C | 2.01 (0.74–5.46) | .17 | 6.74 (2.05–22.2) | .002 |
| G12S | 3.33 (1.22–9.10) | .02 | 4.91 (1.52–15.8) | .01 |
| G13D | 1.36 (0.75–2.47) | .31 | 1.34 (0.67–2.70) | .41 |
Abbreviation: NA, not applicable.
Discussion
Various scoring systems that include different clinicopathologic variables have been proposed to stratify patient prognosis after hepatic resection for CRLM.25,26 However, owing to inconsistent predictive power and lack of reproducibility, these scoring systems are becoming less relevant in clinical practice.27 Attention has turned to the use of biological and molecular markers to predict the long-term outcome of patients undergoing surgery for CRLM.4 Currently, one of the most commonly available biomarkers in the treatment of patients with CRLM is KRAS. The frequency and prognostic impact of KRAS mutation status have been described in both primary and metastatic colorectal cancer4; however, the prognostic implications of specific mutations of the KRAS gene are still not well defined. Given this, we sought to evaluate the impact of different KRAS point mutations on recurrence and survival of patients undergoing curative intent liver resection for CRLM. To our knowledge, this is the first study to evaluate the prognostic effects of different point mutations in KRAS codons among patients who have undergone CRLM resection. Using a comprehensive analysis of 331 patients who underwent liver resection for CRLM, we reported worse OS among patients with KRAS codon 12 mutations and, specifically, those patients with either a KRAS G12V or a G12S mutation. In contrast, patients with a KRAS codon 13 mutation did not have a worse prognosis. These findings suggest that KRAS mutation status has prognostic implications. More importantly, stratification of patients undergoing liver resection for CRLM based on specific KRAS mutations may be even more informative with regard to long-term outcome.
Similar to previous studies reporting an incidence of KRAS mutations ranging from 35% to 45%,5,28–30 we found KRAS mutations in 27.5% of tumor specimens from patients undergoing hepatic resection for CRLM. KRAS codon 12 and 13 mutations were detected in about one-third of patients undergoing resection of CRLM, with KRAS codon 12 mutations being more common (20.2%) than KRAS codon 13 mutations (7.3%). Among patients with KRAS codon 12 mutations, G12V and G12D were noted in about one-quarter of specimens, while mutations in G12C, G12S, and G12A were much less common, occurring in less than 10% of patients. Among those patients who did have a KRAS codon 13 mutation, G13D was the most commonly observed mutation, occurring in about 30% of patients. Interestingly, the incidence of different KRAS codon mutations noted in the current study was consistent with previous data reported by Thierry and colleagues.31 Specifically, Thierry et al31 reported on specific KRAS point mutations among patients with metastatic colorectal cancer and noted G12V (20.5%), G12D (25.6%), and G13D (28.2%) as the most common point mutations in metastatic colorectal cancer, whereas G12A(7.7%),G12C(5.1%), and G12S(12.8%) were less common. Similar frequencies of KRAS mutations have been reported in the Sanger COSMIC database: G12V (21.9%–24.4%), G12D (33.5%–34.4%), G13D (18.9%–19.2%), G12A (6.2%–6.6%), G12C (7.9%), and G12S (4.9%–5.7%).32
In the present study, we found that patients with KRAS codon 12 mutations had a roughly 70% increased risk for long-term death compared with patients with wtKRAS (HR, 1.70; 95% CI, 1.13–2.55; P = .01). Specifically, among the 6 most common KRAS codon 12 and 13 mutations, G12V (n = 22) and G12S (n = 7) mutations were particularly associated with higher mortality (G12V: HR, 1.78; 95% CI, 1.00–3.17; P = .05 and G12S: HR, 3.33; 95% CI, 1.22–9.10; P = .02). In addition, in the subgroup of patients who recurred after curative intent liver resection, the effect of KRAS mutation was even more pronounced among patients with G12V (HR, 2.96; 95% CI, 1.32–6.61; P = .01), G12C (HR, 6.74; 95% CI, 2.05–22.2; P = .002), and G12S (HR, 4.91; 95% CI, 1.52–15.8; P = .01) mutations. These findings are consistent with those by Imamura et al,22 who showed that KRAS codon 12 mutations and, especially, G12V are associated with a worse OS compared with wtKRAS. In fact, the authors suggested that G12V was associated with a more aggressive tumor phenotype in metastatic colorectal cancer in general. The potential differential effects of specific KRAS mutations on clinical outcomes have also been demonstrated in a large multicenter study of 2721 patients with metastatic colorectal cancer.5 Specifically, Andreyev et al5 reported that only the G12V point mutation was a predictive factor of worse OS among patients with advanced colorectal cancer, suggesting a more aggressive behavior of tumors with G12V mutation.5 The aggressive behavior of the G12V mutation has been explained by Al-Mulla et al,33 who showed that the G12V mutation produces proteins that behave differently than other mutated KRAS proteins. The G12V mutation leads to a reduction of GT-Pase activity and affinity for GTPase-activating proteins, which in turn prevents their activation.33,34 Moreover, Guerrero et al35 reported that KRAS codon 12 mutations confer a more aggressive tumor phenotype than codon 13 mutations by an alteration in the threshold for apoptosis induction. Further experimental data suggest that, among the different KRAS codon 12 mutations, G12V mutation is characterized by more potent transforming ability than the others and is associated with a more aggressive biologic phenotype.36
The current study had several limitations that should be considered when interpreting the findings. As with all retrospective studies, undoubtedly there was some selection bias. Although fairly representative as evidenced by the specific mutation incidences, the sample size was relatively small for some groups. Although patients who received perioperative anti–epidermal growth factor receptor agents were excluded from the study, a remote use of those agents cannot be definitely excluded. Finally, we performed KRAS testing forsomatic mutations involving only codons 12 and 13 and wedid not investigate for less commonmutations involving codons 61 and 146.15,37
Conclusions
KRAS codon 12 mutations, and specifically G12V and G12S mutations, were associated with worse prognosis after resection of CRLM, especially among those patients who experienced a recurrence following surgery. Specific mutations in codon 12, such as G12V and G12S, may be helpful in improving clinical decision-making and prognostic models based on both well-known clinicopathological characteristics and molecular features. In addition, our data demonstrate that different mutations, even in a single gene, can shape distinctive biologic behaviors, further supporting the unique tumor principle.38,39
Supplementary Material
Footnotes
Conflict of Interest Disclosures: None reported.
Disclaimer: Dr Pawlik is a deputy editor of JAMA Surgery. He was not involved in the evaluation or decision to accept this article for publication.
Author Contributions: Dr Pawlik had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drs Margonis and Kim contributed equally to this article.
Study concept and design: Margonis, Kim, Spolverato, Gupta, Cosgrove, Anders, Choti, Pawlik.
Acquisition, analysis, or interpretation of data: Margonis, Kim, Spolverato, Ejaz, Gupta, Karagkounis, Choti.
Drafting of the manuscript: Margonis, Spolverato, Gupta.
Critical revision of the manuscript for important intellectual content: Margonis, Kim, Spolverato, Ejaz, Cosgrove, Anders, Karagkounis, Choti, Pawlik. Statistical analysis: Kim, Spolverato, Ejaz.
Administrative, technical, or material support: Spolverato, Gupta, Cosgrove, Karagkounis, Pawlik. Study supervision: Anders, Choti, Pawlik.
References
- 1.de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009;250(3):440–448. doi: 10.1097/SLA.0b013e3181b4539b. [DOI] [PubMed] [Google Scholar]
- 2.Pawlik TM, Choti MA. Surgical therapy for colorectal metastases to the liver. J Gastrointest Surg. 2007;11(8):1057–1077. doi: 10.1007/s11605-006-0061-3. [DOI] [PubMed] [Google Scholar]
- 3.Gleisner AL, Choti MA, Assumpcao L, Nathan H, Schulick RD, Pawlik TM. Colorectal liver metastases: recurrence and survival following hepatic resection, radiofrequency ablation, and combined resection-radiofrequency ablation. Arch Surg. 2008;143(12):1204–1212. doi: 10.1001/archsurg.143.12.1204. [DOI] [PubMed] [Google Scholar]
- 4.Spolverato G, Ejaz A, Azad N, Pawlik TM. Surgery for colorectal liver metastases: the evolution of determining prognosis. World J Gastrointest Oncol. 2013;5(12):207–221. doi: 10.4251/wjgo.v5.i12.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” Study. J Natl Cancer Inst. 1998;90(9):675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 6.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 7.Finkelstein SD, Sayegh R, Christensen S, Swalsky PA. Genotypic classification of colorectal adenocarcinoma: biologic behavior correlates with K-ras-2 mutation type. Cancer. 1993;71(12):3827–3838. doi: 10.1002/1097-0142(19930615)71:12<3827::aid-cncr2820711207>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 8.Linardou H, Briasoulis E, Dahabreh IJ, et al. All about KRAS for clinical oncology practice: gene profile, clinical implications and laboratory recommendations for somatic mutational testing in colorectal cancer. Cancer Treat Rev. 2011;37(3):221–233. doi: 10.1016/j.ctrv.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 9.McDermott U, Longley DB, Johnston PG. Molecular and biochemical markers in colorectal cancer. Ann Oncol. 2002;13(suppl 4):235–245. doi: 10.1093/annonc/mdf665. [DOI] [PubMed] [Google Scholar]
- 10.Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119(23):4137–4144. doi: 10.1002/cncr.28347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose JS, Serna DS, Martin LK, et al. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinoma. Cancer. 2012;118(24):6243–6252. doi: 10.1002/cncr.27666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stremitzer S, Stift J, Gruenberger B, et al. KRAS status and outcome of liver resection after neoadjuvant chemotherapy including bevacizumab. Br J Surg. 2012;99(11):1575–1582. doi: 10.1002/bjs.8909. [DOI] [PubMed] [Google Scholar]
- 13.Teng HW, Huang YC, Lin JK, et al. BRAF mutation is a prognostic biomarker for colorectal liver metastasectomy. J Surg Oncol. 2012;106(2):123–129. doi: 10.1002/jso.23063. [DOI] [PubMed] [Google Scholar]
- 14.Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2013;258(4):619–626. doi: 10.1097/SLA.0b013e3182a5025a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edkins S, O’Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5(8):928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata Y, Abe M, Kobayashi K, et al. Glycine to aspartic acid mutations at codon 13 of the c-Ki-ras gene in human gastrointestinal cancers. Cancer Res. 1990;50(3):480–482. [PubMed] [Google Scholar]
- 17.Shaw P, Tardy S, Benito E, Obrador A, Costa J. Occurrence of Ki-ras and p53 mutations in primary colorectal tumors. Oncogene. 1991;6(11):2121–2128. [PubMed] [Google Scholar]
- 18.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205(12):858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Ye Y, Sun H, Shi G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother Pharmacol. 2013;71(1):265–272. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 20.Peeters M, Douillard JY, Van Cutsem E, et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31(6):759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 21.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30(29):3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 22.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couinaud C. Liver anatomy: portal (and suprahepatic) or biliary segmentation. Dig Surg. 1999;16(6):459–467. doi: 10.1159/000018770. [DOI] [PubMed] [Google Scholar]
- 24.Mayo SC, Shore AD, Nathan H, et al. Refining the definition of perioperative mortality following hepatectomy using death within 90 days as the standard criterion. HPB (Oxford) 2011;13(7):473–482. doi: 10.1111/j.1477-2574.2011.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordlinger B, Guiguet M, Vaillant JC, et al. Association Française de Chirurgie. Surgical resection of colorectal carcinoma metastases to the liver: a prognostic scoring system to improve case selection, based on 1568 patients. Cancer. 1996;77(7):1254–1262. [PubMed] [Google Scholar]
- 26.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230(3):309–318. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George B, Kopetz S. Predictive and prognostic markers in colorectal cancer. Curr Oncol Rep. 2011;13(3):206–215. doi: 10.1007/s11912-011-0162-3. [DOI] [PubMed] [Google Scholar]
- 28.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 30.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS Trial. J Clin Oncol. 2009;27(35):5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 31.Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 32.Wellcome Trust Sanger Institute. [Accessed September 26, 2014];COSMIC v72. http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 33.Al-Mulla F, Milner-White EJ, Going JJ, Birnie GD. Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J Pathol. 1999;187(4):433–438. doi: 10.1002/(SICI)1096-9896(199903)187:4<433::AID-PATH273>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 34.Bollag G, McCormick F. Intrinsic and GTPase-activating protein-stimulated Ras GTPase assays. Methods Enzymol. 1995;255:161–170. doi: 10.1016/s0076-6879(95)55020-8. [DOI] [PubMed] [Google Scholar]
- 35.Guerrero S, Casanova I, Farré L, Mazo A, Capellà G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60(23):6750–6756. [PubMed] [Google Scholar]
- 36.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312(5989):71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 37.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12(6):621–628. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.