Abstract
Background
The periodontal disease is caused by a set of inflammatory disorders characterized by periodontal pocket formation that lead to tooth loss if untreated. The proteomic profile and related molecular conditions of pocket tissue in periodontally-affected patients are not reported in literature. To characterize the proteomic profile of periodontally-affected patients, their interproximal periodontal pocket tissue was compared with that of periodontally-healthy patients. Pocket-associated and healthy tissue samples, harvested during surgical therapy, were treated to extract the protein content. Tissues were always collected at sites where no periodontal-pathogenic bacteria were detectable. Proteins were separated using two-dimensional gel electrophoresis and identified by liquid chromatography/mass spectrometry. After identification, four proteins were selected for subsequent Western Blot quantitation both in pathological and healty tissues.
Results
A significant unbalance in protein expression between healthy and pathological sites was recorded. Thirty-two protein spots were overall identified, and four proteins (S100A9, HSPB1, LEG7 and 14-3-3) were selected for Western blot analysis of both periodontally-affected and healthy patients. The four selected proteins resulted over-expressed in periodontal pocket tissue when compared with the corresponding tissue of periodontally-healthy patients. The results of Western blot analysis are congruent with the defensive and the regenerative reaction of injured periodontal tissues.
Conclusions
The proteomic analysis was performed for the first time directly on periodontal pocket tissue. The proteomic network highlighted in this study enhances the understanding of periodontal disease pathogenesis necessary for specific therapeutic strategies setting.
Keywords: Proteome analysis, Periodontitis, Two-dimensional gel electrophoresis, Protein identification, LC-MS/MS
Background
The periodontal ligament surrounds the tooth root, connects it to its bony socket constituting a fibrous joint named gomphosis. It contains a large variety of cells and tissues, including immune and stem cells [1, 2]. The periodontitis is a set of inflammatory disorders characterized by gingival and periodontal inflammation, periodontal attachment loss, and alveolar bone resorption, following to periodontal pocket development [3–6].
The periodontal-pathogenic microbiome ecosystems are the most proven risk factor of periodontal disease. Pathogens adhere to and grow on the tooth surfaces, and the inappropriate inflammatory response causes the loss of periodontal attachment and alveolar bone, giving rise to the periodontal pocket, the typical expression of periodontitis, that lead to tooth loss, if untreated.
Periodontitis diagnosis is based on clinical assessment only, inspecting the soft gum tissues around the teeth with a probe (i.e. a clinical examination) in order to detect interproximal attach level-loss and periodontal pocket depth, in the absence of a reliable pathogenic check based on appropriate interpretation of inflammation [5, 7, 8].
Recently, the role of cytokines and other protein mediators of inflammation erupted in all their importance [9]. A modern pathogenetic model incorporating gene, protein, and metabolite data into dynamic biological processes is based on a multilevel framework that include disease-initiating and -resolving mechanisms that are regulated by innate and environmental factors [10].
Now, we can describe more effectively the basic elements of a new model of pathogenesis using emerging genomic, proteomic, and metabolomic techniques [10, 11]. Recent progress in tissue isolation, protein separation, quantification and sequence analysis utilizing novel proteomic techniques promises to bring periodontal physiology and pathology into a new era. A list of inflammation-involved proteins, cytokines, matrix expression and cellular proteins in the periodontal tissues is not currently available. Studies on periodontal diseases utilizing proteomic analysis have been performed on saliva or crevicular fluid samples [12–15], peripheral blood [16–18] or periodontal plaque samples [19], but not on the pathologic tissue of the periodontal pocket, which is the key lesion of the periodontal disease.
To overcome the lack of data, studies assessing the proteomic profile of periodontal pocket tissue and evaluating the molecular characteristics of the periodontally affected patient, are needed.
The aim of this work was to compare the proteomic profile of the pathologic interproximal gingival pocket tissue with that of interproximal gingival healthy tissue, obtained from sites where no periodontal-pathogenic bacteria were detectable.
Results
Fifteen subjects (T, test group), 3 males and 12 females, ranging in age from 20 to 64 years, average 42.82 ± 13.2 (m ± SD), and fifteen periodontally healthy subjects (C, control group), 6 males and 9 females, ranging in age from 19 to 60 years, average 44.90 ± 11.55, fulfilled study requirements (Table 1). T subjects underwent to the periodontal resective treatment, while C subjects underwent to the crown lengthening surgical treatment. All subjects followed a stringent post-operative supportive care program and achieved satisfactory clinical outcomes.
Table 1.
Systemic and specific adopted inclusion criteria
| Systemic Inclusion Criteria | Local Inclusion Criteria |
|---|---|
| Absence of relevant medical conditions: Medical history of good health (particularly ruling out bone disease, uncontrolled or poorly controlled diabetes, unstable or life-threatening conditions, or requiring antibiotic prophylaxis were excluded). |
Defect anatomy: Presence of at least one intrabony defect in patient with periodontal disease. Excessive gingival display or gingival margin asymmetries required a surgical correction in periodontally-healthy patients |
| Smoking status: Non-smokers and without a story of alcohol abuse. | Good oral hygiene: full-mouth plaque score (FMPS) ≤20 % |
| Compliance: only patients showing high levels of compliance (as assessed during the cause-related phase of therapy) were selected. | Low level of residual infection: full-mouth bleeding score (FMBS) ≤20 %. |
| Pregnancy or lactation and underage were excluded. | Endodontic status: Experimental teeth had to be vital or properly treated with root canal therapy. |
2DE (Two-dimensional gel electrophoresis) and LC-MS/MS (Liquid Chromatography-tandem mass spectrometry) analysis were performed for T and C gingival tissue samples. Figure 1 shows representative 2DE gel images of T and C tissues and the 32 protein spots identified. Primary accession number, entry name of UniProt database, MW (Molecular Weight), highest score, matches, sequences and emPAI (Exponentially Modified Protein Abundance Index) for each identified protein are reported in Table 2. Web-based bioinformatics tools (iPROClass and CateGOrize) were employed to investigate all potential localizations, molecular functions and biological processes of the identified proteins (Fig. 2). Intracellular proteins represented the most abundant population (77.3 %). Among them, 18.6 % of protein annotations were recognized as belonging to cytoplasm, 5.6 % to cytoskeleton, and nucleus 8.8 %. Other protein annotations were recognized, with smaller fractions, as belonging to other cellular component such as mitochondrion, vacuole or plasma membrane (Fig. 2a). The reported function of the identified proteins suggests that they are mainly involved in binding (46.8 %), protein binding (15.3 %), and catalytic activity (8.8 %). Other relevant functions are nucleic acid binding (6.6 %), enzyme regulator activity (4.4 %), structural molecule activity (4.4 %), antioxidant activity, lipid binding (2.9 %), nucleotide binding (2.2 %) and transferase activity (1.5 %) (Fig. 2b). In summary, the identified proteins are mainly involved in metabolism (31,9 %), transport (13,3 %) and cell organization and biogenesis (12 %) (Fig. 2c).
Fig. 1.
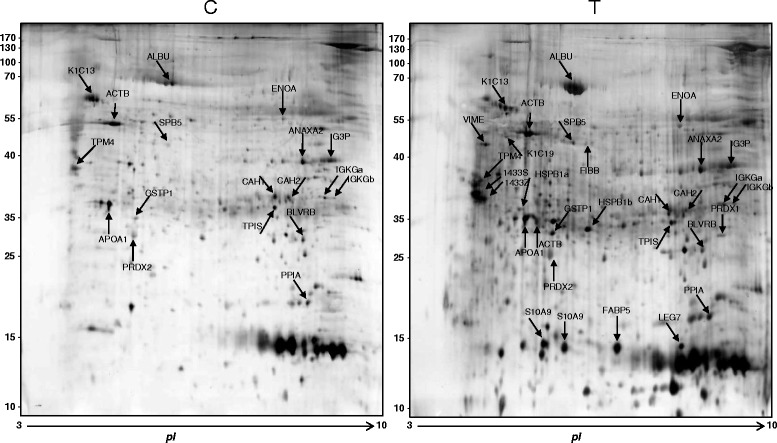
2DE gel of Control and Test tissues. Proteins identified by LC-MS/MS are indicated by Entry name (ID) and pointed by arrows. Control = healthy gingival tissue, Test = periodontal pocket tissues
Table 2.
List of identified proteins in periodontal pocket tissues
| AC Numbera) | Entry Nameb) | Score | Mass (kDa) | pI | Matchesc) | Significant matches | Sequencesd) | Significant sequences | emPAIe) | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| P02768 | ALBU_HUMAN | 5019 | 71317 | 5.92 | 435 | 310 | 43 | 40 | 6.59 | Serum albumin |
| P13646 | K1C13_HUMAN | 2745 | 49900 | 4.91 | 247 | 140 | 31 | 23 | 4.99 | Keratin, type I cytoskeletal 13 |
| P19013 | K2C4_HUMAN | 1509 | 57649 | 6.25 | 176 | 101 | 25 | 18 | 2.21 | Keratin, type II cytoskeletal 4 |
| Q01469 | FABP5_HUMAN | 1050 | 15497 | 6.60 | 95 | 44 | 21 | 12 | 16.08 | Fatty acid-binding protein, epidermal |
| P01834 | IGKC_HUMAN | 829 | 11773 | 5.58 | 65 | 51 | 5 | 5 | 6.66 | Ig kappa chain C region |
| P01834 | IGKC_HUMAN | 771 | 11773 | 5.58 | 62 | 47 | 5 | 5 | 5.11 | Ig kappa chain C region |
| P06733 | ENOA_HUMAN | 757 | 47481 | 7.01 | 67 | 40 | 22 | 15 | 2.43 | Alpha-enolase |
| P31947 | 1433S_HUMAN | 592 | 27871 | 4.68 | 88 | 49 | 19 | 12 | 3.12 | 14-3-3 protein sigma |
| P08727 | K1C19_HUMAN | 492 | 44079 | 5.05 | 51 | 27 | 14 | 9 | 0.78 | Keratin, type I cytoskeletal 19 |
| P00915 | CAH1_HUMAN | 403 | 28909 | 6.59 | 46 | 29 | 8 | 6 | 1.39 | Carbonic anhydrase 1 |
| P07355 | ANXA2_HUMAN | 382 | 38808 | 7.57 | 31 | 17 | 12 | 7 | 0.74 | Annexin A2 |
| P04406 | G3P_HUMAN | 377 | 36201 | 5.43 | 20 | 19 | 5 | 4 | 0.69 | Glyceraldehyde-3-phosphate dehydrogenase |
| Q06830 | PRDX1_HUMAN | 358 | 22324 | 8.27 | 53 | 26 | 13 | 6 | 1.10 | Peroxiredoxin-1 |
| P63104 | 1433Z_HUMAN | 317 | 27899 | 4.7 | 43 | 17 | 14 | 8 | 2.31 | 14-3-3 protein zeta/delta |
| P67936 | TPM4_HUMAN | 282 | 28619 | 4.67 | 51 | 27 | 19 | 14 | 3.41 | Tropomyosin alpha-4 chain |
| P02647 | APOA1_HUMAN | 260 | 30759 | 5.56 | 45 | 22 | 16 | 9 | 5.56 | Apolipoprotein A-I |
| P32119 | PRDX2_HUMAN | 223 | 22049 | 5.66 | 24 | 15 | 9 | 8 | 2.10 | Peroxiredoxin-2 |
| P06702 | S10A9_HUMAN | 217 | 13291 | 5.71 | 42 | 20 | 8 | 4 | 2.94 | Protein S100-A9 |
| P47929 | LEG7_HUMAN | 207 | 15123 | 7.03 | 59 | 22 | 7 | 7 | 3.74 | Galectin-7 |
| P62937 | PPIA_HUMAN | 197 | 18229 | 7.68 | 11 | 7 | 4 | 3 | 0.97 | Peptidyl-prolyl cis-trans isomerase A |
| P02675 | FIBB_HUMAN | 183 | 55892 | 8.54 | 5 | 3 | 5 | 3 | 0.27 | Fibrinogen beta chain |
| P04792 | HSPB1_HUMAN | 177 | 22826 | 5.98 | 27 | 10 | 9 | 5 | 0.83 | Heat shock protein beta-1 |
| P60709 | ACTB_HUMAN | 144 | 42052 | 5.29 | 55 | 23 | 13 | 7 | 0.67 | Actin, cytoplasmic 1 |
| P04792 | HSPB1_HUMAN | 132 | 22826 | 5.98 | 38 | 14 | 7 | 3 | 0.69 | Heat shock protein beta-1 |
| P09211 | GSTP1_HUMAN | 124 | 23569 | 5.43 | 6 | 3 | 2 | 1 | 0.30 | Glutathione S-transferase P |
| P60174 | TPIS_HUMAN | 118 | 31057 | 6.45 | 17 | 9 | 5 | 4 | 0.43 | Triosephosphate isomerase |
| P30043 | BLVRB_HUMAN | 118 | 22219 | 7.13 | 9 | 5 | 3 | 1 | 0.28 | Flavin reductase (NADPH) |
| P36952 | SPB5_HUMAN | 80 | 42530 | 5.72 | 4 | 3 | 2 | 2 | 0.16 | Serpin B5 |
| Q9BYX7 | ACTBM_HUMAN | 76 | 41989 | 5.29 | 2 | 2 | 2 | 2 | 0.24 | Putative beta-actin-like protein 3 |
| P08670 | VIME_HUMAN | 71 | 53676 | 5.06 | 37 | 5 | 10 | 3 | 0.20 | Vimentin |
| P06702 | S10A9_HUMAN | 54 | 13291 | 5.71 | 17 | 6 | 3 | 2 | 0.83 | Protein S100-A9 |
| P00918 | CAH2_HUMAN | 53 | 29285 | 8.15 | 12 | 6 | 3 | 2 | 0.33 | Carbonic anhydrase 2 |
a)Primary accession number from UniProt database
b)Primary entry name from UniProt database
c)The number of peptides that matched the identified proteins
d)The number of distinct sequences
e)Exponentially Modified Protein Abundance Index
Fig. 2.
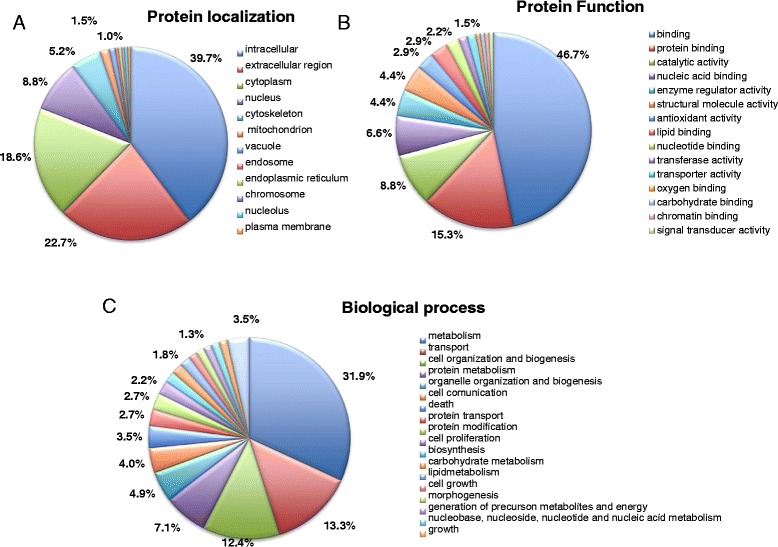
Pie charts describe the distribution of protein localization (a), main biological functions (b) and biological processes (c) of identified proteins of periodontal pocket tissue by GO analysis
PDQuest spot intensity quantification identified four proteins expressed only in the pathological tissue, as shown in Fig. 3, that were selected for subsequent Western blot analysis in both T and C subjects.
Fig. 3.
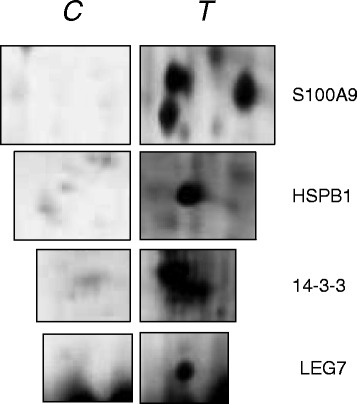
Detailed protein spots, differentially expressed in C and T specimens
Representative Western blot images for S100A9, HSPB1, 14-3-3 and LEG7 and β-actin are reported in Fig. 4. Densitometric analyses of Western blot images, normalized to β-actin expression (Fig. 5) confirmed that in C specimens (control tissue), the S100A9, 14-3-3, LEG7 and HSPB1 proteins were not expressed. In pathological tissue (T specimens), the intensity signal for S100A9 and HSPB1 was much higher than the intensity signal of 14-3-3 and LEG7 proteins.
Fig. 4.
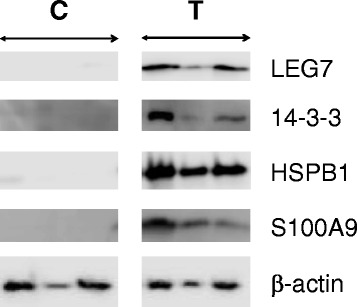
Representative images of Western blot analyses in C and T specimens for LEG7, 14-3-3, HSPB1, S100A9 and β-actin. The β-actin was utilized as reference for protein samples loading and integrity
Fig. 5.
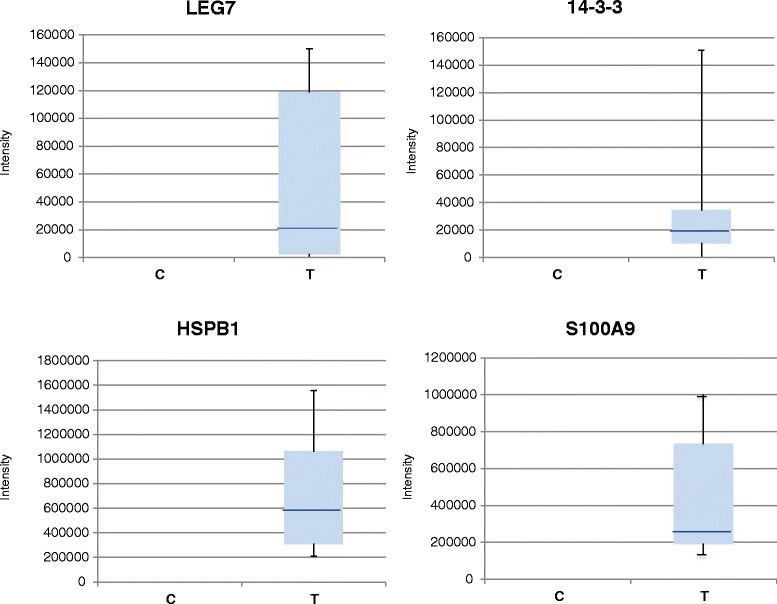
Densitometric analyses of western blot images. Expression level signals are relative to β-actin expression
Discussion
Very little is know on the proteomic analysis of the periodontal pocket tissue [6]. Most proteomic studies have been performed on gingival crevicular fluid, saliva or blood serum, since the non-invasive nature of the collection and availability [20–23]. These studies assume the existence of a direct correlation between the periodontal disease and the biological source to be tested, however there is no direct evidence supporting such assumption. Actually, the periodontal pocket is certainly the anatomo-pathological lesion signifying for an active periodontal disease [7, 8]. Therefore, the periodontal pocket tissue is the only biologic material in which proteomic analysis enables the correct molecular assessment of the periodontal disease. Despite genomic, transcriptomic and proteomic research performed up to now, there are no available biomarkers for periodontitis diagnosis, prognosis and treatment indication giving assistance to the clinician in the disease management. Excluding the studies performed on biologic material different from periodontal tissue, some genomic studies carried out on the gingival tissue showed an association of specific gene polymorphism to periodontitis [24–26].
In our study, proteomics was applied to comparatively analyze the protein content of interproximal tissues of healthy and periodontally affected patients, with the aim to identify differentially expressed proteins. The presence of common periodontopathogenic bacteria was excluded, focusing on disease progression related to the prevailing stimulus, avoiding in this way the possible straining effect caused by periodontopathogenic bacteria on the local protein content. In this way we could discriminate the individual component of the periodontal disease.
The differential expression level observed in 2DE, in pathological and healthy tissues, of 4 proteins, S100A9, HSPB1, LEG7 and 14-3-3, was confirmed by Western blot analysis.
S100A9, is a calcium- or zinc-binding protein involved in the regulation of inflammatory processes and immune response [27], predominantly found as calprotectin (S100A8/S100A9 etherodimer). S100A9 contribute to homeostatic processes that include cytoskeletal rearrangements during trans-endothelial migration of pro-inflammatory phagocytes, binding to receptors as Toll-like receptor 4 and receptor for advanced glycation end products, antimicrobial, oxidant-scavenging and apoptosis-inducing activities [28, 29]. In extracellular fluid, increased levels of S100A9/S100A8 etherodimer were reported in numerous inflammation-associated conditions, such as rheumatoid arthritis, Crohn’s Disease, colorectal cancer and in GCF (Gingival Crevicular Fluid) of patients suffering from gingivitis and periodontitis [30, 31]. The S100A9/S100A8 etherodimer concentration was correlated with periodontal markers of inflammation such as pocket probing depth or gingival bleeding [32, 33]. S100A9 was identified in saliva and proposed as a potential marker to monitor the progression of orthodontic treatment [33]. However, the Authors showed an apparent down-regulation of S100A9 protein, suggesting that this protein could not be involved during bone resorption in orthodontic tooth movement but it was implicated in inflammation. This protein also promotes apoptosis and modulate the inflammatory response in periodontal ligament cells so its down-regulation could suggest a suppression of inflammation [33, 34].
HSPB1 synthesis increases in response to a variety of stresses (e.g. elevated temperatures, heavy metals, toxins, oxidants, bacterial and viral infections) in order to minimize the deleterious consequences of these stimuli and provide the maximal cytoprotective effect [35–39]. In oral tissues, HSPB1 was localized in fibroblasts, odontoblasts, osteoblasts, epithelial cells, endothelial cells of the vascular wall of the dental pulp, and cells of the periodontal ligament [37, 40]. HSPB1 is also a procollagen-binding protein involved in the biosynthesis of type I collagen and major bone extracellular matrix [41]. HSPB1 could be a potential target for the periodontal regeneration process related to cell migration, cytoskeleton maintaining and tissue preservation, also through the modulation of the immune system, and its under-expression induces differentiation abortion, in relation to cell death by apoptosis [40].
Galectin-7 (LEG7) is associated with epithelial cell migration and accelerates the re-epithelialization of wounds. In particular, LEG7 expression contributes to the tissue remodeling processes following tissue damage that involves apoptotic cell death [42]; a defective LEG7 expression could impair the healing processes. LEG7 could function in the maintenance of the normal phenotype of epithelial cell and is activated by a wide range of cellular stresses including UV and γ irradiation. LEG7 protects cell from death by functioning inside the cell and interacting with intracellular proteins. Finally, LEG7 belongs to a protein family promoting healing processes and favor immune responses [43]. HSPB1 and LEG7 would seem to be part of a protein network that play an important role in controlling cell and tissue damage, in moderating the inflammation and destructive immune response. The lack of the previously described multi-protective effect of these proteins, particularly in the presence of inflammation, hinders cells to protect themselves against the cytotoxicity of inflammatory mediators [44], increases their susceptibility to necrotic cell death [45] and probably does not allow an efficient immune response against the noxa [46].
The 14-3-3 protein sigma and zeta/delta are regulatory phosphorserine/threonine binding proteins involved in the control of several cellular events, including cell cycle checkpoint, connective tissue remodeling, apoptosis signaling, Toll-like receptor activation and TNF production during inflammation response [47, 48]. In particular the 14-3-3 sigma induces up-regulation of differentiation, down-regulation of cell proliferation and collagenase induction as matrix metalloproteinase-1 [47, 49, 50]. Wu et al. [12] found decreased 14-3-3 protein sigma in saliva of subjects with generalized aggressive periodontitis, and, analyzing the GCF, Huynh et al. [15] found 2–3 time higher level in gingivitis than in chronic periodontitis.
Our results are congruent with an inflammatory response oriented to defense and regeneration of injured tissues. S100A9, HSPB1, LEG7 and 14-3-3 proteins resulted over-expressed in periodontal pocket tissue when compared with healthy patient analogous tissue. A significant unbalancing in protein expression between healthy and pathological sites was recorded. In a previous study [6] we aimed to compare the interproximal pocket tissue with interproximal tissues at sites with normal probing depth in patients affected from periodontal disease and HSPB1, LEG7 and 14-3-3 proteins resulted significantly under-expressed. Also S100A9 resulted under-expressed, but not significantly.
In all likelihood, patients have to be regarded as affected by periodontal disease because a complex pathologic network (composed by genetic structure involving immunology and inflammatory regulation) at the root of periodontitis even if the periodontal disease is being clinically burnt out [51–54].
A limited number of biomolecular studies have been carried out on the periodontal pocket. Hence, inadequate data are available on the pathognomonic lesion of the periodontal disease. Genes related to apoptosis, antimicrobial humoral response, antigen presentation, regulation of metabolism, signal transduction and angiogenesis were found to be differently expressed in patients with periodontitis and healthy subjects, as found in trascriptomic studies [55, 56]. Moreover, with the aid of microarray technologies, cell communication pathways were shown to be down-regulated in periodontitis-affected tissues, either in cell-to-cell communications at the soft tissue level, or in cell-to-tooth signaling as a consequence of the inflammatory status of the periodontium [55, 56].
It is conjecturable that an high inflammatory background is anyway present in healthy gingival tissues of patients with periodontal disease, and the change of the inflammatory and immunologic processes could have a role in periodontal damage with the hindering of the protecting molecular network when the risk factor challenge induces the periodontal pocket.
The identified proteins play a role in increased cell proliferation, decrease in cell tissue differentiation, impairing of metalloproteinases on connective tissue, organized action monocytes/macrophages (e.g. osteoclasts) and osteoblasts or fibroblast together with enhanced humoral and cell-mediated immunity. All these processes are consistent with a defensive response of the organism that also aims to regenerate the lost tissue after exogenous injury. Furthermore, the phagocytosis of opsonized cells, the alternative way by Toll-like receptors 2 and 4 or the T-cell activation to bacterial LPS could contribute to microbiota clearance along with the presence of chemo attractant molecules in the recruitment of immune cells. During these phases, an increasing cell migration could also play an important role in the recovery of damaged tissue as periodontal ligament or bone. It is also to consider that HSPs, whose synthesis is increased in response to a variety of stresses, and S100A9 play an important role in controlling cell and tissue damage, and in moderating inflammation and destructive immune response. HSPB1 and S100A9 resulted significantly increased, in all the tested pathological sites, while LEG7 and 14-3-3 proteins, although not expressed in normal tissue, exhibited fluctuation in expression level in periodontal lesions. These results need further studies to understand if the lack in expression of these proteins may worsen the tissue damage.
Conclusions
Most proteomic studies have been performed on different biological samples but not on periodontal pocket tissue that is the anatomo-pathological lesion signifying for the clinical diagnosis of the periodontal disease. Therefore, the periodontal pocket tissue is the only biologic material in which proteomic analysis enables the correct molecular assessment of this pathology. The establishment of a robust proteomic expression profile database for periodontal pathology would be highly desirable both to understand the pathogenesis and for periodontitis therapeutic strategies [13]. This is the first work that compares the proteomic content of periodontal pocket lesions and healthy gingival tissue of healthy subjects. The results of this study pointed out a network of proteins, differently expressed in the pathological tissue compared with the healthy one and contribute to the establishment of a proteomic expression profile database for periodontal pathology. These data are highly desirable both to understand the pathogenesis and for periodontitis therapeutic strategies, though further population studies are required to correlate the proteomic data with clinical data, as disease progression and aggressiveness.
Methods
Sample collection
Systemic healthy subjects, with and without periodontal chronic disease, examined and treated in a private dental office, were enrolled in this study, according to the protocol described in Table 1. Italian law does not require any ethical committee authorization for clinical trials performed in private dental offices, while such authorization is required for public dental health centers (DM 18/3/1998 published in the Official Gazette, GU n. 122 of 28-05-1998). Therefore, for the purposes of this study all enrolled subjects signed an informed consent form detailing the study procedures. The research was carried out in full accordance with the ethical principles of the WMA Helsinki Declaration [57].
Fifteen periodontally-healthy patients underwent crown lengthening surgery, where gingiva in excess or gingival margin asymmetries required a surgical correction [58, 59]. Periodontally-affected subjects presented at least a shallow intrabony defect suitable for treatment by osseous resective surgery.
Samples for microbial analysis were obtained from patients immediately before surgery, in order to exclude periodontopathogenic bacteria. The samples were analyzed by PCR-RT technique (GABA International AG, Lorrach, Germany), to confirm the absence of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythensis, Treponema denticola, Fusobacterium nucleatum ssp, and Prevotella intermedia.
The surgery was performed in both periodontally-healthy and -affected patients following the completion of a preliminary cause-related treatment required for the surgical approach and having reached a full-mouth plaque score and full-mouth bleeding score lower than 20 %.
Tissue specimens of interproximal healthy tissues were harvested in fifteen periodontally-healthy patients (C, control) at sites with normal probing depth [58–60]. Tissue specimens of interproximal pocket-associated tissue were harvested in subjects affected by chronic periodontitis (T, test). The harvested tissues were immediately frozen at −80 °C, for proteomic analyses.
Sample preparation for proteomic analysis
Tissue samples were ground in small pieces in a mortar with liquid nitrogen and collected in tubes. Tissue lysate was performed incubating the samples for 1 h with a buffer containing 7 M urea, 2 M thiourea, 3 % CHAPS, 40 mM Tris pH 8.3, 1 % ampholytes pH 3–10, protease inhibitors at room temperature. After incubation, tissues were further disrupted with an ultrasonic homogenizer (Sonoplus HD 2070, Bandelin electronic, Germany), and centrifuged at 10,000 x g for 10 min at +4 °C. Supernatant was precipitated by the addition of cold acetone (dilution ratio 1:12 vol/vol) and incubated at −20 °C overnight. After centrifugation at 14,000 x g for 15 min at +4 °C, the pellet was re-suspended and the protein concentration was determined according to the Bradford method. Three pooled samples, both for pathological and healthy gingival tissues, were obtained by mixing equal protein amount of 5 different subjects and were analyzed in duplicate.
2-Dimension electrophoresis
Tissue extracted proteins were separated by 2DE following a protocol previously described [61]. Briefly, for the first dimension, 80 μg of total proteins were loaded onto IPG strips, 17 cm long, pH range 3–10 (ReadyStripTM, Bio-Rad, USA). Afterwards, the second-dimension separation was carried out at 10 °C using 12 % polyacrylamide gels. Between these two separation steps, strips were reduced with 1 % DTT, and later alkylated with 2.5 % iodoacetamide in an equilibration buffer (6 M urea, 1 % DTT, 50 mM Tris–HCl pH 8.8, 30 % glycerol, 2 % SDS). After 2-DE, the protein spots in the gels were visualized following a silver nitrate staining protocol, as previously described [62]. The silver-stained gel images were acquired using a GS-800 Calibrated Densitometer (Bio-Rad, USA) and analysed with the PDQuest 2-D Image software program, version 7.3.1 (Bio-Rad, USA).
Protein identification by LC-MS/MS analysis
Protein spots excised manually from the gels were subjected to the “in-gel” tryptic digestion as previously described [63]. Dried samples were then re-suspended in 97 % Water/3 % ACN added of 1 % formic acid (Buffer A) and analyzed by a Nano LC-CHIP-MS system, consisting of the Agilent 6520 ESI-Q-TOF, coupled with a 1200 Nano HPLC-Chip microfluidic device (Agilent Technologies Inc., USA). Four microliters of each sample were loaded into the system and transported to the Chip enrichment column (Zorbax C18, 4 mm x 5 μm i.d., Agilent Technologies) by a capillary pump, with a loading flow of 4 μL/min, using 95 % ACN/5 % water added of 0.1 % formic acid (buffer B) as mobile phase. Nitrogen was used as the nebulizing gas. A separation column (Zorbax C18, 43 mm x 75 μm i.d., Agilent Technologies), at flow rate of 0.4 μL/min, was used for peptide separation. Protein-identification peak lists were generated using MASCOT search engine (http://mascot.cigs.unimo.it/mascot) against the UniProt Knowledgebase database (UniProt.org), specifying the following parameters: Homo sapiens taxonomy, parent ion tolerance ± 20 ppm, MS/MS error tolerance ± 0.12 Da, alkylated cysteine as fixed modification and oxidized methionine as variable modification, and two potential missed trypsin cleavages. Proteins that were identified with at least 2 or more significant peptides sequences and with the highest score hits among MASCOT search results. “High-scoring” corresponded to proteins that were above the significant threshold in Mascot searches (5 % probability of false match for each proteins above this score).
Protein functional analysis
The protein functional analysis and classification of identified proteins were performed using the iPROClass integrated database (http://pir.georgetown.edu/pirwww/dbinfo/iproclass.shtml accessed on March, 2015) for protein annotation and GO Terms Classifications Counter (CateGOrize, http://www.animalgenome.org/bioinfo/tools/countgo/ accessed on March, 2015) for clusterization according to the Gene Ontology (GO) hierarchy.
Western blot analysis
For Western blotting experiments, 3 μg of gingival tissue protein extracts were solubilized in Laemmli’s buffer 1X and denatured for 5 min at 95 °C. Protein content was resolved by 12 % or 15 % SDS-PAGE and the proteins were transferred onto a nitrocellulose membrane by electroblotting. Membranes were blocked at 4 °C overnight with 5 % non-fat dry milk in PBS containing 0.1 % Tween 20. The membranes were incubated with the relevant antibodies for 2 h (1:1000 dilution), washed, and incubated with HRP coniugated Polyclonal Goat Anti-rabbit secondary antibody (1:2000 dilution, DakoCytomation, Denmark) for 1 h. The proteins were visualized using WesternSure Premium Chemiluminescent substrate (LI-COR, USA) and C-Digit Blot scanner (LI-COR, USA) according to the manufacturer’s instructions. Acquired images were analyzed and compared using Image Studio Software 4.0.21 (LI-COR, USA). The sources of primary antibodies were as follows: anti-galectin 7 (ab10482), anti-HspB1 (ab1426) and anti-14-3-3 all isoforms (ab9063) were from Abcam (UK); anti- S100A9 (PA1-46489) was from Thermo scientific (USA). β-actin (ab8227, Abcam UK) was used for normalization of western blot analysis.
Acknowledgement
We thank Dr. Filippo Genovese (C.I.G.S., University of Modena and Reggio Emilia) for technical assistance during LC-MS/MS analysis. We are grateful to the “Fondazione Cassa di Risparmio di Modena, Italy” for have supported the purchase of the ESI-Q-TOF mass spectrometer.
Abbreviations
- 2DE
Two-dimensional electrophoresis
- LC-MS/MS
Liquid Chromatography- Mass/Mass spectrometry
- MW
Molecular weight
- emPAI
Exponentially modified protein abundance index
- HSPB1
Heat shock protein ß-1
- LEG7
Galectin-7
- 1433S
14-3-3 Protein sigma
- 1433Z
14-3-3 protein zeta/delta
- S100A9
Protein S100-A9
- GCF
Gingival Crevicular Fluid
- CHAPS
3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate
- DTT
Dithiothreitol
- ACN
Acetonitrile
- ESI-Q-TOF
Electrospray ionization quadrupole time-of-flight mass
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- HRP
Horseradish peroxidase
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EM designed the study, performed sample preparation, western blotting analysis and drafted the manuscript; AC carried out ESI-QTOF-MS analysis and performed protein identification; EB and SB performed bi-dimensional gel electrophoresis and in gel digestions; AL participated to the sample collections; AT and DZ provided useful advices to improve the study and revised the manuscript; PC participated in the design of the study; CB conceived the study, participated in its design and coordination, enrolled all subject of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Emanuela Monari, Phone: ++39-0594224792, Email: emanuela.monari@unimore.it.
Aurora Cuoghi, Email: aurora.cuoghi@unimore.it.
Elisa Bellei, Email: elisa.bellei@unimore.it.
Stefania Bergamini, Email: stefania.bergamini@unimore.it.
Andrea Lucchi, Email: dottlucchiandrea@gmail.com.
Aldo Tomasi, Email: aldo.tomasi@unimore.it.
Pierpaolo Cortellini, Email: sandro@cortellini.org.
Davide Zaffe, Email: davide.zaffe@unimore.it.
Carlo Bertoldi, Email: carlo.bertoldi@unimore.it.
References
- 1.Bertoldi C, Bencivenni D, Lucchi A, Consolo U. Augmentation of keratinized gingiva through bilaminar connective tissue grafts: a comparison between two techniques. Minerva Stomatol. 2007;56:3–20. [PubMed] [Google Scholar]
- 2.Tripodo C, Di Bernardo A, Ternullo MP, Guarnotta C, Porcasi R, Ingrao S, et al. CD146(+) bone marrow osteoprogenitors increase in the advanced stages of primary myelofibrosis. Haematologica. 2009;94:127–30. doi: 10.3324/haematol.13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemmig TF. Periodontitis. Ann Periodontol. 1999;4:32–8. doi: 10.1902/annals.1999.4.1.32. [DOI] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. The nature of periodontal diseases. Ann Periodontol. 1997;2:3–10. doi: 10.1902/annals.1997.2.1.3. [DOI] [PubMed] [Google Scholar]
- 5.Bertoldi C, Pellacani C, Lalla M, Consolo U, Pinti M, Cortellini P, et al. Herpes Simplex I virus impairs regenerative outcomes of periodontal regenerative therapy in intrabony defects: a pilot study. J Clin Periodontol. 2012;39:385–92. doi: 10.1111/j.1600-051X.2012.01850.x. [DOI] [PubMed] [Google Scholar]
- 6.Bertoldi C, Bellei E, Pellacani C, Ferrari D, Lucchi A, Cuoghi A, et al. Non-bacterial protein expression in periodontal pockets by proteome analysis. J Clin Periodontol. 2013;40:573–82. doi: 10.1111/jcpe.12050. [DOI] [PubMed] [Google Scholar]
- 7.Tonetti MS, Claffey N. Advances in the progression of periodontitis and proposal of definitions of a periodontitis case and disease progression for use in risk factor research. Group C consensus report of the 5th European Workshop in Periodontology. J Clin Periodontol. 2005;32(Suppl 6):210–3. doi: 10.1111/j.1600-051X.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- 8.Page RC, Eke PI. Case definitions for use in population-based surveillance of periodontitis. J Periodontol. 2007;78:1387–99. doi: 10.1902/jop.2007.060264. [DOI] [PubMed] [Google Scholar]
- 9.Huang R-Y, Lu S-H, Su K-W, Chen J-K, Fang W-H, Liao W-N, et al. Diacerein: a potential therapeutic drug for periodontal disease. Med Hypotheses. 2012;79:165–7. doi: 10.1016/j.mehy.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J Periodontol. 2008;79:1560–8. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- 11.Gonçalves LDR, Soares MR, Nogueira FCS, Garcia C, Camisasca DR, Domont G, et al. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J Proteomics. 2010;73:1334–41. doi: 10.1016/j.jprot.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Shu R, Luo L-J, Ge L-H, Xie Y-F. Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J Periodontal Res. 2009;44:636–44. doi: 10.1111/j.1600-0765.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 13.Gorr S-U, Abdolhosseini M. Antimicrobial peptides and periodontal disease. J Clin Periodontol. 2011;38(Suppl 1):126–41. doi: 10.1111/j.1600-051X.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 14.Jönsson D, Ramberg P, Demmer RT, Kebschull M, Dahlén G, Papapanou PN. Gingival tissue transcriptomes in experimental gingivitis. J Clin Periodontol. 2011;38:599–611. doi: 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh AHS, Veith PD, McGregor NR, Adams GG, Chen D, Reynolds EC, et al. Gingival crevicular fluid proteomes in health, gingivitis and chronic periodontitis. J Periodontal Res. 2015;50:637–49. doi: 10.1111/jre.12244. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama T, Kobayashi T, Yamamoto K, Yamagata A, Oofusa K, Yoshie H. Proteomic profiling of human neutrophils in relation to immunoglobulin G Fc receptor IIIb polymorphism. J Periodontal Res. 2010;45:780–7. doi: 10.1111/j.1600-0765.2010.01300.x. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno N, Niitani M, Shiba H, Iwata T, Hayashi I, Kawaguchi H, et al. Proteome analysis of proteins related to aggressive periodontitis combined with neutrophil chemotaxis dysfunction. J Clin Periodontol. 2011;38:310–7. doi: 10.1111/j.1600-051X.2010.01693.x. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama T, Kobayashi T, Ito S, Yamagata A, Ishida K, Okada M, et al. Comparative analysis of serum proteins in relation to rheumatoid arthritis and chronic periodontitis. J Periodontol. 2014;85:103–12. doi: 10.1902/jop.2013.120741. [DOI] [PubMed] [Google Scholar]
- 19.Pham TK, Roy S, Noirel J, Douglas I, Wright PC, Stafford GP. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics. 2010;10:3130–41. doi: 10.1002/pmic.200900448. [DOI] [PubMed] [Google Scholar]
- 20.AlMoharib HS, AlMubarak A, AlRowis R, Geevarghese A, Preethanath RS, Anil S. Oral fluid based biomarkers in periodontal disease: part 1. Saliva. J Int oral Heal. 2014;6:95–103. [PMC free article] [PubMed] [Google Scholar]
- 21.Trindade F, Oppenheim FG, Helmerhorst EJ, Amado F, Gomes PS, Vitorino R. Uncovering the molecular networks in periodontitis. Proteomics Clin Appl. 2014;8:748–61. doi: 10.1002/prca.201400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman YA, Sakellari D, Arsenakis M, Floudas CA. Proteomics for the discovery of biomarkers and diagnosis of periodontitis: a critical review. Expert Rev Proteomics. 2014;11:31–41. doi: 10.1586/14789450.2014.864953. [DOI] [PubMed] [Google Scholar]
- 23.Gupta A, Govila V, Saini A. Proteomics - The research frontier in periodontics. J Oral Biol Craniofacial Res. 2015;5:46–52. doi: 10.1016/j.jobcr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–7. doi: 10.1111/j.1600-051X.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 25.Masamatti S, Kumar A, Mehta D, Bhat K, Baron TK. Evaluation of interleukin -1B (+3954) gene polymorphism in patients with chronic and aggressive periodontitis: a genetic association study. Contemp Clin Dent. 2012;3:144. doi: 10.4103/0976-237X.96815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Jia Y, Wu H. Four tumor necrosis factor alpha genes polymorphisms and periodontitis risk in a Chinese population. Hum Immunol. 2013;74:1684–7. doi: 10.1016/j.humimm.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–22. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 28.Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2011;41:821–42. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- 29.Srikrishna G. S100A8 and S100A9: new insights into their roles in malignancy. J Innate Immun. 2012;4:31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kido J, Bando M, Hiroshima Y, Iwasaka H, Yamada K, Ohgami N, et al. Analysis of proteins in human gingival crevicular fluid by mass spectrometry. J Periodontal Res. 2012;47:488–99. doi: 10.1111/j.1600-0765.2011.01458.x. [DOI] [PubMed] [Google Scholar]
- 31.Kojima T, Andersen E, Sanchez JC, Wilkins MR, Hochstrasser DF, Pralong WF, et al. Human gingival crevicular fluid contains MRP8 (S100A8) and MRP14 (S100A9), two calcium-binding proteins of the S100 family. J Dent Res. 2000;79:740–7. doi: 10.1177/00220345000790020701. [DOI] [PubMed] [Google Scholar]
- 32.Andersen E, Dessaix IM, Perneger T, Mombelli A. Myeloid-related protein (MRP8/14) expression in gingival crevice fluid in periodontal health and disease and after treatment. J Periodontal Res. 2010;45:458–63. doi: 10.1111/j.1600-0765.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- 33.Ellias MF, Zainal Ariffin SH, Karsani SA, Abdul Rahman M, Senafi S, Megat Abdul Wahab R. Proteomic analysis of saliva identifies potential biomarkers for orthodontic tooth movement. Sci World J. 2012;2012:1–6. doi: 10.1100/2012/647240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, Hou J, Peng L, Zhang X, Jia L, Wang X, et al. The Pro-Apoptotic and Pro-Inflammatory Effects of Calprotectin on Human Periodontal Ligament Cells. Glogauer M, editor. PLoS One. 2014;9:e110421. doi: 10.1371/journal.pone.0110421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonardi R, Villari L, Caltabiano M, Travali S. Heat shock protein 27 expression in the epithelium of periapical lesions. J Endod. 2001;27:89–92. doi: 10.1097/00004770-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Park J-H, Yoon J-H, Lim Y-S, Hwang H-K, Kim S-A, Ahn S-G, et al. TAT-Hsp27 promotes adhesion and migration of murine dental papilla-derived MDPC-23 cells through beta1 integrin-mediated signaling. Int J Mol Med. 2010;26:373–8. doi: 10.3892/ijmm_00000465. [DOI] [PubMed] [Google Scholar]
- 37.Mitsuhashi M, Yamaguchi M, Kojima T, Nakajima R, Kasai K. Effects of HSP70 on the compression force-induced TNF-α and RANKL expression in human periodontal ligament cells. Inflamm Res. 2011;60:187–94. doi: 10.1007/s00011-010-0253-x. [DOI] [PubMed] [Google Scholar]
- 38.Sreedharan R, Riordan M, Thullin G, Van Why S, Siegel NJ, Kashgarian M. The maximal cytoprotective function of the heat shock protein 27 is dependent on heat shock protein 70. Biochim Biophys Acta. 2011;1813:129–35. doi: 10.1016/j.bbamcr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koromantzos PA, Makrilakis K, Dereka X, Offenbacher S, Katsilambros N, Vrotsos IA, et al. Effect of non-surgical periodontal therapy on C-Reactive Protein, Oxidative Stress, and Matrix Metalloproteinase (MMP)-9 and MMP-2 levels in patients with type 2 diabetes: a randomized controlled study. J Periodontol. 2012;83:3–10. doi: 10.1902/jop.2011.110148. [DOI] [PubMed] [Google Scholar]
- 40.Kwon S-M, Kim S-A, Yoon J-H, Ahn S-G. Transforming growth factor beta1-induced heat shock protein 27 activation promotes migration of mouse dental papilla-derived MDPC-23 cells. J Endod. 2010;36:1332–5. doi: 10.1016/j.joen.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Chung E, Rylander MN. Response of preosteoblasts to thermal stress conditioning and osteoinductive growth factors. Cell Stress Chaperones. 2012;17:203–14. doi: 10.1007/s12192-011-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuwabara I, Kuwabara Y, Yang R-Y, Schuler M, Green DR, Zuraw BL, et al. Galectin-7 (PIG1) exhibits pro-apoptotic function through JNK activation and mitochondrial cytochrome c release. J Biol Chem. 2002;277:3487–97. doi: 10.1074/jbc.M109360200. [DOI] [PubMed] [Google Scholar]
- 43.Saussez S, Kiss R. Galectin-7. Cell Mol Life Sci. 2006;63:686–97. doi: 10.1007/s00018-005-5458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polla BS, Cossarizza A. Stress proteins in inflammation. EXS. 1996;77:375–91. doi: 10.1007/978-3-0348-9088-5_25. [DOI] [PubMed] [Google Scholar]
- 45.Leonardi R, Pannone G, Magro G, Kudo Y, Takata T, Lo ML. Differential expression of heat shock protein 27 in normal oral mucosa, oral epithelial dysplasia and squamous cell carcinoma. Oncol Rep. 2002;9:261–6. [PubMed] [Google Scholar]
- 46.Alford KA, Glennie S, Turrell BR, Rawlinson L, Saklatvala J, Dean JLE. Heat shock protein 27 functions in inflammatory gene expression and transforming growth factor-beta-activated kinase-1 (TAK1)-mediated signaling. J Biol Chem. 2007;282:6232–41. doi: 10.1074/jbc.M610987200. [DOI] [PubMed] [Google Scholar]
- 47.Asdaghi N, Kilani RT, Hosseini-Tabatabaei A, Odemuyiwa SO, Hackett T-L, Knight DA, et al. Extracellular 14-3-3 from human lung epithelial cells enhances MMP-1 expression. Mol. Cell. Biochem. 2012;360:261–70. doi: 10.1007/s11010-011-1065-1. [DOI] [PubMed] [Google Scholar]
- 48.Ben-Addi A, Mambole-Dema A, Brender C, Martin SR, Janzen J, Kjaer S, et al. IκB kinase-induced interaction of TPL-2 kinase with 14-3-3 is essential for Toll-like receptor activation of ERK-1 and −2 MAP kinases. Proc Natl Acad Sci U S A. 2014;111:E2394–403. doi: 10.1073/pnas.1320440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Np63 Phosphoprotein Binds the p21 and 14-3-3 Promoters In Vivo and Has Transcriptional Repressor Activity That Is Reduced by Hay-Wells Syndrome-Derived Mutations. Mol Cell Biol. 2003;23:2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh J-E, Jang DH, Kim H, Kang HK, Chung C-P, Park WH, et al. Alpha3Beta1 integrin promotes cell survival via multiple interactions between 14-3-3 isoforms and proapoptotic proteins. Exp Cell Res. 2009;315:3187–200. doi: 10.1016/j.yexcr.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Dowsett SA, Archila L, Foroud T, Koller D, Eckert GJ, Kowolik MJ. The effect of shared genetic and environmental factors on periodontal disease parameters in untreated adult siblings in Guatemala. J Periodontol. 2002;73:1160–8. doi: 10.1902/jop.2002.73.10.1160. [DOI] [PubMed] [Google Scholar]
- 52.Kinane DF, Lappin DF. Immune processes in periodontal disease: a review. Ann Periodontol. 2002;7:62–71. doi: 10.1902/annals.2002.7.1.62. [DOI] [PubMed] [Google Scholar]
- 53.Dasanayake AP. Periodontal disease is related to local and systemic mediators of inflammation. J Evid Based Dent Pract. 2010;10:246–7. doi: 10.1016/j.jebdp.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Taba M, de Souza SLS, Mariguela VC. Periodontal disease: a genetic perspective. Braz Oral Res. 2012;26(Suppl 1):32–8. doi: 10.1590/S1806-83242012000700006. [DOI] [PubMed] [Google Scholar]
- 55.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–24. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abe D, Kubota T, Morozumi T, Shimizu T, Nakasone N, Itagaki M, et al. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. J Periodontal Res. 2011;46:345–53. doi: 10.1111/j.1600-0765.2011.01349.x. [DOI] [PubMed] [Google Scholar]
- 57.Puri KS, Suresh KR, Gogtay NJ, Thatte UM. Declaration of Helsinki, 2008: implications for stakeholders in research. J Postgrad Med. 2009;55:131–4. doi: 10.4103/0022-3859.52846. [DOI] [PubMed] [Google Scholar]
- 58.Pontoriero R, Carnevale G. Surgical crown lengthening: a 12-month clinical wound healing study. J Periodontol. 2001;72:841–8. doi: 10.1902/jop.2001.72.7.841. [DOI] [PubMed] [Google Scholar]
- 59.Ribeiro FV, Hirata DY, Reis AF, Santos VR, Miranda TS, Faveri M, et al. Open-flap versus flapless esthetic crown lengthening: 12-month clinical outcomes of a randomized controlled clinical trial. J Periodontol. 2014;85:536–44. doi: 10.1902/jop.2013.130145. [DOI] [PubMed] [Google Scholar]
- 60.Nethravathy R, Vinoth SK, Thomas AV. Three different surgical techniques of crown lengthening: a comparative study. J Pharm Bioallied Sci. 2013;5:S14–6. doi: 10.4103/0975-7406.113281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellei E, Monari E, Bergamini S, Ozben T, Tomasi A. Optimizing protein recovery yield from serum samples treated with beads technology. Electrophoresis. 2011;32:1414–21. doi: 10.1002/elps.201000699. [DOI] [PubMed] [Google Scholar]
- 62.Bellei E, Rossi E, Lucchi L, Uggeri S, Albertazzi A, Tomasi A, et al. Proteomic analysis of early urinary biomarkers of renal changes in type 2 diabetic patients. Proteomics Clin Appl. 2008;2:478–91. doi: 10.1002/prca.200780109. [DOI] [PubMed] [Google Scholar]
- 63.Bellei E, Bergamini S, Monari E, Fantoni LI, Cuoghi A, Ozben T, et al. High-abundance proteins depletion for serum proteomic analysis: concomitant removal of non-targeted proteins. Amino Acids. 2011;40:145–56. doi: 10.1007/s00726-010-0628-x. [DOI] [PubMed] [Google Scholar]


