Abstract
Background
Immunosuppressive viruses are frequently found as co-infections in the chicken industry, potentially causing serious economic losses. Because traditional molecular biology methods have limited detection ability, a rapid, high-throughput method for the differential diagnosis of these viruses is needed. The objective of this study is to develop a GenomeLab Gene Expression Profiler Analyser-based multiplex PCR method (GeXP-multiplex PCR) for simultaneous detection of eight immunosuppressive chicken viruses.
Results
Using chimeric primers, eight such viruses, including Marek's disease virus (MDV), three subgroups of avian leucosis virus (ALV-A/B/J), reticuloendotheliosis virus (REV), infectious bursal disease virus (IBDV), chicken infectious anaemia virus (CIAV) and avian reovirus (ARV), were amplified and identified by their respective amplicon sizes. The specificity and sensitivity of the optimised GeXP-multiplex PCR assay were evaluated, and the data demonstrated that this technique could selectively amplify these eight viruses at a sensitivity of 100 copies/20 μl when all eight viruses were present. Among 300 examined clinical specimens, 190 were found to be positive for immunosuppressive viruses according to this novel assay.
Conclusion
The GeXP-multiplex PCR assay is a high-throughput, sensitive and specific method for the detection of eight immunosuppressive viruses and can be used for differential diagnosis and molecular epidemiological surveys.
Keywords: GeXP analyser, Multiplex PCR, Immunosuppressive viruses
Background
Immunosuppression causes major economic losses in poultry farming because immunosuppressed chickens are more susceptible to viral and bacterial pathogens, respond poorly to vaccination, and display lower feed conversion efficiency as well as growth retardation. Immunosuppressive chicken viruses include Marek’s disease virus (MDV), avian leucosis virus (ALV), reticuloendotheliosis virus (REV), infectious bursal disease virus (IBDV), chicken infectious anaemia virus (CIAV) and avian reovirus (ARV), all of which affect immune function in chickens and lead to immunosuppression [1–7]. The typical symptoms elicited by these viruses differ, and some symptoms are not readily observable. Immunosuppression in chickens infected with MDV, ALV or REV occurs much earlier than does tumour development and death [1, 2]. Although early infection with IBDV in chicks less than 3 weeks old may not result in the typical symptoms of IBD, this infection nonetheless causes serious immunosuppression [8]. Chickens older than 3 weeks of age are resistant to anaemia after infection with CIAV yet remain susceptible to immunosuppression [9]. Furthermore, the possibility of co-infection makes it difficult to differentiate among these immunosuppressive viruses [10–13].
The detection and differential diagnosis of immunosuppressive viruses are important for the poultry industry. However, conventional methods, such as virus isolation and serum neutralisation tests, are typically time-consuming and labour-intensive procedures [14]. Molecular methods have been used to rapidly detect immunosuppression in chickens, but they are limited by their ability to detect only a few pathogens per reaction [15–20]. Therefore, a rapid, cost-effective, and high-throughput detection technique is needed for the clinical diagnosis of immunosuppressive viral infection in chickens.
The GenomeLab Gene Expression Profiler genetic analysis system (GeXP) is a new multi-target, high-throughput detection platform that integrates RT-PCR or PCR with a labelled, amplified product in a multiplex RT-PCR/PCR assay followed by fluorescence capillary electrophoresis separation based on the sizes of the amplified products [21]. The GeXP profiler utilises gene-specific primers containing 5′-universal adaptor sequences [21]: the chimeric primers consist of a universal sequence fused to the 5′-end of a gene-specific sequence. The forward primer consists of a universal dye-labelled sequence fused to the 5′-end of a gene-specific sequence, whereas the reverse primer consists of a universal unlabelled sequence fused to the 5′-end of a gene-specific sequence. Products differing by 7–10 bp in size are separated by capillary electrophoresis. This technique has been used to identify various diseases in humans, including 11 genotypes of HPV [22]; 9 serotypes of hand, foot, and mouth disease [23]; cancer [21, 24]; 7 enteric viruses [25]; and H1N1 [26]. The GeXP genetic analysis system has also been successfully utilised to simultaneously detect 9 avian respiratory pathogens in clinical samples, 8 swine reproductive and respiratory pathogens and 11 duck viruses [27–29].
In this study, a GeXP-multiplex PCR assay was developed and optimised to simultaneously detect eight immunosuppressive chicken viruses: MDV, ALV (three subgroups of ALV, ALV-A/B/J), REV, IBDV, CIAV and ARV.
Results
Specificity of the GeXP-multiplex PCR assay
The concentrations of the GeXP-multiplex PCR-specific primers (listed in Table 1) were optimised, and DNA/cDNA from the immunosuppressive viruses described in Table 2 was used individually as a template for evaluating the specificity of the GeXP-multiplex PCR assay. The expected size of each immunosuppressive viral amplicon was determined. A single peak for the complex PCR was detected using the GeXP analyser system (Fig. 1 a-i), and no mispriming with or non-specific amplification of other avian pathogens or the chicken genome was observed.
Table 1.
Primer sequences and PCR product sizes
| Virus | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) | Amplicon size (bp) | Target region | Primer concentration (μM) |
|---|---|---|---|---|---|
| MDV | AGGTGACACTATAGAATAAGGGAGCAGACGTACTATGTAGACAA | GTACGACTCACTATAGGGATGGTAAGCAGTCCAAGGGTCA | 227 | meq | 0.16 |
| ALV-A | AGGTGACACTATAGAATACAAGGGGTTCCTTGGTATCT | GTACGACTCACTATAGGGATGTGCCTATCCGCTGTCA | 155 | gp85 | 0.2 |
| ALV-B | AGGTGACACTATAGAATATCAATCACGATTCTCCCACC | GTACGACTCACTATAGGGATGTGACGCTTCGTTTACGTCTT | 285 | gp85 | 0.2 |
| ALV-J | AGGTGACACTATAGAATACTGATGCAACAACCAGGAAA | GTACGACTCACTATAGGGAGCAGTAACATTAGTGACATACCC | 204 | gp85 | 0.2 |
| REV | AGGTGACACTATAGAATAGACCAGGCGAGCAAAATC | GTACGACTCACTATAGGGAGGTGTAATAGGTAGGTATGGAGGA | 182 | gp90 | 0.2 |
| IBDV | AGGTGACACTATAGAATAGGGTCAGGGCTAATTGTCTT | GTACGACTCACTATAGGGATCTGTCAGTTCACTCAGGCTTC | 294 | VP2 | 0.2 |
| CIAV | AGGTGACACTATAGAATAAAAGGCGAACAACCGATGA | GTACGACTCACTATAGGGATGCCCTGGAGGAAAAGACC | 269 | VP1 | 0.2 |
| ARV | AGGTGACACTATAGAATAGGACCCCTACTTCTGTTCTCA | GTACGACTCACTATAGGGAATTTCCCGTGGACGACAT | 215 | S1 | 0.16 |
| Universal primers | AGGTGACACTATAGAATA | GTACGACTCACTATAGGGA | 0.25 |
Universal primers sequences are underlined. Chimeric primers were synthesised using universal primers and specific primers
Table 2.
Sources of pathogens
| Pathogen | Source | Pathogen | Source |
|---|---|---|---|
| Reference viruses | Other viruses | ||
| Marek’s disease virus KC453972, KC453973, GX130112, GX140301, 050118, 070123, 090201, 100428 | GVRI | Inactivated H5N1 avian influenza virus Re-1 | HVRI |
| Avian leucosis virus subgroup A isolate RSV-1 | CVCC | Avian influenza virus H9N6/Duck/HK/147/77 | HKU |
| Avian leucosis virus subgroup A isolate GX110521, GX110522, ALVA01, ALVA02, ALVA03 | GVRI | Avian influenza virus H7N2/chicken PA/3979/97 | PU |
| Avian leucosis virus subgroup B isolate RSV-2 | CVCC | Newcastle disease virus F48E9 | GVRI |
| Avian leucosis virus subgroup B isolate GX111230, GX130401, ALVB15, ALVB23, ALVB28 | GVRI | Newcastle disease virus GX6/02 | GVRI |
| Avian leucosis virus subgroup J isolate KC453974, KC453975, GX090201, GX090521, GX110110, GX120081, GX130018, GX140010 | GVRI | Infectious bronchitis virus Massachusetts 41 | GVRI |
| Reticuloendotheliosis virus AV235 | CVCC | Infectious laryngotracheitis virus AV1231 | CIVDC |
| Reticuloendotheliosis virus KC453976, KC453977, GX120825, GX131118 | GVRI | Mycoplasma synoviae CAU0748 | CVCC |
| Avian reovirus S1133, 1733, 526, C78, GuangxiR1, GuangxiR2, GX110058 | GVRI | ||
| Infectious bursal disease virus CA, AV162, AV144 | CVCC | ||
| Infectious bursal disease virus AV6 | CIVDC | ||
| Infectious bursal disease virus 070124, 080113, 090053, 100008, 110110, 130223 | GVRI | ||
| Chicken infectious anaemia virus CAU0728, CAU0729, CAU0730, CAU0731, CAU0732 | CVCC | ||
| Chicken infectious anaemia virus GXC060821 | GVRI |
a) HVRI = Harbin Veterinary Research Institute, China
b) HKU = The University of Hong Kong, China
c) GVRI = Guangxi Veterinary Research Institute, China
d) CIVDC = China Institute of Veterinary Drug Control, China
e) PU = University of Pennsylvania
f) CVCC = China Veterinary Culture Collection Centre
Fig. 1.
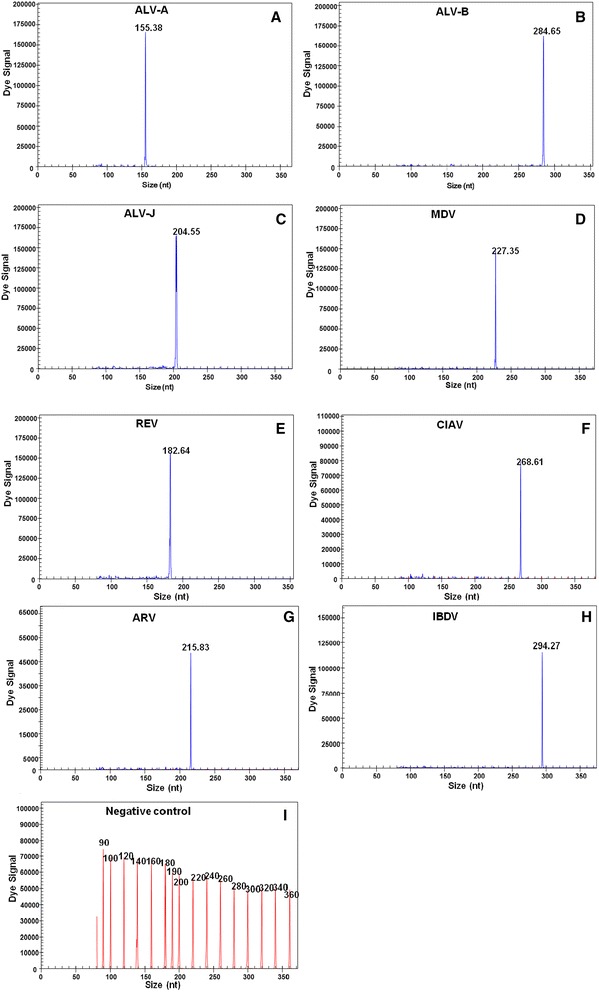
GeXP-multiplex PCR assay specificity. The GeXP-multiplex PCR assay was performed using a single template and mixed primers for the following: ALV-A: 155.38 bp (a); ALV-B: 284.65 bp (b); ALV-J: 204.55 bp (c); MDV: 227.35 bp (d); REV: 182.64 bp (e); CIAV: 268.61 bp (f); ARV: 215.83 bp (g); IBDV: 294.27 bp (h). DNA from the thymus, spleen and bursa of SPF chickens was used as a negative control (I). The x axes represent the sizes of PCR products in bp, and the y axes represent the dye signal in absorbance units (A.U.). Blue peaks denote specific amplification peaks, and red peaks denote marker peaks
Sensitivity of the GeXP-PCR assay
The GeXP-multiplex PCR assay achieved the following minimum sensitivity levels in the detection of each of the eight detectable immunosuppressive viruses using eight sets of primers and either plasmid or in vitro ssRNA transcripts: 10 copies for the ALV-A, ALV-J, REV, CIAV and ARV viruses; and 100 copies for the MDV, ALV-B and IBDV viruses (electropherograms not shown). Tenfold serial dilutions of specific DNA-containing plasmids and in vitro ssRNA transcripts from the eight immunosuppressive viruses, i.e., 105 (Fig. 2a), 104 (Fig. 2b), 103 (Fig. 2c) and 102 (Fig. 2d) copies per reaction, were prepared and amplified using an equal amount of template. When all of the pre-mixed, specific DNA-containing plasmids and in vitro ssRNA transcripts corresponding to the eight immunosuppressive viruses were present, the GeXP-multiplex PCR assay achieved a minimum sensitivity of 100 copies.
Fig. 2.
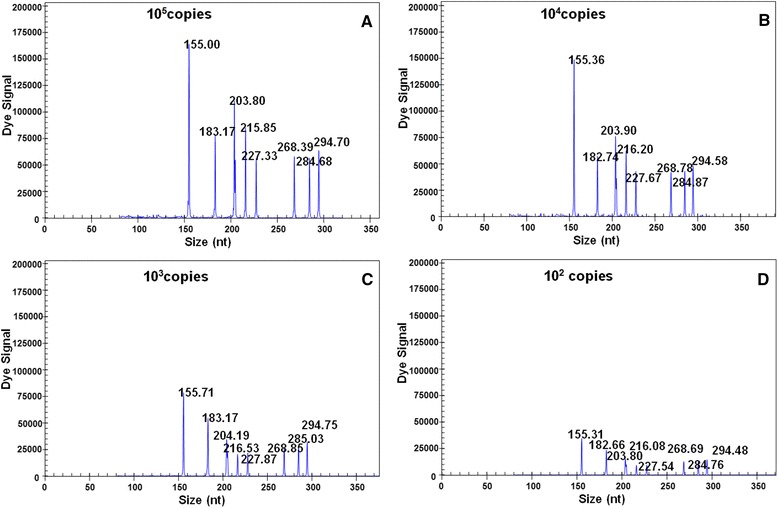
GeXP-multiplex PCR assay sensitivity. The GeXP-multiplex PCR assay was performed with mixed primers using equal amounts of specific DNA-containing plasmid template and in vitro-transcribed ssRNA corresponding to eight immunosuppressive viruses at concentrations of 105 (a), 104 (b), 103 (c) or 102 (d) copies per reaction. The x axes represent the sizes of PCR products in bp, and the y axes represent the dye signal in absorbance units (A.U.)
Artificial mixtures and interference assays
To test the differentiation ability of the GeXP-multiplex PCR assay, samples previously deemed positive for avian immunosuppressive viruses were randomly mixed; DNA/RNA was then extracted, and the appropriate specific amplification peaks were observed (Fig. 3). When either ARV and IBDV or ALV-J and ALV-B were mixed, two specific amplification peaks were observed (ARV, 216.16 bp; IBDV, 294.55 bp; ALV-J, 203.93 bp; and ALV-B, 284.83 bp), and when DNA/cDNA from all eight immunosuppressive viruses was mixed, eight specific amplification peaks were observed (ALV-A, 155.37 bp; REV, 182.69 bp; ALV-J, 203.71 bp; ARV, 216.01 bp; MDV, 227.49 bp; CIAV, 268.60 bp; ALV-B, 284.65 bp; and IBDV, 294.55 bp).
Fig. 3.
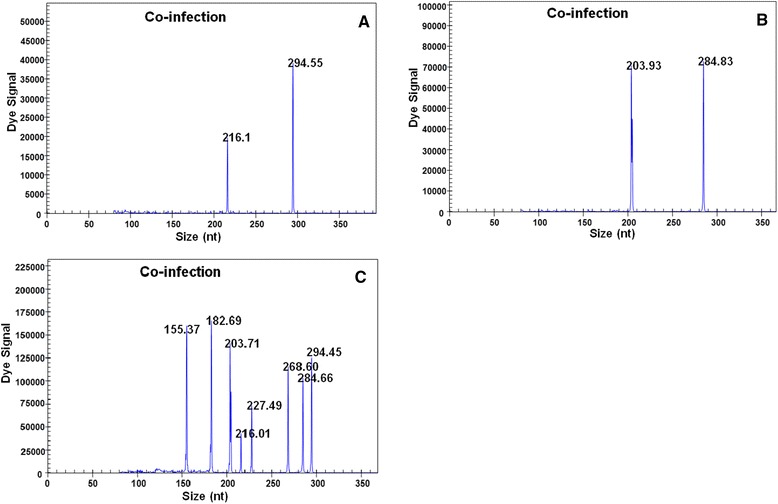
Artificially mixed templates for the GeXP-multiplex PCR assay. The GeXP-multiplex PCR assay was performed using artificially mixed templates and mixed primers for ARV and IBDV (a), ALV-J and ALV-B (b), or eight immunosuppressive viruses (c). The x axes represent the sizes of PCR products in bp, and the y axes represent the dye signal in absorbance units (A.U.)
Specific amplification peaks were also observed when 107 copies of ALV-J, 102 copies of MDV and 103 copies of CIAV were mixed in a single reaction, and similar amplification peaks were observed when 107 copies of ALV-J or 102 copies of MDV were individually tested (Fig. 4). These results demonstrate minimal to no interference by mixed infections in the GeXP-multiplex PCR assay.
Fig. 4.
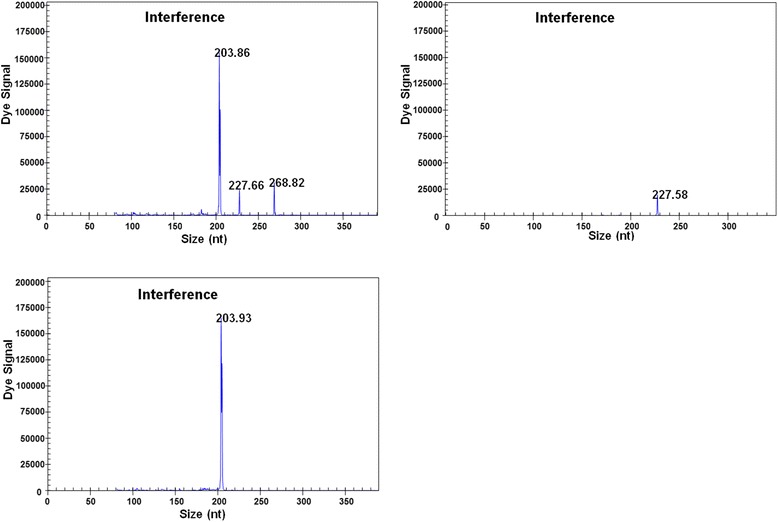
GeXP-multiplex PCR interference assays. The GeXP-multiplex PCR assay was performed using templates for ALV-J, MDV and CIAV (a), MDV (b), or ALV-J (c). The x axes represent the sizes of PCR products in bp, and the y axes represent the dye signal in absorbance units (A.U.)
Evaluation of the GeXP-PCR assay using clinical specimens
A total of 300 clinical specimens were collected from diseased farm chickens and tested using the GeXP-multiplex PCR assay. The chickens ranged from 50 to 135 days old, and the cohort included layer, broiler and breeding birds. The results of the GeXP-multiplex PCR assay are provided in Table 3. Independent real-time PCR/RT-PCR and sequencing to identify true positives and negatives confirmed the GeXP-multiplex PCR results, with the positive results being 100 % comparable to those for real-time PCR/RT-PCR and sequencing. Among a total of 190 positive results, 119 specimens displayed a single infection with an immunosuppressive virus. A total of 110 specimens were negative for the eight immunosuppressive viruses. A Kappa value of 1 was found with regard to consistency among GeXP-multiplex PCR, real-time PCR/RT-PCR and sequencing results.
Table 3.
Detection results for clinical specimens
| Single infection | Co-infection | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Rate (%) | Real-time PCR/RT-PCR results | Sequencing results | Measures of agreement Kappa values | Number | Rate (%) | Real-time PCR/RT-PCR results | Sequencing results | Measures of agreement Kappa values | ||
| MDV | 26 | 8.7 % | 26 | 26 | 1 (p < 0.001) | MDV + ALV-A | 4 | 1.3 % | 4 | 4 | 1 (p < 0.001) |
| ALV-A | 6 | 2.0 % | 6 | 6 | 1 (p < 0.001) | MDV + ALV-J | 25 | 8.3 % | 25 | 25 | 1 (p < 0.001) |
| ALV-B | 2 | 0.7 % | 2 | 2 | 1 (p < 0.001) | ALV-B + ALV-J | 1 | 0.4 % | 1 | 1 | 1 (p < 0.001) |
| ALV-J | 21 | 7.0 % | 21 | 21 | 1 (p < 0.001) | MDV + ALV-J + REV | 6 | 2 % | 6 | 6 | 1 (p < 0.001) |
| REV | 15 | 5 % | 15 | 15 | 1 (p < 0.001) | MDV + CIAV | 5 | 1.6 % | 5 | 5 | 1 (p < 0.001) |
| IBDV | 17 | 5.7 % | 17 | 17 | 1 (p < 0.001) | MDV + REV | 6 | 2 % | 6 | 6 | 1 (p < 0.001) |
| CIAV | 9 | 3 % | 9 | 9 | 1 (p < 0.001) | ALV-J + REV | 3 | 1.5 % | 3 | 3 | 1 (p < 0.001) |
| ARV | 23 | 7.7 % | 23 | 23 | 1 (p < 0.001) | IBDV + ALV-J | 5 | 1.6 % | 5 | 5 | 1 (p < 0.001) |
| MDV + CIAV + ALV-J | 4 | 1.3 % | 4 | 4 | 1 (p < 0.001) | ||||||
| REV + ARV | 3 | 1 % | 3 | 3 | 1 (p < 0.001) | ||||||
| ALV-J + ARV | 6 | 2 % | 6 | 6 | 1 (p < 0.001) | ||||||
| ALV-J + IBDV | 3 | 1 % | 3 | 3 | 1 (p < 0.001) | ||||||
Discussion
MDV, ALV (three subgroups found predominantly in China, ALV-A/B/J), REV, IBDV, CIAV and ARV are the major immunosuppressive viruses causing economic losses to the chicken industry. Furthermore, the possibility of co-infection increases the difficulty of differentially diagnosing individual viral infections [16, 30–33]. However, conventional diagnostic methods are time consuming, and molecular methods are limited by their ability to detect only a few pathogens per reaction.
The advantages of the GeXP-multiplex PCR assay include its specificity and its high-throughput ability to differentiate eight immunosuppressive viruses. These advantages stem from the use of chimeric and universal primers in a 3-step PCR procedure with different annealing temperatures: the first step amplifies gene-specific sequences within specific regions of the chimeric primers; the second step utilises the entire chimeric primer; and the last step uses universal primers for amplification. As non-specific amplification is minimised by using chimeric primers in the second step at a temperature 13 °C higher than the temperature in step one, only specific amplicons are produced. Furthermore, false-positive reactions are minimised via capillary electrophoresis separation to confirm the identity of the bands [21]. Based on electropherograms, amplicon sizes were found to deviate from their theoretical size by approximately +/-1 bp; for example, amplicons of 154–156 bp indicated a positive result for ALV-A. In contrast, conventional multiplex PCR and multiplex real-time PCR are able to identify only two to four pathogens in one reaction [16, 34–36].
Another advantage of this GeXP-multiplex PCR assay is improved detection sensitivity [27]. In this study, the minimum detection limit of the GeXP-multiplex PCR assay was 100 copies (DNA plasmids or in vitro ssRNA transcripts) for all mixed templates, and the minimum absorbance unit (A.U.) was 9000; by default, a reaction was considered positive when A.U. >2000. Further evaluation of clinical specimens confirmed that the positive results were 100 % comparable to the results of sequencing.
The third advantage of this GeXP-multiplex PCR assay is the ability to avoid interference due to the use of universal primers in the last step, ensuring equal amplification efficiency of each target gene, regardless of differences in pathogen concentration.
Of the viruses evaluated in this study, MDV and CIAV are DNA viruses, and IBDV, ALV, REV and ARV are RNA viruses. ALV and REV are retroviruses, which insert their proviral DNA into the host genome for viral replication [37]. In these cases, we recommend using a DNA/RNA kit to extract DNA/RNA together, followed by the procedure described above. All of these immunosuppressive viruses affect the immune organs of chickens, including the thymus, spleen and bursa, and these organs should be collected from diseased chickens for diagnosis.
An analysis of 300 specimens using the GeXP-multiplex PCR assay revealed MDV as the most prevalent single infection, followed by ARV. MDV + ALV-J was the most prevalent co-infection, yet different co-infections were also present. Our data demonstrate that MDV and ALV-J are the main pathogens that co-infect with other immunosuppressive viruses and that various co-infections are common in chickens in south China. Chickens infected with one immunosuppressive virus are more susceptible to attack by another immunosuppressive virus, and vaccine contamination might be a cause of common co-infections [38, 39].
As no assay can be 100 % accurate, there are some limitations to this assay. Virus genomes vary; thus, some primers will likely not be suitable for future use, and additional primers may be required for different variations. Furthermore, the GeXP instrument is expensive for clinical applications; therefore, the availability of less expensive machines will be important.
Conclusion
In conclusion, the GeXP-multiplex PCR assay is a high-throughput, sensitive and specific method for detecting eight immunosuppressive viruses in chickens. Accordingly, this assay is a potentially useful tool for the detection and differentiation of immunosuppressive viruses and for molecular epidemiologic testing, especially in situations in which chicken flocks are not performing well and the underlying cause may be immunosuppressive viruses. Identifying the immunosuppressive viruses responsible for these infections will be helpful for designing better disease-control programmes.
Methods
Ethics statement
This study was approved by the Animal Care and Use Committee (IACUC) of the Guangxi Veterinary Research Institute. Specimen collection from diseased chickens and SPF chickens, the use of chicken embryo fibroblasts (CEFs) and the collection of allantoic fluid from SPF chicken embryos were performed in accordance with the IACUC protocol to minimise animal suffering. The CEFs and embryos were then used for virus propagation.
Viruses and clinical specimens
MDV, ALV (three subgroups of ALV, A/B/J), REV, IBDV, CIAV, ARV and the other viruses used in this study are listed in Table 2. MDV, ALV-A/B/J, and REV were propagated in chicken embryo fibroblasts (CEFs), and IBDV and ARV were propagated in SPF chicken embryos. CIAV was propagated using a homogenate of positive samples of chicken liver and bone marrow to infect SPF 1-day-old broilers, from which the bone marrow was collected at 10 days after infection. Clinical tissue specimens of the thymus, bursa, spleen, bone marrow, blood and liver were collected from diseased chickens at poultry farms.
RNA/DNA extraction and RNA reverse transcription
MDV DNA from infected CEFs, CIAV DNA from the bone marrow of affected chickens, REV and ALV-A/B/J genomic proviral DNA from infected CEFs, and IBDV and ARV RNA from allantoic fluid were extracted using the E.Z.N.A.® Total DNA/RNA Isolation Kit (OMEGA, Norcross, GA, USA). The extracted DNA/RNA was eluted in DNase- and RNase-free dH2O and stored at −80 °C. The RNA was used to generate cDNA via reverse transcription, as described previously [36].
GeXP-multiplex PCR primer design
Gene-specific primers were designed based on sequence information obtained from GenBank using Primer premier 5.0 software (Premier, Palo Alto, USA) and NCBI Primer-Blast. ALV-A/B/J primers were designed to correspond to a specific region of the envelope gene gp85, MDV primers to a specific region of the gene encoding Marek’s EcoRI-Q protein (meq), REV primers to a specific region of the envelope gene gp90, IBDV primers to a specific region of the capsid protein gene VP2, CIAV primers to a specific region of the capsid protein gene VP1, and ARV primers to a specific region of the S1 gene. The primer sequences, the sizes of the resulting amplicons, and the target regions are listed in Table 2. The chimeric primers consisted of a universal sequence fused to the 5′-end of a gene-specific sequence. The forward universal primer was labelled at the 5′-end with the fluorescent dye Cy5. All chimeric primers and universal primers were synthesised and purified by polyacrylamide gel electrophoresis (Invitrogen, Shanghai, China).
GeXP-multiplex PCR assay
GeXP-multiplex PCR assays were performed using the Genome Lab GeXP Starter Kit (Beckman Coulter, Brea, USA) in a 20-μl volume containing 4 μl of 5× buffer, 0.25 μM (final concentration) universal forward and reverse primers, 2 μl of MgCl2 (25 μM), 1 μl of chimeric primer mixture, 0.35 μl of Thermo-Start DNA polymerase, 1 μl of cDNA/DNA, and nuclease-free H2O. The concentration of the chimeric primers was optimised according to the amplification efficiency of the GeXP-multiplex PCR assay.
The GeXP-multiplex PCR assay was performed via a three-step amplification procedure after a 5-min incubation at 95 °C: 10 cycles of 30 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C; 10 cycles of 30 s at 94 °C, 30 s at 68 °C, and 30 s at 72 °C; 20 cycles of 30 s at 94 °C, 30 s at 50 °C, and 30 s at 72 °C; and 5 min at 72 °C. The reactions were then held at 4 °C in the thermal cycler (Thermo, Milford, USA).
Separation by capillary electrophoresis and fragment analysis
After amplification, 1 μl of PCR product was added to 38.75 μl of sample loading solution along with 0.25 μl of DNA size standard 400 (Beckman Coulter, Brea, USA). The fluorescently labelled amplicons were separated into distinct peaks on a electropherogram via GeXP high-resolution capillary electrophoresis and then identified by their respective sizes. The dye signal strength of each peak was measured in A.U. of optical fluorescence and was defined as the fluorescence signal minus the background signal. The data were imported into the analysis module of eXpress Profiler software (Beckman Coulter, Brea, USA) as a tab-delimited file for subsequent analyses.
Specificity and sensitivity of the GeXP-multiplex PCR assay
The assay specificity for each immunosuppressive viral target was individually tested with a mixture of 8 sets of chimeric primers in a multiplex PCR assay after optimisation. Other conventional chicken viruses, including the H5/H7/H9 serotypes of avian influenza virus (AIV), Newcastle disease virus (NDV), infectious bronchitis virus (IBV) and infectious laryngotracheitis virus (ILTV), were used as negative controls. DNA from the thymus, spleen and bursa of SPF chickens was also used as a negative control.
Specific PCR amplicons for each virus were individually cloned into the pGEM-T vector (Promega, Madison, USA), and the plasmids were purified and sequenced. The sequence data were analysed and compared with the corresponding sequence data in GenBank. The IBDV and ARV plasmids were linearised with SpeI (Takara, Dalian, China) and then in vitro transcribed into ssRNA using the RiboMAXLarge Scale RNA Production Systems SP6/T7 Kit (Promega, Madison, USA). After DNase I digestion, ssRNA was purified with TRIzol (Invitrogen, Shanghai, China) and chloroform. The concentration of plasmid DNA and transcribed ssRNA was measured at 260 nm using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, USA), and the copy number was calculated [40]. Plasmid DNA and transcribed ssRNA were diluted to a final concentration ranging from 105 copies/μl to 1 copy/μl and then subjected to the GeXP-multiplex PCR assay with 8 sets of chimeric primers, both individually and in pre-mixed solutions. The sensitivity of the GeXP-multiplex PCR assay was re-evaluated three times on three different days.
Artificial mixture and interference assays
To simulate co-infection, thymus, spleen, bursa, bone marrow, blood and liver samples from chickens previously diagnosed with the immunosuppressive viruses were randomly chosen and mixed together in various amounts; DNA/RNA was then extracted. cDNA was generated from RNA as described above. GeXP-multiplex PCR assays were performed using the mixed DNA/cDNA.
Different concentrations of specific DNA-containing plasmids and in vitro ssRNA transcripts (107 copies/μl to 102 copies/μl) were used as templates for GeXP-multiplex PCR assays to evaluate interference, and the results were compared with those of single-template GeXP-multiplex PCR assays.
Evaluation of the GeXP-multiplex PCR assay with clinical specimens
A total of 300 clinical specimens, including the thymus gland, spleen, bursa, bone marrow, blood and liver of diseased chickens, were collected from farms. These diseased chickens showed various symptoms, with most exhibiting depression and anepithymia and some showing extreme emaciation. One-third of the diseased chickens were diarrheic, and approximately half showed respiratory symptoms; some chickens displayed both diarrhea and respiratory symptoms. Approximately one-fourth of the diseased chickens exhibited a poor reaction to vaccination or was prone to bacterial infection. Mortality ranged from 2 % to 30 %. Tumours in the liver, heart, spleen or skin were observed in only approximately 20 chickens and not always in those showing extreme emaciation.
DNA/RNA was extracted, and RNA was reverse transcribed as described above. The GeXP-multiplex PCR assay was performed using DNA/cDNA as the template, and the results were confirmed using independent real-time PCR/RT-PCR and sequencing to determine true positives. Independent real-time PCR/RT-PCR was performed using the primers described above but without the universal sequence at the 5′-end. The results of GeXP-multiplex PCR, real-time PCR/RT-PCR and sequencing were analysed by the Kappa statistical method using SPSS software (IBM, New York, USA).
Acknowledgements
The authors thank Dr. Mazhar I Khan for his critical review and editing of the manuscript. This work was supported by the National Natural Science Foundation (31160512), the Guangxi Science and Technology Bureau (1222003-2) and the Guangxi Government Senior Scientist Foundation (2011B020).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TT Zeng designed the primers, established the PCR assay and drafted the manuscript. ZX Xie conceived the study and participated in its design. LJ Xie and XW Deng participated in the specificity and sensitivity analyses of the GeXP-multiplex PCR assay. ZQ Xie, SS Luo, L Huang and JL Huang prepared the viruses and specimens. All the authors read and approved the final manuscript.
References
- 1.Calnek B. Pathogenesis of Marek’s disease virus infection. In: Kanji Hirai editors. Marek’s Disease. Berlin: Springer; 2001. p. 25-55. [DOI] [PubMed]
- 2.Enrietto PJ, Wyke JA. The Pathogenesis of Oncogenic Avian Retrovirusesxs. Adv Cancer Res. 1983;39:269–314. doi: 10.1016/S0065-230X(08)61037-4. [DOI] [PubMed] [Google Scholar]
- 3.Cui Z, Sun S, Wang J. Reduced serologic response to Newcastle disease virus in broiler chickens exposed to a Chinese field strain of subgroup J avian leukosis virus. Avian Dis. 2006;50(2):191–5. doi: 10.1637/7409-071305R1.1. [DOI] [PubMed] [Google Scholar]
- 4.Fadly A, Witter R, Lee L. Effects of chemically or virus-induced immunodepression on response of chickens to avian leukosis virus. Avian diseases. 1985;29(1):12–25. doi: 10.2307/1590690. [DOI] [PubMed] [Google Scholar]
- 5.Sharma JM, Kim I-J, Rautenschlein S, Yeh H-Y. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev. Comp. Immunol. 2000;24(2):223–35. doi: 10.1016/S0145-305X(99)00074-9. [DOI] [PubMed] [Google Scholar]
- 6.Smyth J, Moffett D, Connor T, McNulty M. Chicken anaemia virus inoculated by the oral route causes lymphocyte depletion in the thymus in 3-week-old and 6-week-old chickens. Avian Pathol. 2006;35(3):254–9. doi: 10.1080/03079450600717349. [DOI] [PubMed] [Google Scholar]
- 7.Roessler DE. Studies on the pathogenicity and persistence of avian reovirus pathotypes in relation to age resistance and immunosuppression: University of Delaware. 1987. [Google Scholar]
- 8.Faragher J, Allan W, Wyeth P. Immunosuppressive effect of infectious bursal agent on vaccination against Newcastle disease. Vet. Rec. 1974;95(17):385–8. doi: 10.1136/vr.95.17.385. [DOI] [PubMed] [Google Scholar]
- 9.McConnell C, Adair B, McNulty M. Effects of chicken anemia virus on cell-mediated immune function in chickens exposed to the virus by a natural route. Avian diseases. 1993;37(2):366–74. doi: 10.2307/1591660. [DOI] [PubMed] [Google Scholar]
- 10.Dong X, Ju S, Zhao P, Li Y, Meng F, Sun P, et al. Synergetic effects of subgroup J avian leukosis virus and reticuloendotheliosis virus co-infection on growth retardation and immunosuppression in SPF chickens. Vet Microbiol. 2014. [DOI] [PubMed]
- 11.Diao X, Zhang L, Cheng Z, Zhu G, Wang G, Meng X, et al. Dynamic pathology and antigen location study on broiler breeders with coinfection of MDV and REV. Sci. Agric. Sin. 2008;41(6):1838–44. [Google Scholar]
- 12.Cloud S, Lillehoj H, Rosenberger J. Immune dysfunction following infection with chicken anemia agent and infectious bursal disease virus. I. Kinetic alterations of avian lymphocyte subpopulations. Vet Immunol Immunopathol. 1992;34(3):337–52. doi: 10.1016/0165-2427(92)90174-O. [DOI] [PubMed] [Google Scholar]
- 13.McNeilly F, Smyth J, Adair B, McNulty M. Synergism between chicken anemia virus (CAV) and avian reovirus following dual infection of 1-day-old chicks by a natural route. Avian diseases. 1995;39(3):532–7. doi: 10.2307/1591806. [DOI] [PubMed] [Google Scholar]
- 14.Zuo T, Zhao Z, Wei P, Wei X, Li Y, Mo M. Isolation and identification of a field isolate of Marek’s disease virus with acute oncogenicity. 2007. [Google Scholar]
- 15.Drén C, Farkas T, Németh I. Serological survey on the prevalence of chicken anaemia virus infection in Hungarian chicken flocks. Vet Microbiol. 1996;50(1):7–16. doi: 10.1016/0378-1135(96)00002-8. [DOI] [PubMed] [Google Scholar]
- 16.Gopal S, Manoharan P, Kathaperumal K, Chidambaram B, Divya KC. Differential detection of avian oncogenic viruses in poultry layer farms and turkeys by use of multiplex PCR. J Clin Microbiol. 2012;50(8):2668–73. doi: 10.1128/JCM.00457-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackwood D, Nielsen C. Detection of infectious bursal disease viruses in commercially reared chickens using the reverse transcriptase/polymerase chain reaction-restriction endonuclease assay. Avian diseases. 1997;41(1):137–43. doi: 10.2307/1592453. [DOI] [PubMed] [Google Scholar]
- 18.Ke GM, Cheng HL, Ke LY, Ji WT, Chulu JL, Liao MH, et al. Development of a quantitative Light Cycler real-time RT-PCR for detection of avian reovirus. J Virol Methods. 2006;133(1):6–13. doi: 10.1016/j.jviromet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Kim H-R, Kwon Y-K, Bae Y-C, Oem J-K, Lee O-S. Molecular characterization of chicken infectious anemia viruses detected from breeder and broiler chickens in South Korea. Poult Sci. 2010;89(11):2426–31. doi: 10.3382/ps.2010-00911. [DOI] [PubMed] [Google Scholar]
- 20.Caterina KM, Frasca S, Jr, Girshick T, Khan MI. Development of a multiplex PCR for detection of avian adenovirus, avian reovirus, infectious bursal disease virus, and chicken anemia virus. Mol Cell Probes. 2004;18(5):293–8. doi: 10.1016/j.mcp.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Rai AJ, Kamath RM, Gerald W, Fleisher M. Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal Bioanal Chem. 2009;393(5):1505–11. doi: 10.1007/s00216-008-2436-7. [DOI] [PubMed] [Google Scholar]
- 22.Yang MJ, Luo L, Nie K, Wang M, Zhang C, Li J, et al. Genotyping of 11 human papillomaviruses by multiplex PCR with a GeXP analyzer. J Med Virol. 2012;84(6):957–63. doi: 10.1002/jmv.23275. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Zhang Y, Zhou X, Xu B, Yang M, Wang M, et al. Simultaneously typing nine serotypes of enteroviruses associated with hand, foot, and mouth disease by a GeXP analyzer-based multiplex reverse transcription-PCR assay. J Clin Microbiol. 2012;50(2):288–93. doi: 10.1128/JCM.05828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drew JE, Mayer CD, Farquharson AJ, Young P, Barrera LN. Custom design of a GeXP multiplexed assay used to assess expression profiles of inflammatory gene targets in normal colon, polyp, and tumor tissue. J Mol Diagn. 2011;13(2):233–42. doi: 10.1016/j.jmoldx.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Xu ZQ, Zhang Q, Jin M, Yu JM, Li JS, et al. Simultaneous detection of seven enteric viruses associated with acute gastroenteritis by a multiplexed Luminex-based assay. J Clin Microbiol. 2012;50(7):2384–9. doi: 10.1128/JCM.06790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin M, Wang DY, Huang F, Nie K, Qu M, Wang M, et al. Detection of pandemic influenza A H1N1 virus by multiplex reverse transcription-PCR with a GeXP analyzer. J Virol Methods. 2010;168(1-2):255–8. doi: 10.1016/j.jviromet.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Xie Z, Luo S, Xie L, Liu J, Pang Y, Deng X, et al. Simultaneous typing of nine avian respiratory pathogens using a novel GeXP analyzer-based multiplex PCR assay. J Virol Methods. 2014;207(0):188–95. doi: 10.1016/j.jviromet.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Y-f Z, Xie Z-x, Xie L-j, Deng X-w, Xie Z-q, Luo S-s, et al. GeXP analyzer-based multiplex reverse-transcription PCR assay for the simultaneous detection and differentiation of eleven duck viruses. BMC Microbiol. 2015;15(1):247. doi: 10.1186/s12866-015-0590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Xie Z, Xie L, Deng X, Xie Z, Luo S, et al. Simultaneous detection of eight swine reproductive and respiratory pathogens using a novel GeXP analyser-based multiplex PCR assay. J Virol Methods. 2015;224:9–15. doi: 10.1016/j.jviromet.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 30.H-h Z, Liu Q, Qiu B, G-z L, Z-q C. Mixed Infection of ALV-J and MDV in a Flock of Shandong Free Range Chickens [J] Chinese J Anim. Vet. Sci. 2009;8:018. [Google Scholar]
- 31.L-t Q, Pan W, Gao Y-l, X-y D, Li K, Sun F-f, et al. Investigation of co-infection of ALV-J with REV, MDV, CAV in layer chicken flocks in some regions of China. Chinese J Prev. Vet. Sci. 2010;2:006. [Google Scholar]
- 32.Z-z C, S-s M, S-j J, J-p W. Serological surveys of chicken anemia virus, avian reticuloendotheliosis virus and avian reovirus infections in white meattype chickens in China. Acta Veterinaria et Zootechnica Sinica. 2006;37(2):152. [Google Scholar]
- 33.Ducatez M, Owoade A, Abiola J, Muller C. Molecular epidemiology of chicken anemia virus in Nigeria. Arch Virol. 2006;151(1):97–111. doi: 10.1007/s00705-005-0609-7. [DOI] [PubMed] [Google Scholar]
- 34.Xie Z, Pang Y-s, Liu J, Deng X, Tang X, Sun J, et al. A multiplex RT-PCR for detection of type A influenza virus and differentiation of avian H5, H7, and H9 hemagglutinin subtypes. Mol Cell Probes. 2006;20(3):245–9. doi: 10.1016/j.mcp.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, Xie L, Fan Q, Pang Y, Deng X, Xie ZQ, et al. A duplex quantitative real-time PCR assay for the detection of Haplosporidium and Perkinsus species in shellfish. Parasitol Res. 2013;112(4):1597–606. doi: 10.1007/s00436-013-3315-5. [DOI] [PubMed] [Google Scholar]
- 36.Xie Z, Xie L, Pang Y, Lu Z, Xie Z, Sun J, et al. Development of a real-time multiplex PCR assay for detection of viral pathogens of penaeid shrimp. Arch Virol. 2008;153(12):2245–51. doi: 10.1007/s00705-008-0253-0. [DOI] [PubMed] [Google Scholar]
- 37.Telesnitsky A, Goff S. Reverse transcriptase and the generation of retroviral DNA. 1997. [PubMed] [Google Scholar]
- 38.Zavala G, Cheng S. Detection and characterization of avian leukosis virus in Marek’s disease vaccines. Avian Dis. 2006;50(2):209–15. doi: 10.1637/7444-092405R.1. [DOI] [PubMed] [Google Scholar]
- 39.Sharma J. Effect of infectious bursal disease virus on protection against Marek’s disease by turkey herpesvirus vaccine. Avian diseases. 1984;28(3):629–40. doi: 10.2307/1590231. [DOI] [PubMed] [Google Scholar]
- 40.Chen N-H, Chen X-Z, Hu D-M, Yu X-L, Wang L-L, Han W, et al. Rapid differential detection of classical and highly pathogenic North American Porcine Reproductive and Respiratory Syndrome virus in China by a duplex real-time RT-PCR. J Virol Methods. 2009;161(2):192–8. doi: 10.1016/j.jviromet.2009.06.007. [DOI] [PubMed] [Google Scholar]


