Abstract
Background
In most bacteria, various jumping genetic elements including insertion sequences elements (IS elements) cause a variety of genetic rearrangements resulting in harmful effects such as genome and recombinant plasmid instability. The genetic stability of a plasmid in a host is critical for high-level production of recombinant proteins, and in this regard, the development of an IS element-free strain could be a useful strategy for the enhanced production of recombinant proteins. Corynebacterium glutamicum, which is a workhorse in the industrial-scale production of various biomolecules including recombinant proteins, also has several IS elements, and it is necessary to identify the critical IS elements and to develop IS element deleted strain.
Results
From the cultivation of C. glutamicum harboring a plasmid for green fluorescent protein (GFP) gene expression, non-fluorescent clones were isolated by FACS (fluorescent activated cell sorting). All the isolated clones had insertions of IS elements in the GFP coding region, and two major IS elements (ISCg1 and ISCg2 families) were identified. By co-cultivating cells harboring either the isolated IS element-inserted plasmid or intact plasmid, it was clearly confirmed that cells harboring the IS element-inserted plasmids became dominant during the cultivation due to their growth advantage over cells containing intact plasmids, which can cause a significant reduction in recombinant protein production during cultivation. To minimize the harmful effects of IS elements on the expression of heterologous genes in C. glutamicum, two IS element free C. glutamicum strains were developed in which each major IS element was deleted, and enhanced productivity in the engineered C. glutamicum strain was successfully demonstrated with three models: GFP, poly(3-hydroxybutyrate) [P(3HB)] and γ-aminobutyrate (GABA).
Conclusions
Our findings clearly indicate that the hopping of IS elements could be detrimental to the production of recombinant proteins in C. glutamicum, emphasizing the importance of developing IS element free host strains.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0401-7) contains supplementary material, which is available to authorized users.
Keywords: Corynebacterium glutamicum, IS element, FACS screening, Poly(3-hydroxybutyrate), γ-aminobutyrate
Background
Corynebacterium glutamicum is a Gram-positive bacterium which is non-pathogenic, non-sporulating, and generally-regarded as safe (GRAS) [1, 2]. Due to its safety and process performance, it has been used as one of the best amino acid producers in industry and for the production of various bio-based chemicals such as poly(3-hydroxybutyrate) [P(3HB)], γ-aminobutyrate, cadaverine, and putrescine [3, 4]. Recently, C. glutamicum has also been regarded as a potential emerging host for the production of many recombinant proteins because it can secrete proteins in a non-pathogenic background, which can be advantageous in biopharmaceutical protein production with subsequent downstream protein purification processes [5, 6]. However, although C. glutamicum is an efficient host strain, several properties remain to be improved. In particular, mobile genetic elements (MGEs) are problematic for bacterial genome and plasmid stability [7, 8]
After the discovery of the transposon in 1950 [9], mobile genetic elements (MGEs) have been studied in many organisms including bacteria. MGEs are the key factors in genome rearrangement and evolution, and they have important roles in the adaptations of bacteria to their environment by inversion, recombination, and insertion [10, 11]. There are various classes of bacterial MGEs: insertion sequence elements (IS elements), prophages, bacterial interspersed mosaic elements, miniature inverted-repeat transposable elements, repetitive extragenic palindromic elements sequences, etc. [12]. In C. glutamicum, a few studies on the effects of MGEs on the bacterial cells and their protein productivities have been reported. The C. glutamicum prophage CGP3 has an increased copy number under stress conditions including UV light, genotoxic agent, or DNA damage, and bacterial cells containing multiple prophage DNA molecules were more prone to cell lysis [7]. In addition, the enhanced production of recombinant proteins and high transformation efficiency were achieved in prophage-free C. glutamicum [8]. Also, there was an attempt to construct chassis strain of C. glutamicum by removing 36 unnecessary gene clusters which include some of IS elements [13].
As another important factor, IS elements have been studied in many bacterial species [14, 15]. IS elements usually encode transposases and terminal inverted repeats (TIR) which determine the IS family. IS elements move by encoding transposases and insert into specific sequences by the TIR [16]. Most IS elements have no activity by themselves, but when they are inserted into a gene, they change the gene expression by activation or disruption [17]. They induce large duplication and inversion, and large genomic rearrangement making the genome unstable [18–20]. Additionally, IS elements can cause a significant instability of plasmid that carry heterologous genes, which cause the reduced production of the encoded proteins. For example, in the production of a DNA vaccine which is produced from a plasmid in E. coli, the insertion of IS elements from the chromosome into the plasmid reduced the productivity [21, 22]. In another case, cells harboring IS inserted plasmids had a better growth rate than that of cells harboring non IS inserted plasmids, which encoded the target recombinant protein, and this difference in growth rate induced about a 25 % reduction in productivity [23]. Thus, a stable and efficient host strain that is free from the adverse effects of IS elements is highly desirable for both laboratory and industrial applications.
In this study, we first sought to identify the major IS elements in C. glutamicum, of which insertion can cause the disruption of genes in plasmid. Using a FACS-based high throughput screening system with the green fluorescent protein (GFP) as the reporter protein, the major IS element could be identified. Next, we tried to engineer C. glutamicum in which the major IS element was deleted in the chromosomal DNA and, using the engineered C. glutamicum, we demonstrated the enhanced production of recombinant proteins, poly(3-hydroxybutyrate) [P(3HB)], and γ-aminobutyrate (GABA).
Results
Isolation of IS elements by FACS and their identification
To isolate the major IS elements in C. glutamicum, a GFP reporter system (pCES-H36-GFP) was constructed in which GFP expression was under a strong constitutive promoter (PH36). If the IS element is inserted in the GFP coding sequence, cells cannot produce functional GFPs, and those cells can be selectively sorted by a high–speed FACS sorter. After flask cultivation, the fluorescence intensity of the C. glutamicum harboring either pCES-NMCS or pCES-H36-GFP was analyzed by flow cytometry. Cell harboring pCES-NMCS which was used as negative control had no fluorescence intensity (Fig. 1a). Most cells harboring pCES-H36-GFP showed high fluorescence intensities (mean fluorescent intensity ≅8000); however, there was also a small population (about ~0.6 %) with no fluorescence (Fig. 1b). To identify the cells in this region, cells with no fluorescence were selectively sorted by the FACS sorter. In the first round of sorting, approximately 3.10 × 107 cells were screened, and the bottom 0.61 % of cells (~1.9 × 105 cells) were sorted. The sorted cells were cultivated and used for the next round of sorting. After the first round of sorting, the cell population showing no fluorescence was enriched (about 50 % of the total population) (Fig. 1c), and these non-fluorescent cells (~2.1 × 105 cells) were also selectively sorted. After the second round of sorting, most of the population was found to have no fluorescent signal (Fig. 1d), and we concluded that the non-fluorescent cells were successfully enriched.
Fig. 1.
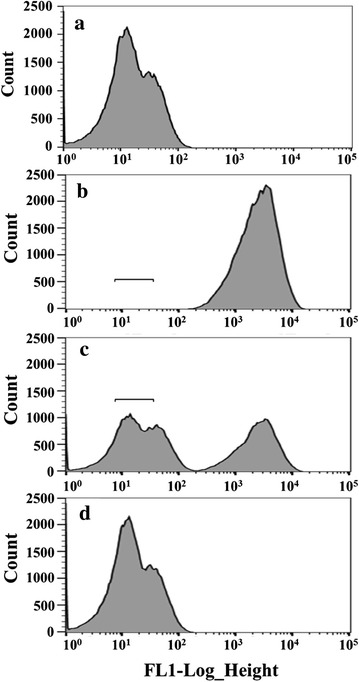
FACS sorting of low fluorescent cells in the cultivation of wild type C. glutamicum harboring pCES-H36-GFP. a The histogram of wild type C. glutamicum harboring pCES-NMCS (negative control). b, c, d The histogram of wild type C. glutamicum harboring pCES-H36-GFP (b), the 1st round sorted cells (c) and the 2nd round sorted cells (d). The bars indicate the sorting region
In each round of FACS screening, plasmids were prepared from the sorted cells, and after digestion with the BamHI restriction enzyme, they were analyzed on agarose gels. In the original population, a single band for the plasmid was clearly observed for which the size coincided well with that of pCES-H36-GFP (~6.6 kb) (lane 1 of Fig. 2a). However, in the samples after the first round of sorting, another distinct band was also observed at approximately 8 kb. The ratio of the densities between the two bands was about 50:50 (lane 2 of Fig. 2a). After the second round of sorting, it was clearly observed that the 8 kb-long plasmid became dominant in the sorted cells (lane 3 of Fig. 2a). After the second round of sorting, 14 clones were randomly selected, and GFP expression in each clone was analyzed by SDS-PAGE. As expected, all 14 clones did not produce GFP (Additional file 1: Figure S1). We performed sequencing experiment for the DNA from 72-bp upstream of H36 promoter to 71-bp downstream of GFP stop codon. The sequences of the GFP coding regions for the 14 clones were determined by sequencing which showed that all 14 clones had insertions of IS elements at various positions in the GFP coding region (Fig. 2b). However, we could not find any insertion of IS element in promoter region. From the sequencing analysis, it was found that two types of IS elements were inserted, and the IS elements were identified to be part of the ISCg1 and ISCg2 family, by searching the C. glutamicum genome database. Among the 14 clones, 11 clones contained insertions from the ISCg2 family, and the rest (3 clones) contained insertions from the ISCg1 family (Fig. 2b).
Fig. 2.
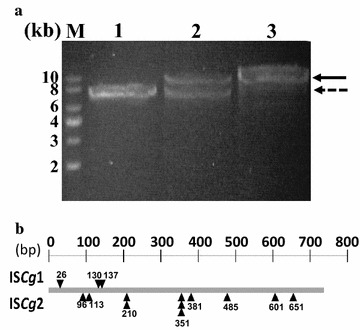
Analysis of the isolated cells by FACS screening. a Confirmation of plasmids by agarose gel electrophoresis. Lane M represents DNA size markers (kb). Lanes 1 represents plasmid from the original cell cultivation harboring pCES-H36-GFP. Lanes 2 and 3 represent plasmids from the 1st round sorted cells and the 2nd round sorted cells. The dashed and solid arrows indicate the correct pCES-H36-GFP and IS element-inserted plasmid, respectively. b Location of IS element insertion in egfp. Gray bar indicates the open read frame of egfp. Arrows indicate the IS element insertion site. Upper and lower region represent ISCg1 and ISCg2, respectively. Double or triple triangles means double or triple clones
Effect of gene deletion by IS element insertion on cell growth
Cells containing IS element-inserted plasmid cannot produce the target protein, and in many reports, it is well recognized that cells producing recombinant proteins have slower cell growth (very poor growth in some cases) compared to that of non-producing cells due to the metabolic burden or other problems including the toxicity of the proteins to the host cells, etc. [23]. Particularly, in large-scale production which generally needs long operation time, the fast growth of non-producing cells can be a very serious issue due to a significant decrease in protein productivity by the overgrowth of non-producing cells. To confirm this deleterious effect of IS element insertion on cell growth, two expression systems were constructed: (1) α-Amylase gene expression under the constitutive PH36 promoter (pCES-H36-porBss-Amy) and (2) IS element-inserted version (pCES-H36-porBss-IS-Amy) in which ISCg1 was inserted in the middle of signal sequence. C. glutamicum harboring each plasmid were cultivated in flasks, and the growth of each strain was compared. Additionally, IS element-inserted C. glutamicum cells were mixed with C. glutamicum producing α-amylase cells at different ratios (1:103, 1:104, and 1:105), and their growth rates were also compared. As shown in Fig. 3, C. glutamicum harboring the pCES-H36-porBss-Amy grew slowly (specific growth rate, μ = 0.268 ± 0.052 h−1) after a long lag phase and reached stationary phase around 36 h. In contrast, the IS element-inserted version (C. glutamicum harboring pCES-H36-porBss-IS-Amy) grew much faster (μ = 0.552 ± 0.012 h−1) after a short lag phase and reached stationary phase much earlier (around 12 h). In the co-culture with the IS element-inserted cells and α-amylase-producing cells, the overall growth rates were also increased as the ratio of the IS element-inserted cells increased from 1:105 to 1:103 (μ = 0.305 ± 0.021, μ = 0.405 ± 0.041, and μ = 0.498 ± 0.039, respectively). In the co-culture experiments, the change in population during cultivation was analyzed by PCR with individual clones. At the end of each cultivation (at 36 h), cells were spread onto an agar plate, and 30 colonies from each plate were randomly selected, and the plasmids were confirmed by PCR experiment. For both ratios of 1:103 and 1:104, all the colonies were IS element-inserted clones, and for the ratio of 105, IS element-inserted clones were also the major population (86 %) (data not shown). We also checked α-amylase activity in all cultivations at 36 h time-point, and we found that the cultivation of C. glutamicum harboring pCES-H36-porBss-Amy (without mixing with IS element-inserted clones) showed much higher activity (6.14 ± 0.44 U/mL) than those of other mixed cultures (Fig. 3b). In the cultivation of cells containing pCES-H36-porBss-Amy only, the possible insertion of IS element into plasmid was also analyzed by PCR but we could not find any insertion in the plasmid during the cultivation (data not shown). These results clearly show that the insertion of the IS element causes a significant decrease in protein productivity by the overgrowth of non-producing cells, and it is postulated that the removal of the IS element in the chromosome could be an important strategy for enhancing the production of target molecules in C. glutamicum.
Fig. 3.
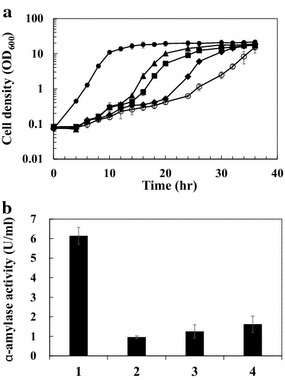
Determination of cell population change during co-culture of C. glutamicum wild type harboring pCES-H36-porBss-Amy and pCES-H36-porBss-IS-Amy. a Growth curves during the cultivation. Open circles and closed circles represent C. glutamicum harboring pCES-H36-porBss-Amy and C. glutamicum harboring pCES-H36-porBss-IS-Amy, respectively. Triangles, squares and diamonds represent co-culture of C. glutamicum (pCES-H36-porBss-IS-Amy) and C. glutamicum (pCES-H36-porBss-Amy) with the ratio of 1:103, 1:104, and 1:105, respectively. b α-Amylase activity of the co-culture. 1, 2, 3, and 4 represent enzyme activity in the cultivation of C. glutamicum (pCES-H36-porBss-Amy), co-culture with the ratio of 1:103, 1:104, and 1:105, respectively
Deletion of the IS families (ISCg1 and ISCg2)
It has been reported that four copies of ISCg1 (ISCg1a, ISCg1b, ISCg1c, and ISCg1d) and five copies of ISCg2 (ISCg2b, ISCg2c, ISCg2d, ISCg2e, and ISCg2f) exist in the chromosomal DNA of C. glutamicum [24]. Each of the IS elements was deleted sequentially, and finally, two strains, WJ004 with the deletion of ISCg1 family and WJ008 with the deletion of ISCg2 family, were developed. The successful deletion of each IS element was clearly confirmed by PCR (Additional file 2: Figure S2). Interestingly, during the deletion of the IS elements, we discovered new information about the copy number and location of the IS elements. First, only four copies of ISCg2 were found in C. glutamicum ATCC 13032. ISCg2e which was reported to exist at position (2317958..2319472; GenBank No. NC_006958.1) was not found in this study. Actually, after deletion of the four IS elements (ISCg2b, ISCg2c, ISCg2d, and ISCg2f) in C. glutamicum, no PCR products for the ISCg2 family members were detected which means ISCg2 was not present in the chromosome any more. Second, we also found that ISCg1c, one of the ISCg1 family members, does not exist in the reported position (611123..612433; GenBank No. NC_006958.1); however, we found a new ISCg1 element at another position (613777..615087; GenBank No. NC_006958.1). This IS element was named as ISCg1e which was also deleted in the WJ004 strain. All IS elements deleted in this study are listed in Table 1. After developing both IS element-deleted mutants, the possible jumping of IS elements to other positions, was also checked by PCR, but we could not find any more ISCg1 (in WJ004) and ISCg2 (in WJ008) (data not shown).
Table 1.
List of deleted IS elements in C. glutamicum ATCC13032 in this study
| IS family | Copy number | IS element | Position |
|---|---|---|---|
| ISCg1 | 4 | ISCg1a | 1115955..1117265 |
| ISCg1b | 2595722..2597032 | ||
| ISCg1d | 2481196..2482506 | ||
| ISCg1e a | 613777..615087 | ||
| ISCg2 b | 4 | ISCg2b | 3005658..3007172 |
| ISCg2c | 2716287..2717801 | ||
| ISCg2d | 2317958..2319472 | ||
| ISCg2f | 192954..194468 |
aNew IS element found in this study. This gene was not reported in both C. glutamicum genome sequences deposited with GenBank Accession numbers NC_003450 and NC_006958.1
bISCg2e in ISCg2 family was absent in C. glutamicum ATCC13032 in our lab stock
Enhanced production of recombinant protein in the IS element-deleted strain
To evaluate the effect of the IS element deletions on the production of recombinant proteins, the expression of GFP in the engineered strains (WJ004 and WJ008) was examined. In flask cultivations, cell growth and fluorescence intensity of both strains harboring pCES-H36-GFP were measured and we found all strains showed similar growth rates as that of wild-type cell harboring same plasmid (Additional file 3: Figure S3). Among the three strains, the WJ004 strain, in which the ISCg1 element was deleted, had almost the same production yield as the wild type C. glutamicum; however, the WJ008 strain, in which the ISCg2 elements were deleted, had an improved production yield as high as 130 % compared with WJ004 and wild type C. glutamicum (Fig. 4a). The higher production of GFP in WJ008 strain than other strains was also confirmed by Western blotting (Fig. 4b). In addition, the portion of non-fluorescent cells in each IS element-deleted strain harboring pCES-H36-GFP were also analyzed by FACS. In this analysis, WJ004 showed similar portion of non-fluorescent cells compared with that of wild type cell, but the portion of non-fluorescent cells in the cultivation of WJ008 decreased relatively by 66 %. These results indicate that the ISCg2 element is a more critical factor causing adverse effects on the productivity of heterologous proteins by insertion into the target gene, and this result is also agreed with the FACS screening result in which the ISCg2–inserted clones were the major population (11 out of 14 clones) in the isolated cells.
Fig. 4.
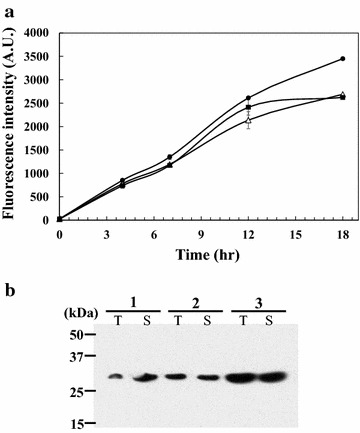
Comparison of GFP production in IS element-deleted mutants. a Analysis of fluorescence intensity in the cell by FACS. C. glutamicum WJ004 harboring pCES-H36-GFP and C. glutamicum WJ008 harboring pCES-H36-GFP are represented by squares and circles, respectively. Open triangles represent wild type C. glutamicum harboring pCES-H36-GFP. b Analysis of GFP production by Western blotting. Lanes 1, 2, and 3 represent the protein sample from wild type C. glutamicum, WJ004, and WJ008 harboring pCES-H36-GFP at 18 h. Lanes T and S represent total protein fraction and soluble protein fraction. Protein sample was prepared from the same cell concentration which was normalized to OD600 of 4
Enhanced production of P(3HB) and GABA in the IS element-deleted strain
The production of useful metabolites in C. glutamicum can be achieved by the expression of biosynthesis genes. However, if the IS element is inserted into the biosynthesis genes, those genes cannot be expressed, and the productivity of the target metabolites subsequently decreases. In this regard, the use of an IS element-deleted mutant can be beneficial for the biosynthesis of metabolites. To demonstrate this beneficial concept, the production of (1) poly(3-hydroxybutyrate) [P(3HB)], biodegradable polymers [25], and (2) gamma-aminobutyric acid (GABA), the bioactive component in various foods and pharmaceutical products [26], were examined with the IS element-deleted mutants. For the biosynthesis of P(3HB) in C. glutamicum, C. glutamicum wild type, WJ004 and WJ008 strains were transformed with the plasmid, pCES-H36-PhaCAB, in which the expressions of three genes (phaC, phaA and phaB) from Ralstonia eutropha were under the constitutive PH36 promoter. In these cultivations, all cells showed almost same growth profiles (Additional file 4: Figure S4). After flask cultivation for 24 h, the P(3HB) content in each strain was analyzed by gas chromatography. As shown in Fig. 5a, the use of the WJ008 strain resulted in a higher content (23.4 ± 0.36 wt%) than that in the WJ004 and wild type C. glutamicum strain (19.1 ± 0.44 wt% and 17.1 ± 0.55 wt%, respectively). For the biosynthesis of GABA in C. glutamicum, the plasmid pHGmut, in which the expression of glutamate decarboxylase (gadB) from Escherichia coli is under the constitutive PH36 promoter, was transformed into the WJ004 and WJ008 strains, and were cultured for 72 h. A wild type C. glutamicum grew a little faster than others, but all strains reached almost same optical density after 36 h cultivation (Additional file 4: Figure S4). The GABA content in each strain was analyzed by liquid chromatography. Like in the previous results, higher production of GABA (9.43 ± 0.52 g/L) could be obtained in the WJ008 strain than in the WJ004 and wild type C. glutamicum strain (8.34 ± 0.62 and 8.17 ± 0.66 g/L, respectively) (Fig. 5b).
Fig. 5.
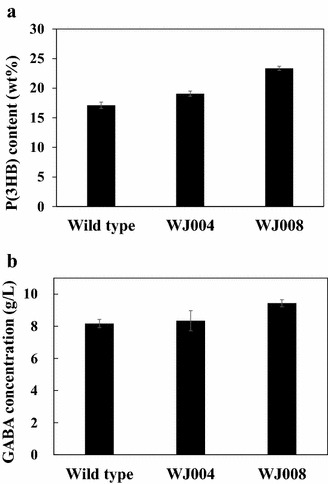
Production of a secondary metabolite. a Production of P(3HB) in the wild type C. glutamicum, WJ004 and WJ008 harboring pCES-H36-PhaCAB. All samples were prepared after 24 h of cultivation. b Production of GABA in the wild type C. glutamicum, WJ004 and WJ008 harboring pHGmut. All samples were prepared after 72 h of cultivation
Effect of IS element deletion on transformation efficiency
An earlier study [8] showed that the deletion of prophages in the chromosome of C. glutamicum (ATCC 13032) resulted in an increase in the transformation efficiency due to the deletion of the restriction modification (RM) system present in the prophage gene. Takahashi N et al. [27] also reported that the movement of the restriction modification system in Escherichia coli was linked with an IS element. In this regard, the deletion of IS elements may also have a positive effect on the transformation efficiency; thus, the effect of the IS element deletion was examined on the transformation efficiency as another characteristic of the IS element-deleted strains. The two IS element deleted mutants (WJ004 and WJ008) and wild-type C. glutamicum strain were transformed with the same plasmid (pCES-H36-GFP), and their transformation efficiencies were compared by checking the colony forming units (CFUs) on selective agar plates. As shown in Fig. 6, the WJ004 and WJ008 strains had about a four- and six-fold higher transformation efficiency (12.2 ± 0.94 × 105 cfu/μg and 19.8 ± 1.2 × 105 cfu/μg) than that of the wild type strain (3.86 ± 0.18 × 105 cfu/μg), respectively.
Fig. 6.
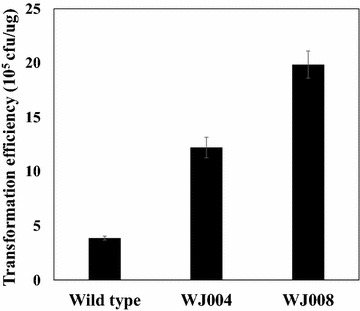
Transformation efficiency of the wild type C. glutamicum, WJ004 and WJ008. All error bars represent standard deviations of six-times repeated experiments
Discussion
Maximizing the production yield of the target product is crucial in the industrial biotechnology field, and for microbial production of recombinant proteins, it is necessary to construct a stable gene expression system. On a few occasions, the disruption of the heterologous gene occurs when IS elements are inserted inside the gene coding region [28, 29]. The disrupted gene cannot be translated into the desired protein, so it can decrease the production yield of the recombinant protein. In addition, the overgrowth of non-producing cells can cause a serious problem in the overall productivity in industrial cultivation. In this study, we isolated the major IS elements with a FACS-based high throughput screening system, and we also developed C. glutamicum strains in which the major IS elements were completely removed. So far, there have been only a few reports on the IS elements in C. glutamicum, and all of them were focused on the isolation of IS elements and identification of their locations in the genome. However, the adverse effects of IS elements on recombinant protein production in C. glutamicum has not been investigated yet. As shown in Fig. 3, C. glutamicum harboring the IS-inserted plasmid, in which the target protein cannot be produced, had a much higher cell growth compared with cells harboring intact plasmids in which the target proteins were overproduced. This difference in cell growth can be very serious in industrial fermentations which generally need long cultivation times. Even though the possibility of an IS element insertion event is very low at about 1 × 10−5 [28, 30, 31], IS element-inserted cells that can grow much faster become dominant during the fermentation, and consequently, the production yield can be significantly reduced. If the target molecules are not favorable to the C. glutamicum cells, this harmful effect on cell growth and productivity can be a much more serious issue.
The deletion of IS elements in a host strain can be one strategy to minimize the harmful effect of IS elements on the production of recombinant proteins. However, in most bacteria including C. glutamicum, IS elements are present in multiple copies [24, 32], and there have been a few attempts to develop a minimized genome strain in which all the IS elements were removed [33, 34]. However, although there has been significant progress in genome editing technology in the last decade, genome minimizing is still laborious and challenging work. Additionally, not all genome-minimized strains have always shown better performance in cell growth and protein productivity compared to the wild-type strains [23, 29]. One research group attempted to prevent IS element insertion using the ‘Clean Genome’ of the E. coli MDS42 strain in which 15 % of the genome including the IS elements is reduced [29]. However, the MDS42 strain had a lower expression level of GFP because the reduced genome made the cells very unstable, and it also took more time to maximize the growth rate given the extra metabolic load [29]. Thus, instead of deleting of all the IS elements, the deletion of specific IS elements is necessary to engineer a C. glutamicum strain with a high growth rate and productivity, and for this purpose, it is necessary to isolate the major IS elements which are most frequently inserted into heterologous genes. For this, FACS screening strategy was introduced, and we could successfully isolate two important IS elements (ISCg1 and ISCg2). Of the two IS elements, the ISCg2 was more dominant than that of ISCg1 (Fig. 2b), and this means ISCg2 has a relatively higher transposable activity than that of ISCg1, and the deletion of ISCg2 can be more effective in the production of target molecules than deletion of ISCg1 as we clearly showed with the three target biomolecules, GFP, P(3HB) and GABA. In the FACS screening, only 0.6 % cells in the total population which showed no fluorescent signals, was sorted (Fig. 1), but the WJ008 strain harboring same plasmid (pCES-H36-GFP) exhibited a very high increase (about 30 %) in the GFP production compared with that of wild type strain (Fig. 4). We consider that the sorting region for FACS screening (0.6 % in the total population) in the Fig. 1b did not cover all non-fluorescent population shown in Fig. 1a. This means that the actual portion of non-producing cells were much higher than 0.6 % in the total population, and the deletion of IS element could eliminate much more non-fluorescent cells than the cells in the sorted region (0.6 %). In addition, it can be considered that the removal of non-fluorescent cells which could grow much faster, might result in the better growth of the fluorescent cells, and much higher production yield than the removed amount of non-fluorescent cells could be obtained. Likewise, the positive effect of IS element deletion on target gene expression has also been reported in E. coli [23]. Even though IS hopping on target genes occurred very rarely (10−8/gene/h), IS-free E. coli showed 120 and 125 % enhanced productivities [23].
Additionally, we would like to emphasize the usefulness of the FACS-based high throughput screening method for the isolation of IS elements. In other methods that using promoterless antibiotics resistance genes [35] and other lethal genes [36, 37] as selection markers, cells can be selected when a copy of an IS element is inserted into the selection markers, and the IS element can be identified from the isolated clone. However, those methods have a limitation in the screening size and is not suitable for the screening of large sized libraries in general. When we consider the low possibility of an IS element insertion event, the FACS-based screening method, which can screen a big population (>108 cells) in a short time, could be a more powerful tool as shown here.
As an additional advantage, we successfully confirmed that the deletion of the IS element had a highly positive effect on the transformation efficiency (Fig. 6). Transformation efficiency has a correlation with a restriction modification system (RM system), and there are reports that IS elements guide RM systems [27, 38, 39]. It is also known that IS element-RM system complexes can integrate into a plasmid or chromosomal DNA, which causes instability in the plasmid [21–23]. By deleting the IS elements, the activity of the RM system can be diminished, and as a consequence, the transformation efficiency can increase through the higher stability of the plasmid.
Conclusions
In this study, we isolated the major IS elements in C. glutamicum and successfully developed two IS element-deleted C. glutamicum strains to minimize the harmful effect of IS elements on the production of target biomolecules. Using FACS-based high throughput screening, the major IS element, which can insert into heterologous genes in a plasmid most frequently, was successfully isolated. Using three different molecules, we also successfully showed that the production yield of the target biomolecules were significantly improved by deletion of the major IS element. To the best of our knowledge, this is the first report on the isolation of major IS elements using FACS screening strategy. Although we did not try in this study, it may be worth to combine the deletions of both IS elements (ISCg1 and ISCg2) in one strain. The development of that mutant and its use for the overproduction of recombinant proteins in the large-scale cultivation will be the future work of ours. Our study presents the IS element could be harmful to the production of recombinant proteins in C. glutamicum, emphasizing the importance of the development of IS-deleted host strains. Thus, we believe that the IS element deleted strain with its enhanced genetic stability can be a robust and appropriate host for the production of recombinant proteins and many other biomolecules.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 2. E. coli XL1-blue was used as the host for gene cloning and plasmid maintenance. C. glutamicum ATCC 13032 was used for isolation of the major IS elements and the production of recombinant proteins. E. coli was cultivated in Luria–Bertani (LB) medium (BD, Franklin Lakes, New Jersey) at 37 oC. C. glutamicum was cultivated at 30 oC in brain heart infusion (BHI) medium (BD) with or without 2 % (w/v) d-glucose and shaking at 200 rpm. Additionally, a defined medium containing 20 g/L d-glucose, 3 g of K2HPO4, 1 g of KH2PO4, 2 g of urea, 10 g of (NH4)2SO4, 2 g of MgSO4, 200 μg of biotin, 5 mg of thiamine, 10 mg of calcium pantothenate, 10 mg of FeSO4, 1 mg of MnSO4, 1 mg of ZnSO4, 200 μg of CuSO4, and 10 mg of CaCl2 per liter [5] and modified GP medium [26] were used. For the co-culture experiment, wild-type C. glutamicum harboring pCES-H36-porBss-Amy or pCES-H36-porBss-IS-Amy were inoculated into separate BHI medium. After 24 h, the cells were harvested at an OD600 of 4.0 and resuspended in 1 mL of phosphate-buffered saline (PBS). The resuspended cells were transferred into fresh defined medium at pCES-H36-porBss-IS-Amy:pCES-H36-porBss-Amy ratios of 1:103, 1:104, and 1:105. Then, cells were cultivated for 27 h at 30 oC. For the heterologous protein production assay, cells were inoculated into BHI medium. After overnight cultivation, fully grown cells were transferred into a 250 mL flask containing 50 mL of fresh BHI medium and cultivated for 24 h. To produce poly(3-hydroxy butyrate) [P(3HB)] and γ -aminobutyric acid (GABA), C. glutamicum was first inoculated into BHI medium, and after cultivation for 24 h, cells were transferred into a 250 mL flask containing 100 mL of fresh BHI medium containing 2 %(w/v) d-glucose or modified GP medium for the production of P(3HB) or GABA, respectively. All experiments were performed in triplicate. In all cultivations, kanamycin (Km, 25 μg/mL) was added to the culture medium as the sole antibiotic.
Table 2.
Bacterial strains and plasmids used in this study
| Relevant characteristics | Reference or source | |
|---|---|---|
| Strain | ||
| E. coli | ||
| XL1-blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proABlacIq ZΔM15 Tn10 (Tet r )] | Stratagenea |
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild type | ATCC |
| WJ001 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg1a) | This study |
| WJ002 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg1a, ISCg1b) | This study |
| WJ003 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg1a, ISCg1b, ISCg1c) | This study |
| WJ004 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg1a, ISCg1b, ISCg1c, ISCg1e) | This study |
| WJ005 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg2b) | This study |
| WJ006 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg2b, ISCg2c) | This study |
| WJ007 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg2b, ISCg2c, ISCg2e) | This study |
| WJ008 | ATCC 13032 with in frame deletion of ISCg2 family (ISCg2b, ISCg2c, ISCg2e, ISCg2f) | This study |
| Plasmids | ||
| pCES208 | E. coli—C. glutamicum shuttle vector, Kmr | [45] |
| pCES-NMCS | pCES208 derivative; MCS and rrn terminator, Kmr | [6] |
| pH36M2 | pCES208 derivative; PH36, PorB signal sequence, codon-optimized M18 scFv gene (opt) | [5] |
| pCES-H36-porBss | pCES208 derivative; PH36, porinB signal sequence, Kmr | This study |
| pCES-H36-porBss-Amy | pCES208 derivative; PH36, porinB signal sequence amylase, Kmr | This study |
| pCES-H36-porBss-IS-Amy | pCES208 derivative; PH36, IS30 inserted porinB signal sequence, amylase, Kmr | This study |
| pCES-H36-GFP | pCES208 derivative; PH36, eGFP, Kmr | [46] |
| pCnCAB | phaCABC gene expression | [41] |
| pCES-H36-PhaC | pCES208 derivative; PH36, phaC, Kmr | This study |
| pCES-H36-PhaCA | pCES208 derivative; PH36, phaCA, Kmr | This study |
| pCES-H36-PhaCAB | pCES208 derivative; PH36, phaCAB, Kmr | This study |
| pHGmut | pCES208 derivative, Glu89Gln/Δ452-466 gene | [26] |
| pK19mobsacB | Mobilizable vector, Kmr | [42] |
| pK19-∆ISCg1a | pK19mobsacB derivative; flanking region of ISCg1a | This study |
| pK19-∆ISCg1b | pK19mobsacB derivative; flanking region of ISCg1b | This study |
| pK19-∆ISCg1c | pK19mobsacB derivative; flanking region of ISCg1c | This study |
| pK19-∆ISCg1e | pK19mobsacB derivative; flanking region of ISCg1e | This study |
| pK19-∆ISCg2b | pK19mobsacB derivative; flanking region of ISCg2b | This study |
| pK19-∆ISCg2c | pK19mobsacB derivative; flanking region of ISCg2c | This study |
| pK19-∆ISCg2e | pK19mobsacB derivative; flanking region of ISCg2e | This study |
| pK19-∆ISCg2f | pK19mobsacB derivative; flanking region of ISCg2f | This study |
| pJET1.2/blunt | Commercial cloning vector | Thermob |
aNew England Biolabs, Beverly, MA, USA
bThermo fisher scientific, Waltham, MA, USA
Fluorescence assay and screening by FACS
C. glutamicum harboring pCES-H36-GFP was cultivated in BHI medium for 24 h. Optical density of cells were measured and amount of OD600 1 cells were harvested by centrifugation at 6,000 rpm for 10 min. at 4 °C. After washing with 1x phosphate-buffered saline (PBS), cells were resuspended in the same buffer, and the fluorescence intensity was measured with a fluorescent activated cell sorter (FACS; MoFlo XDP, Beckman Coulter, Inc., MI, FL, USA). The cells were excited with a laser at 488 nm and detected with 530/40 band-pass filter for the GFP emission spectrum. Cells showing low fluorescence intensity (mean fluorescence value of <30) were sorted and dropped into fresh BHI media. The cells were grown overnight and transferred into BHI media in a 250 mL flask for the next round of screening.
Plasmid construction
The enzymes for the recombinant DNA work were purchased from Enzynomics (Daejeon, Korea). Polymerase chain reaction (PCR) was performed with the C1000™ Thermal Cycler (Bio-Rad, Hercules, CA, USA) and the Prime STAR HS polymerase (Takara Bio Inc., Shiga, Japan). All oligonucleotides used for PCR are listed Additional file 5: Table S1. For construction of the secretory production system, the H36 promoter and PorB signal peptide gene were amplified from pH36M2 by PCR with the primers H36-porB-F and H36-porB-R. The PCR product was digested with SalI and XbaI and cloned into pCES-NMCS digested with the same restriction enzymes yielding pCES-H36-porBss. The α-amylase gene (GenBank No. AB000829.1) was obtained from the chromosomal DNA of Streptococcus bovis by PCR with the primers Amy-F and Amy-R. After digestion of the PCR product with SfiI, the digested product was cloned into pCES-H36-porBss yielding pCES-H36-porBss-Amy. For the production of poly(3-hydroxybutyrate) [P(3HB)], the engineered Pseudomona sp. 6–19 PHA synthase (PhaC1Ps6-19) [40], PhaA, and PhaC genes were amplified from pCnCAB [41] by PCR with three primer sets, ‘PhaC-F and PhaC-R’, ‘PhaA-F and PhaA-R’, and ‘PhaB-F and PhaB-R’, respectively. The PCR product of PhaC was cut with BamHI and XbaI, and ligated into pCES-NMCS digested with the same restriction enzymes to yield pCES-H36-PhaC. The PCR product of PhaA was cut with XbaI and NotI and ligated into pCES-H36-PhaC to yield pCES-H36-PhaCA. The PCR product of PhaB was cut with NotI and ligated into pCES-H36-PhaCA to yield pCES-H36-PhaCAB.
To construct the IS element-inserted vector, the IS element was amplified from the chromosomal DNA of C. glutamicum by PCR with IS-amy-F and IS-amy-R1, and PCR overlapping was performed with IS-amy-F and IS-amy-R2 using this PCR product. The PCR product was cut with BamHI and XbaI and cloned into pCES-H36-porBss-Amy to yield pCES-H36-porBss-IS-Amy. To construct the IS element deletion plasmid pK19-∆ISCg1a, the upstream region (called A region) and downstream region (called B region) were amplified with the oligonucleotide pairs, ‘ISCg1a A-F and ISCg1a A-R’ and ‘ISCg1a B-F and ISCg1a B-R’, respectively. The PCR products were used as templates for overlap PCR with ISCg1a A-F/ISCg1a B-R. The resulting PCR products were digested with SalI and XbaI and cloned into pK19mobsacB to yield pK19-∆ISCg1a. This cloning method was used to construct the other pK19-∆ISCg IS element deletion plasmid series for the rest of ISCg1 and ISCg2 family genes using the corresponding primers.
Protein fractionation and analysis
After cultivation, cells were harvested by centrifugation (6000 rpm, 10 min and 4 °C). The cells pellets were re-suspended in 300 μl of phosphate-buffered saline (PBS, 135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.2) and were disrupted by sonication at 9 min at 50 % pulse and 20 % amplitude (Sonic, Vibra cell, USA). Total protein fraction was collected after sonication. After centrifugation of cell lysates at 10,000 rpm for 10 min at 4 °C, soluble proteins were collected from supernatant. The fractionated protein samples were analyzed by SDS-PAGE and Western blot. In the SDS-PAGE analysis, protein samples were loaded on 12 % polyacrylamide gels and after the gel electrophoresis, the gels were stained with Coomassie brilliant blue [50 % (v/v) methanol, 10 % (v/v) acetic acid, 1 g/L Coomassie brilliant blue R-250] for 1 h and destained using a destaining solution [10 % (v/v) methanol, 10 % (v/v) acetic acid]. For Western blotting, the proteins were transferred on to a polyvinyl difluoride membrane (Roche, Rotkreuz, Switzerland) for 2 h at 70 mA by using Bio-Rad transblot apparatus (Bio-Rad). The membrane was incubated with a blocking solution (TBS-T, Tris-buffered saline, 24.7 mM Tris, 137 mM NaCl, 2.7 mM KCl, and 0.5 % Tween-20) with 5 wt % skim milk for 1 h at room temperature. The membrane was incubated with blocking solution containing a horseradish peroxidease (HRP)-conjugated monoclonal anti-FLAG M2 antibody (Sigma Aldrich, St. Louis, MO, USA) for the immune-detection of FLAG-tagged protein. After the incubation, each membrane was washed four times with TBS-T for 5 min and the ECL kit (GE Healthcare Bio-Science AB, Buckinghamshire, UK) was used for protein detection.
α-Amylase activity assay
After cell cultivation, 1 mL of cultured cell volume was harvested by centrifugation at 13000 rpm for 10 min. Supernatant of cell culture was obtained and diluted 10 or 100 times with PBS. 100 μL of sample was mixed with 100 μL of 200 μg/mL DQ™ starch which is labeled with BODIPY® FL dye (Molecular Probes™, Eugene, OR, USA). After incubation at room temperature in black tube for 20 min, 100 μL of mixture was loaded on 96 well black plate (Corning, Corning, NY, USA). The fluorescence was measured with 495 nm excitation wavelength and 515 nm filter by using a TECAN Infinite M200 Pro ELISA plate reader (Tecan Group Ltd., Männedorf, Switzerland). The fluorescence value was revised with negative control. One unit (U) of α-amylase was defined as the amount of enzyme required to liberate 1.0 mg of maltose from starch in 1 min at pH 6.9 at 20 °C.
Construction of the IS element-deleted strains
Deletion of the IS element was done with the double crossover method [42]. To delete ISCg1a, C. glutamicum was transformed with pK19-∆ISCg1a by electroporation. First, screening was done on a kanamycin BHIS (BHI plus 30 g/L sorbitol) plate to isolate the plasmid-integrated clones. Then, the Km-resistant cells were cultivated in BHI medium, and they were spread on a 10 % (w/v) sucrose LB plate to pop out the integrated plasmid. With the isolated clones, the deletion of the desired IS element was confirmed by colony-PCR with the two primers ISCg1a A-F and ISCg1a B-R. This gene deletion method was used to engineer the other IS element-deleted strains using the corresponding plasmids and primers. After construction of IS element-deleted strains, existence of IS element was confirmed. To check deletion of ISCg1, PCR was performed with two primer sets, Confirm ISCg1 A F, Confirm ISCg1 A R and Confirm ISCg1 B F, Confirm ISCg1 B R. In case of ISCg2, PCR was performed with two primer sets, Confirm ISCg2 A F, Confirm ISCg2 A R and Confirm ISCg2 B F, Confirm ISCg2 B R.
Inverse PCR
The chromosomal DNA of C. glutamicum ATCC 13032 was prepared with the MasterPure™ DNA purification kit (Illumina, SD, CA, USA). The purified chromosomal DNA was cut with the NotI restriction enzyme. Inverse PCR was carried out with the Pfu-X polymerase (Solgent, Daejeon, Korea) and two primers, Confirm-IS-F and Confirm-IS-R. The PCR product was cloned into pJET1.2/blunt using the CloneJET PCR cloning kit (Thermo fisher scientific, Waltham, MA, USA). After transformation into E. coli XL1-blue, the plasmid was prepared, and the sequence of the PCR product in plasmid was determined by DNA sequencing.
Analytical procedures
The content of P(3HB) synthesized in C. glutamicum was measured by gas chromatography (GC) [43]. An Agilent 6890N GC system (Agilent Technologies, PA, CA, USA) equipped with an Agilent 7683 automatic injector, flame ionization detector, and a fused silica capillary column (ATTM-Wax, 30 m, ID 0.53 mm, film thickness 1.20 mm, Alltech, Deerfield, IL, USA) was used. The dried cells were treated for methanolysis with 15 % sulfuric acid and benzoic acid as an internal standard [44]. After extraction of the polymers, the content of the synthesized polymer was measured by gas chromatography (Agilent, SCL, CA, USA). Cell concentration, defined as the dry cell weight (DCW) per liter of culture broth, was determined as previously described [43]. The residual cell concentration was defined as the cell concentration minus the P(3HB) concentration. The P(3HB) content (wt%) was defined as the percentile ratio of the P(3HB) concentration to the cell concentration.
To determine the concentration of GABA, the culture supernatant was applied to reversed phase high-pressure liquid chromatography (HPLC, LC-20 AD, CTO-20A, SPD-20A; Shimadzu, Japan) equipped with Zorbax Eclipse amino acid analysis (AAA) column (4.6 × 150 mm 3.5-Micron; Agilent, USA). Mobile phase A (10 mM Na2HPO4—10 mM Na2B4O7, pH 8.2) and mobile phase B (acetonitrile:MeOH:H2O 45:45:10 by volume) were applied for separation of the samples. The elution condition was as follows: equilibration (1.9 min., 100 % A), gradient (16.2 min, 0–57 % B; 3.7 min, 57–100 % B), and cleaning (4 min, 100 % B). The column temperature was maintained at 40 °C. Samples were detected with UV at 338 nm.
Determination of the transformation efficiency
C. glutamicum was prepared for transformation as follows: 200 mL of RG media (BHI 40 g, glucose 10 g, beef extract 10 g, sorbitol 30 g per 1 L) were inoculated with cells from an overnight culture grown in BHI at 30 °C with shaking at 200 rpm. When the optical density of the cells reached an OD600 of 1, the cells were harvested by centrifugation at 6000 rpm for 10 min. at 4 °C. The cell pellets were washed twice with 10 % ice-chilled glycerol. The final cell suspension in 10 % glycerol was adjusted to an OD600 of 10. To determine the transformation efficiency, the following procedure was used [8]. pCES-H36-GFP (500 ng prepared from C. glutamicum) was mixed with 150 μL of cell suspension on ice. Electroporation was carried out in a 1 mm cuvette at 1.8 kV, 25 μF, and 200 Ω using a Gene Pulser system (BioRad, Hercules, CA, USA). After the electroporation, 1 mL of RG medium was immediately added to the cells, and the cells were cultivated at 30 °C and 200 rpm for 2 h, and then the cells were plated on RG agar plates containing 25 μg/mL kanamycin. The colony forming units (CFUs) were counted after one day of incubation at 30 °C, and the number of colonies was calculated as the CFU per μg DNA.
Authors’ contributions
JWC, SSY and MJK performed all experiments. JWC prepared the draft of manuscript. KJJ initiated and coordinated the study and contributed to the experimental design, data interpretation, and reviewing the manuscript. All authors have read and approved the final manuscript.
Acknowledgements
This work was supported by the Intelligent Synthetic Biology Center of Global Frontier Project (Grant no. 2014M3A6A8066443) and by the National Research Foundation of Korea (Grant no. NRF-2015R1A2A2A01007674) funded by the Ministry of Science, ICT and Future Planning (MSIP).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- IS element
insertion element
- FACS
fluorescence cell sorter
- GABA
γ-aminobutyrate
- P(3HB)
poly(3-hydroxybutyrate)
Additional files
10.1186/s12934-015-0401-7 SDS-PAGE analysis of the sorted cells by FACS.
10.1186/s12934-015-0401-7 Confirmation of IS element deletion by agarose gel electrophoresis of PCR samples
10.1186/s12934-015-0401-7 Growth profile of cells harboring pCES-H36-GFP.
10.1186/s12934-015-0401-7 Growth profile of cells harboring pCES-H36-PhaCAB or pHGmut.
10.1186/s12934-015-0401-7 List of primers used in the PCR experiments.
Contributor Information
Jae Woong Choi, Email: jwoongci@gmail.com.
Sung Sun Yim, Email: vidalsungsun@gmail.com.
Min Jeong Kim, Email: mjflorakim@gmail.com.
Ki Jun Jeong, Phone: +82-42-350-3934, Email: kjjeong@kaist.ac.kr.
References
- 1.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Becker J, Wittmann C. Systems and synthetic metabolic engineering for amino acid production–the heartbeat of industrial strain development. Curr Opin Biotechnol. 2012;23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Woo HM, Park J-B. Recent progress in development of synthetic biology platforms and metabolic engineering of Corynebacterium glutamicum. J Biotechnol. 2014;180:43–51. doi: 10.1016/j.jbiotec.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol. 2013;6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yim SS, An SJ, Choi JW, Ryu AJ, Jeong KJ. High-level secretory production of recombinant single-chain variable fragment (scFv) in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;98:273–284. doi: 10.1007/s00253-013-5315-x. [DOI] [PubMed] [Google Scholar]
- 6.Yim SS, Choi JW, Lee RJ, Lee YJ, Lee SH, Kim SY, Jeong KJ: Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol Bioeng. 2015. [DOI] [PubMed]
- 7.Frunzke J, Bramkamp M, Schweitzer J-E, Bott M. Population heterogeneity in Corynebacterium glutamicum ATCC 13032 caused by prophage CGP3. J Biotechnol. 2008;190:5111–5119. doi: 10.1128/JB.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumgart M, Unthan S, Rückert C, Sivalingam J, Grünberger A, Kalinowski J, Bott M, Noack S, Frunzke J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl Environ Microbiol. 2013;79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler M, Mahillon J: Mobile DNA II. Insertion Sequences revisited American Society for Microbiology. Washington DC 2002. p. 305–66.
- 11.de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/S0966-842X(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 12.Darmon E, Leach DR. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unthan S, Baumgart M, Radek A, Herbst M, Siebert D, Brühl N, Bartsch A, Bott M, Wiechert W, Marin K. Chassis organism from Corynebacterium glutamicum–a top-down approach to identify and delete irrelevant gene clusters. Biotechnol J. 2015;10:290–301. doi: 10.1002/biot.201400041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig NL. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 17.Ooka T, Ogura Y, Asadulghani M, Ohnishi M, Nakayama K, Terajima J, Watanabe H, Hayashi T. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 2009;19:1809–1816. doi: 10.1101/gr.089615.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 2010;6:e1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haack KR, Roth JR. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daveran-Mingot M-L, Campo N, Ritzenthaler P, Le Bourgeois P. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J Bacteriol. 1998;180:4834–4842. doi: 10.1128/jb.180.18.4834-4842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Heijden I, Gomez-Eerland R, van den Berg J, Oosterhuis K, Schumacher T, Haanen J, Beijnen J, Nuijen B. Transposon leads to contamination of clinical pDNA vaccine. Vaccine. 2013;31:3274–3280. doi: 10.1016/j.vaccine.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Prather KLJ, Edmonds MC, Herod JW. Identification and characterization of IS1 transposition in plasmid amplification mutants of E. coli clones producing DNA vaccines. Appl Microbiol Biotechnol. 2006;73:815–826. doi: 10.1007/s00253-006-0532-1. [DOI] [PubMed] [Google Scholar]
- 23.Park MK, Lee SH, Yang KS, Jung S-C, Lee JH, Kim SC. Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences. Appl Microbiol Biotechnol. 2014;98:6701–6713. doi: 10.1007/s00253-014-5739-y. [DOI] [PubMed] [Google Scholar]
- 24.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 25.Avella M, Martuscelli E, Raimo M. Review properties of blends and composites based on poly (3-hydroxy) butyrate (PHB) and poly (3-hydroxybutyrate-hydroxyvalerate)(PHBV) copolymers. J Mater Sci. 2000;35:523–545. doi: 10.1023/A:1004740522751. [DOI] [Google Scholar]
- 26.Choi JW, Yim SS, Lee SH, Kang TJ, Park SJ, Jeong KJ. Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb Cell Fact. 2015;14:21. doi: 10.1186/s12934-015-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi N, Ohashi S, Sadykov MR, Mizutani-Ui Y, Kobayashi I. IS-linked movement of a restriction-modification system. PLoS One. 2011;6:e16554. doi: 10.1371/journal.pone.0016554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalrymple B. Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. Mol Gen Genet. 1987;207:413–420. doi: 10.1007/BF00331609. [DOI] [PubMed] [Google Scholar]
- 29.Yang S, Sleight SC, Sauro HM. Rationally designed bidirectional promoter improves the evolutionary stability of synthetic genetic circuits. Nucleic Acids Res. 2013;41:e33. doi: 10.1093/nar/gks972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sousa A, Bourgard C, Wahl LM, Gordo I. Rates of transposition in Escherichia coli. Biol Lett. 2013;9:20130838. doi: 10.1098/rsbl.2013.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiss J, Nagy Z, Toth G, Kiss GB, Jakab J, Chandler M, Olasz F. Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol Microbiol. 2007;63:1731–1747. doi: 10.1111/j.1365-2958.2007.05621.x. [DOI] [PubMed] [Google Scholar]
- 32.Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology. 2007;153:1042–1058. doi: 10.1099/mic.0.2006/003657-0. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M, Uchiyama T, Ōmura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat Biotechnol. 2002;20:1018–1023. doi: 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- 35.Szeverényi I, Hodel A, Arber W, Olasz F. Vector for IS element entrapment and functional characterization based on turning on expression of distal promoterless genes. Gene. 1996;174:103–110. doi: 10.1016/0378-1119(96)00407-6. [DOI] [PubMed] [Google Scholar]
- 36.Jäger W, Schäfer A, Kalinowski J, Fühler A. Isolation of insertion elements from Gram-positive Brevibacterium,Corynebacterium and Rhodococcus strains using the Bacillus subtilis sacB gene as a positive selection marker. FEMS Microbiol Lett. 1995;126:1–6. doi: 10.1111/j.1574-6968.1995.tb07381.x. [DOI] [PubMed] [Google Scholar]
- 37.Vertes AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H. Isolation and characterization of IS31831, a transposable element from Corynebacterium glutamicum. Mol Microbiol. 1994;11:739–746. doi: 10.1111/j.1365-2958.1994.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome–restriction–modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/S0959-437X(99)00026-X. [DOI] [PubMed] [Google Scholar]
- 40.Park SJ, Jang Y-A, Lee H, Park A-R, Yang JE, Shin J, Oh YH, Song BK, Jegal J, Lee SH. Metabolic engineering of Ralstonia eutropha for the biosynthesis of 2-hydroxyacid-containing polyhydroxyalkanoates. Metab Eng. 2013;20:20–28. doi: 10.1016/j.ymben.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Yang TH, Kim TW, Kang HO, Lee SH, Lee EJ, Lim SC, Oh SO, Song AJ, Park SJ, Lee SY. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase. Biotechnol Bioeng. 2010;105:150–160. doi: 10.1002/bit.22547. [DOI] [PubMed] [Google Scholar]
- 42.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 43.Braunegg G, Sonnleitner By, Lafferty R. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. doi: 10.1007/BF00500854. [DOI] [Google Scholar]
- 44.Jacquel N, Lo C-W, Wei Y-H, Wu H-S, Wang SS. Isolation and purification of bacterial poly (3-hydroxyalkanoates) Biochem Eng J. 2008;39:15–27. doi: 10.1016/j.bej.2007.11.029. [DOI] [Google Scholar]
- 45.Park JU, Jo JH, Kim YJ, Chung SS, Lee JH, Lee HH. Construction of heat-inducible expression vector of Corynebacterium glutamicum and C. ammoniagenes: fusion of lambda operator with promoters isolated from C. ammoniagenes. J Microbiol Biotechnol. 2008;18:639–647. [PubMed] [Google Scholar]
- 46.Yim SS, An SJ, Kang M, Lee J, Jeong KJ. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng. 2013;110:2959–2969. doi: 10.1002/bit.24954. [DOI] [PubMed] [Google Scholar]


