Abstract
Non-viral methods have been explored as the replacement of viral systems for their low toxicity and immunogenicity. However, they have yet to reach levels competitive to their viral counterparts. In this paper, we combined physical and chemical methods to improve the performance of polyplex delivery of DNA and siRNA. Specifically, gold nanoparticles (AuNPs) were used to carry polyplex (a chemical approach) while electroporation (a physical approach) was applied for fast and direct cytosolic delivery. In this hybrid approach, cationic polymer molecules condense and/or protect genetic probes as usual while AuNPs help fix polycations to reduce their cytotoxicity and promote the transfection efficiency of electroporation. AuNPs of various sizes were first coated with polyethylenimine (PEI), which were further conjugated with DNA plasmids or siRNA molecules to form AuNPs-polyplex. The hybrid nanoparticles were then mixed with cells and introduced into cell cytosol by electroporation. The delivery efficiency was evaluated with both model anchor cells (i.e., NIH 3T3) and suspension cells (i.e., K562), together with their impact on cell viability. We found that AuNP-polyplex showed 1.5~2 folds improvement on the transfection efficiency with no significant increase of toxicity when compared to free plasmid delivery by electroporation alone. Such a combination of physical and chemical delivery concept may stimulate further exploration in the delivery of various therapeutic materials for both in vitro and in vivo applications.
Keywords: Electroporation, Polyplex, Gene Delivery, Gold Nanoparticles
1. Introduction
Gene induction and/or inhibition provide an invaluable approach to understand gene function1, control cellular signals2, and develop new therapeutic technologies3. Having safe and effective delivery tools is the key to achieving its full potential. Viral transduction is stable and efficienct4–6, but still has high risk of oncogenesis and inflammation7,8. This stimulates the rapid growth of nonviral delivery systems. In chemical-mediated nonviral delivery systems, potent therapeutic molecules are condensed and/or protected by cationic chemicals via forming polymer-DNA complexes (polyplex) or lipid-DNA complexes (lipoplex) to overcome multiple delivery barriers. They serve as the favorable alternative to their virus-mediated counterparts and have been successfully tested for both in vitro and in vivo delivery of plasmids, oligonucleotides, ribozyme, and small interfering RNAs9–21. However, many of these systems still suffer insufficient delivery efficiency and cell viability, which often ties with their poor nanoparticle quality, slow and inefficient cellular uptake and endosome escape, and serious cytotoxicity from free cationic molecules after the unpacking of lipoplex or polyplex. As captured cationic molecules are found much less toxic than their free counterparts, nanoparticles have been introduced to help fix cationic polymer22. This was also found helpful to produce nanoparticles with much narrow size distribution. Gold nanoparticles (AuNPs) are favored in these applications for their good biocompatibility and multiple functionalities (i.e., targeting, therapeutic, and imaging)22–28. However, issues like ineffective cellular internalization remain.
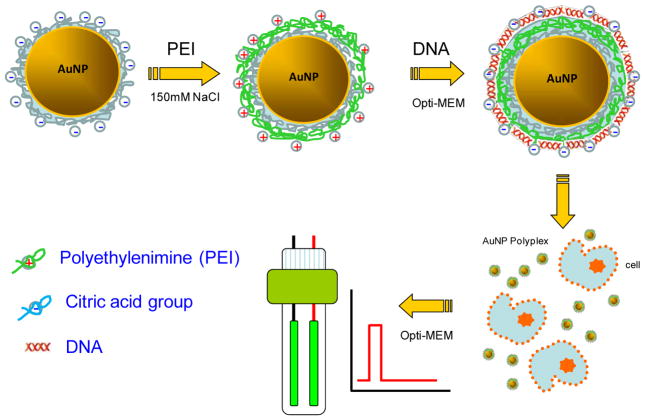
Herein we introduce the use of electroporation to bypass the slow and inefficient endocytosis process by directly delivering therapeutic probes into cell cytosol. Electroporation is a physical delivery approach in which cells are imposed with short electrical pulses to create temporary pathways on the cell membrane to facilitate the cellular uptake29. It has been widely used to either evaluate the therapeutic performance of exogenous probes or study their trafficking inside cells29–46. A simple combination of lipoplex nanoparticles and electroporation has been explored early in the delivery of oligonucleotides in the format of lipoplex47, 48. However, negative impacts on both the delivery efficiency and the cell viability were found47. It was believed that the destroyed complex structure during electroporation released a large number of free cationic molecules, which significantly lower the overall cell viability. To avoid similar situation, we first immobilized cationic polymer on AuNPs and then allowed conjugation with negatively charged therapeutic probes to form AuNPs-polyplex complex. In addition to the help on retaining cationic polymer on the surface, the presence of AuNPs also enhances the electroporation performance with focused electric pulses and localized poration49, which was proved beneficial for not only the recovery of treated cells to gain high cell viability, but also the uptake of probes from multiple sites to facilitate the cytosolic delivery. Specifically, cationic polymer, polyethylenimine (PEI), was immobilized on AuNPs by electrostatic interactions (Figure 1). DNA plasmids or siRNA probes were then conjugated with PEI molecules to form AuNPs-polyplex. The complex nanoparticles were then mixed with cells for electroporation. The delivery enhancement was evaluated by the cell viability and the transfection efficiency.
Figure 1.
Schematic illustration on the procedure of AuNPs-polyplex synthesis and delivery.
2. Materials and Methods
2.1 Materials and reagents
Branched PEI (MW=25kDa), gold nanoparticles of 5–40 nm were obtained from Sigma-Aldrich. The concentration of 1X AuNPs refers to the stock solution, which has 0.01 wt% of Au (0.1 mg/ml) while the actual particle number varies with the size of AuNPs. Other concentrations of AuNPs were prepared by either concentrating or diluting from the stock solution. DNA plasmids with gWiz™ GFP and gWiz™ Luc reporter genes were purchased from Aldevron, Inc. (Fargo, ND). Small interfering RNA (siRNA) used for silencing GFP (expressed by pmaxGFP purchased from Lonza) and Luciferase genes were synthesized by Thermo Scientific (Pittsburgh, PA) and the sequences were as follows: siRNA for GFP silence, sense strand, 5′-CGCAUGACCAACAAGAUGAUU-3′; antisense strand, 5′-UCAUCUUGUUGGUCAUGCGGC-3′; Luciferase GL3 Duplex (Luc-siRNA), sense strand, 5′-CUUACGCUGAGUACUUCGA-3′; antisense strand, 5′-UCGAAGUACUCAGCGUAAG-3′. All other chemicals were purchased from Sigma-Aldrich and the cell culture reagents were purchased from Life Technologies (Carlsbad, CA) unless specified.
2.2 Preparation of AuNPs-polyplex
To prepare AuNPs/PEI polyplex, 500μl 0.01wt% of AuNPs was added into 500μl 0.5mg/ml PEI (pH7.0). The original citric acid terminated surface of AuNPs facilitates the deposition of PEI molecules through electrostatic interactions. The incubation was performed at room temperature for 20 min and the extra PEI was removed by centrifuging at 15000×g for 10 min. The PEI coated AuNPs were resuspended in desirable amount of PBS (pH=7.0) and 5μl of nucleic acid solution (with a concentration of 5mg/mL) was added to AuNPs/PEI of varying concentrations. The resulting mixture was mixed by pipetting and further incubated at room temperature for 20 min.
2.3 NIH/3T3 and K562 cell culture
NIH/3T3 cells (ATCC, CRL-1658) were grown and maintained in high glucose DMEM supplemented with 10% newborn calf serum (NCS), 1% penicillin and streptomycin, 1% L-glutamine and 1% sodium pyruvate. K562 cells (ATCC, CCL-243) were routinely cultured in RPMI 1640 supplemented with 10% NCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 100 μg/mL L-glutamine. All cultures were maintained at 37° C with 5% CO2 and 100% relative humidity.
2.4 Electroporation setup and procedure
NIH/3T3 or K562 cells were first centrifuged and resuspended in fresh OPTI-MEM I (a serum free medium) at a density of 0.5×106 cells/ml. Samples were then mixed with polyplexes of various concentrations and sizes. Cell electroporation was done with a commercial instrument (ECM 830, Harvard Apparatus) in cuvettes with a 2-mm gap, each containing a 100 μL sample solution. The electroporation conditions are established from previous work as follows: 125V, 10 ms with a single unipolar pulse50. After electroporation, samples were transferred to 6-well cell culture plates, incubated in fresh medium for another 24 hr and then harvested for analysis.
2.5 AuNPs-polyplex delivery efficiency and cell viability
The transfection efficiency of gWiz™ GFP plasmids was evaluated both qualitatively by visualizing the number of cells with green fluorescence within a representative area selected from the entire culture surface under an inverted fluorescence microscope (Olympus, Japan) and quantitatively by counting cells using a four-color flow cytometry system (FACS Calibur, BD Biosciences, CA) 24 hr post transfection. Briefly, an amount of 1.5×106 cells/mL was collected and the percentage of GFP-positive cells was calculated quantitatively via flow cytometer. The unstained samples were run first to adjust the voltage setting and compensation of the flow cytometer. Then the tested samples were processed by CellQuest. At least 10,000 events were collected for each sample.
The GFP down regulation efficiency was determined by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The fluorescence intensity of GFP was measured using Cell Assay Module with live cells stained with carboxy-naphthofluorescein (CBNF). The results were analyzed with Agilent 2100 Expert Software and 500–1,500 events were counted for each sample. The Luc-siRNA down regulation efficiency was quantified by One-Glo™ Luciferase assay system (Promega, Madison, WI). 100ul One-Glo™ reagent was added to the cell growth in 100ul of medium in 96-well plate. Luminescence was measured with a plate reader (FLUOstar OPTIMA, BMG LABTECH, Germany) after 10 min incubation at room temperature for complete cell lysis.
The cell viability was evaluated by an MTS cell proliferation assay (Promega, Madison, WI). Briefly, the cells in 100 μL/well of medium were transferred to a 96-well plate and incubated. 20 μl of CellTiter 96 AQueous One solution (Promega, Madison, WI) was added to each well and cells were incubated at 37°C for another 4 hr. Absorbance was measured at 492 nm on an automated plate reader (Elx 800, Biotek, VT). Data points were represented as the mean ± standard deviation (SD) of triplicates, unless otherwise indicated.
2.6 Cellular uptake of AuNPs-polyplex nanoparticles
The distribution of AuNPs-polyplex in 3T3 cells were examined using an inverted fluorescent microscopy. As AuNPs are well known to quench the fluorescent signal from proximal fluroprobes, a sandwich design of fluorophore-labeled AuNPs (FNP, from Nanopartz, Inc, having Alexa Fluor 546) with fluorophores separated from the gold surface by polymer spacers, were used to circumvent this problem. Plasmids were stained with YOYO-1 iodide (Life Technology) with a ratio of 100bp/dye. The mixture of cells with nucleic acids, AuNPs or AuNPs-polyplex were washed twice with 1X PBS (pH7.0), followed by fixation with 4% paraformaldehyde for 30 min. Nuclei were stained with 20 μM of DAPI for 10 min at room temperature. Cells from each sample were then mounted on cover glass slides. Images of phase contrast, green (nucleic acids), red (AuNPs) and blue (nuclei) fluorescence channels were taken on an Olympus 1×51 inverted microscope (Olympus, Japan) with a 100x objectives.
Note: In our nomenclature, symbols like “A/B” or “A−B” means materials A and B are conjugated together through electrostatic interactions after incubation; “A+B” means A and B are simply mixed without incubation before further treatment.
3. Results and discussions
3.1 AuNPs-polyplex size and size distribution
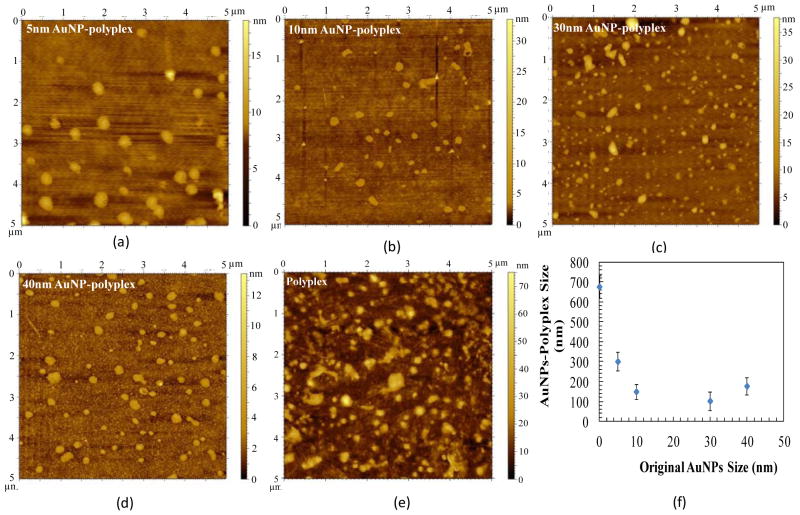
Current polyplex delivery vehicles have not yet shown competitive delivery advantages over natural virus-based counterparts. This is at least partially attributed to their heterogeneous assembly conditions and poor synthetic quality of nanoparticles (i.e., relatively large variations in size, structure, and component quantity). As in AuNPs-polyplex synthesis, cationic polymer molecules (e.g., PEI) were first immobilized on the surface of AuNPs, their amount in individual AuNPs-polyplex should be more uniform than those synthesized through dynamic complexation of free charged agents. This further determines the total dosage of genetic probes which are condensed on AuNPs-polyplex later on. Therefore, the introduction of AuNPs in polyplex not only helps fix free or dissociated polycations on a solid surface, but also provides better management on molecule assembly and multiple-agent packaging. As the consequence, nanoparticles of better quality are produced. As shown in AFM images in Figure 2, more homogeneous morphology was found for AuNPs-polyplex than polyplex synthesized via vortex mixing (Figure 2e)10. Their size was also much smaller and more uniform, which was further confirmed with quantitative measurements using dynamic light scattering (Figure 2f). Except for AuNPs-polyplex synthesized from 5 nm AuNPs, the average size o AuNPs-polyplex with various original sizes fell between 100–200 nm, an appropriate size range of nanoparticles for efficient cellular uptake. As the size of DNA plasmids used in this study is much bigger than that of AuNPs, the same DNA molecules are suspected to interact with multiple AuNPs-PEI nanoparticles simultaneously (as shown schematically in supplemental Figure S1). Therefore, clusters (or aggregates from conjugation networking) of AuNPs-PEI-DNA, instead of many individual AuNPs-polyplex nanoparticles with assembly structure schematically shown in Figure 1, are more likely formed. With smaller size and higher mobility, AuNPs of 5 nm allow easier occurrence of such conjugation networking than other AuNPs with larger original size. As the results, such stable clusters might become the dominated population when small AuNPs are used in AuNPs-polyplex synthesis.
Figure 2.
The AFM images of AuNPs-polyplex morphology with the original size of AuNPs of (a) 5nm, (b) 10 nm, (c) 30 nm, (d) 40 nm. Panel (e) is the traditional polyplex synthesized through vortex mixing approach10. Panel (f) is the quantitative DLS particle size measurement.
3.2 Cellular uptake of AuNPs-polyplex via electroporation
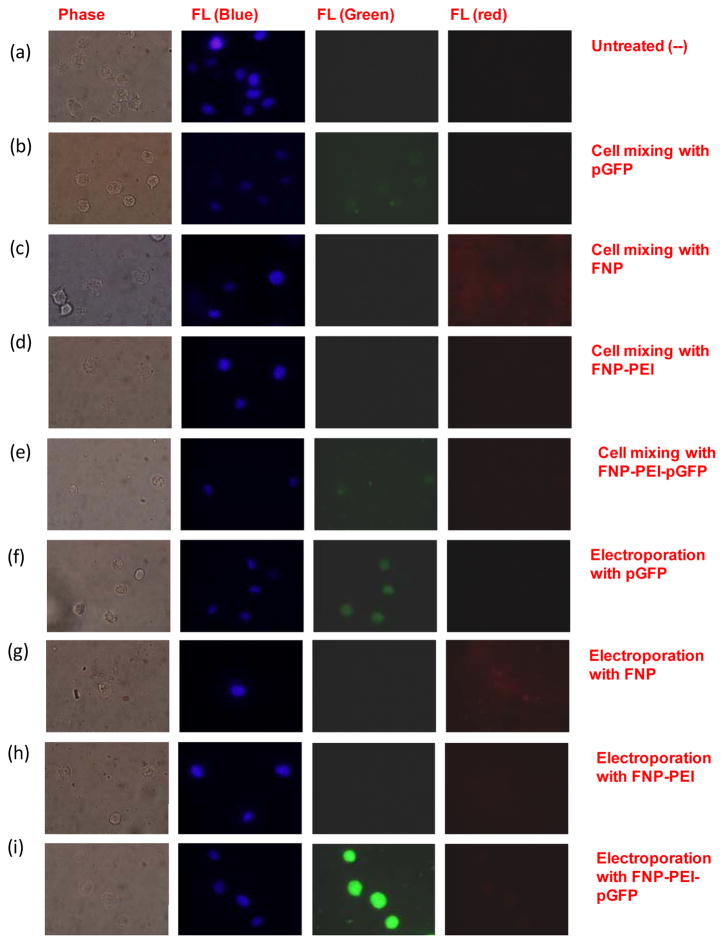
To verify that PEI molecules retain on AuNPs during electroporation, fluorescence probes were tagged to track the locations and fate of AuNPs-polyplex during the cellular uptake. As AuNPs quench fluorescent signal from proximal fluroprobes, fluorophore-labeled AuNPs (FNP) with polymer spacer separating fluorophores from the gold surface were used. These nanoparticles are also carboxylated to match similar interaction capacity as those with citric acid terminated surface. After conjugating with PEI molecules, fluoroprobes are pushed back to the gold surface and therefore, the fluorescent signal of FNP is quenched again unless most immobilized PEI molecules are gone. When DNA plasmids are condensed on FNP, the PEI layer underneath serves as the new thick spacer so that the YOYO-1 labeled DNA probes become visible and are used to track the uptake of AuNPs-polyplex.
Compared to the untreated sample (Figure 3a), samples of simply mixing cells and YOYO-1 labeled DNA plasmids (Figure 3b) or FNP (Figure 3c) have weak fluorescence spots visible. This is attributed to their tiny particle size and limited fluorescence signal when staying as individual nanoparticles. After capped with a layer of PEI, even such weak fluorescence signals disappeared (Figure 3d), which confirmed that the original fluorophores were pushed back close to the gold surface. As the PEI layer serves as the new spacer layer, the condensed DNA plasmids labeled with YOYO-1 exhibited similar fluorescent signal as free DNA plasmids (showing weak green fluorescence spots in Figure 3e), indicating that the location of AuNPs-polyplex was mainly outside cells. After electroporation, stronger fluorescent signals were generally seen in electroporated cells with naked plasmids and FNP as nanoparticles accumulated in cells (Figures 3f–3g). No fluorescence signal was observed for the sample using PEI-coated FNP (Figure 3h), though similar accumulation of AuNPs-PEI nanoparticles were clearly observed in the phase contrast image. This suggests that the electric pulses did not break the interactions between AuNPs and PEI molecules. For YOYO-1 labeled AuNPs-PEI-DNA polyplex, strong green fluorescence signal was shown inside cells (Figure 3i). As samples were fixed immediately after electroporation, this clearly indicates the similar quick and direct cytosolic delivery of plasmids by electroporation.
Figure 3.
Phase contrast and fluorescence microscopic images of distribution and fate of AuNPs-polyplex when mixing with NIH 3T3 cells (a–e) and immediately after electroporation (f–i): (a) untreated samples (negative control), (b) mixture of cells and naked DNA plasmids (green); (c) mixture of cells and FNP (red); (d) mixture of cells with FNP/PEI nanoparticles; (e) mixture of cells with FNP/PEI/DNA; (f) electroporation with DNA alone (green); (g) electroporation with FNP (red); (h) electroporation with FNP/PEI; and (i) electroporation with FNP/PEI/DNA.
3.3 Plasmid DNA delivery in NIH 3T3 cells by AuNPs-polyplex
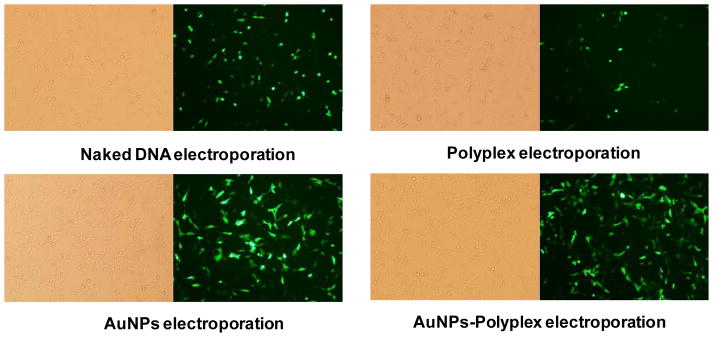
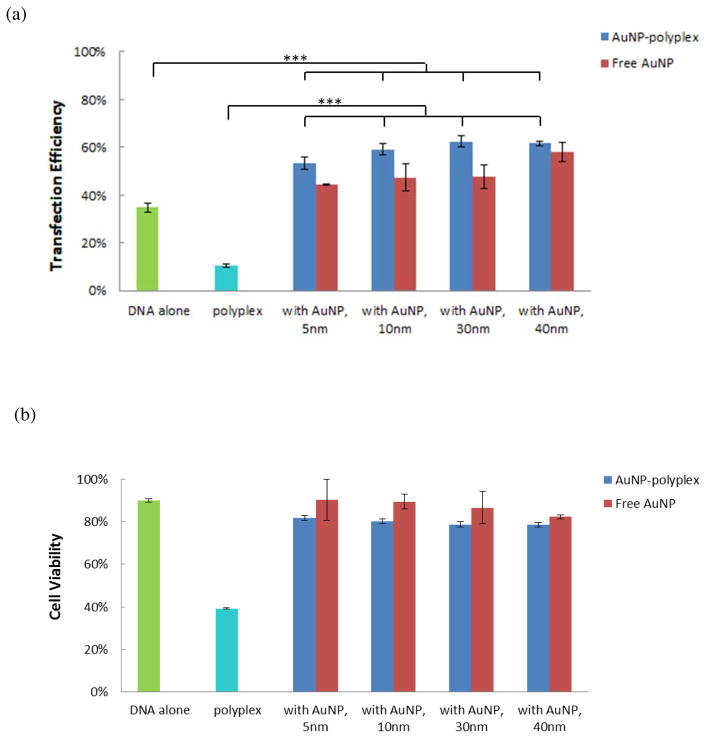
We further explored the delivery of DNA plasmids from AuNPs-polyplex by electroporation. The electroporation was done with NIH 3T3 cells with a BTX system using pWizGFP plasmids and the following pulse scheme was applied: 125 V (625 V/cm), single 10 ms pulse. Successful transfection was observed 24 hr after electroporation in all four cases: electroporation with DNA alone (no AuNPs), with AuNPs+DNA, with polyplex (no AuNPs), and with AuNPs-polyplex, as shown in Figure 4. However, a simple combination of electroporation and polyplex showed significantly negative impact on both the delivery efficiency and cell viability (Figure 4b), which is consistent with an early observation for lipoplex delivery using a similar approach47. The poor-quality and loose structure of polyplex might have been destroyed to release a large number of free cationic PEI molecules. These free positively charged macromolecules, together with additional harsh electric pulses, further lowered the overall cell viability and transfection when compared to the electroporation of naked plasmids (Figure 4a). Electroporation delivery of plasmids together with AuNPs and AuNPs-polyplex showed better GFP expression (Figures 4c–4d), which confirmed again our early observation the enhancement of AuNPs to electroporation performance49. More quantitative comparison was done by counting the percentage of GFP-positive cells using flow cytometry (Figure 5). Efficiency of pGFP transfection from AuNPs-polyplex (using 5 nm AuNPs at a concentration of 1X or 0.1 mg/ml) was about one and half folds of that using naked plasmids and a simple mixture of DNA and AuNPs (DNA alone: 34.8%±2.0%; 5nm AuNPs and DNA: 44.4%; 5 nm AuNPs-polyplex: 53.4%). When AuNPs of larger size (10–40 nm for their original size) were used, the transfection efficiency was further enhanced to about two folds of that using naked DNA alone. As for comparison, the GFP transfection using electroporation with polyplex was only about one third of that standard electroporation. Some loss on the cell viability (i.e., ~10%) was observed in AuNPs-polyplex electroporation samples than that using naked DNA. But it is worth-while the sacrifice for using electroporation to bypass the endocytosis delivery route with direct cytosol delivery and 1.5–2 folds increase on the transfection efficiency. When comparing to a cell viability of ~40% (i.e., less than half of the standard electroporation of naked DNA, ~90%) that using polyplex in electroporation, our approach of introducing AuNPs to fix free or dissociated PEI is effective. This observation was also endorsed with further complex cytotoxicity analysis without electroporation (see supplemental Figure S2).
Figure 4.
Fluorescence and phase contrast microscopic images of pGFP plasmid transfection to NIH 3T3 cells by electroporation naked DNA plasmids, a mixture of AuNPs and DNA plasmids, polyplex, and AuNPs-polyplex.
Figure 5.
Quantitative measurement of electroporation enhanced AuNPs-polyplex delivery performance on 3T3 cells: (a) the flow cytometry results on transfection efficiency and (b) the cell viability via MTS assay. As comparison, results from electroporation with DNA alone, polyplex, and samples of a simple mixing of AuNPs and DNA are also shown. The error bars correspond to triplicate experiments made with independently produced batches. (***P<0.001)
In this delivery improvement, the AuNPs core helps enhance the electroporation performance from two different aspects49: (1) reducing the resistance of the electroporation buffer solution so that the local pulse strength on cells is enhanced; (2) serving as virtual microelectrodes to locally porate cells with limited area from many different sites. Because of their high conductivity (~4.5×106 S/m), AuNPs dispersed in buffer and cytoplasm (the conductivity is ~0.3–1.5 S/m) help greatly reduce the potential drop there so that the majority of the electric voltage is imposed on the cell membrane indeed. The cell membrane disruption could therefore be done more effectively without altering the cell physiological conditions (i.e., salt concentration) or losing cell viability. With the electric field converges in the vicinity of AuNPs, they work like many virtual nanoelectrodes to cause localized poration. Different from the traditional bulk electroporation with two large breakdown locations, multiple small poration sites are formed after adding AuNPs that benefits not only the recovery of the cell membrane, but also the cytosolic delivery of plasmids from multiple sites.
The contribution of AuNPs-polyplex to the transfection improvement of DNA plasmids is also multifactorial: (1) they help fix PEI on the surface of AuNPs to significantly reduce the toxicity caused by the presence of free and/or dissociated cationic polymer molecules in polyplex (see supplemental Figure S2); (2) they also effectively produce polyplex nanoparticles with smaller average size than the naked DNA plasmids and narrower size distribution when compared to that from vortex mixing synthesis (Figure 2f); (3) the PEI molecules in AuNPs-polyplex help protect DNA plasmids and condense them near the vicinity of cell membrane to promote the cytosolic delivery and also later nuclear transport. These facts offer AuNP-polyplexes advantages over the use of the mixture of AuNP and naked DNA in electroporation (Figure 5) as well as many traditional transfection approaches (see supplemental Figure S3) on the transfection efficiency and cell viability. The slight loss on cell viability (in Figure 5b) probably results from the presence of some free PEI molecules to the electroporated cells.
3.4 Small Interfering RNA delivery in K562 cells by AuNPs-polyplex
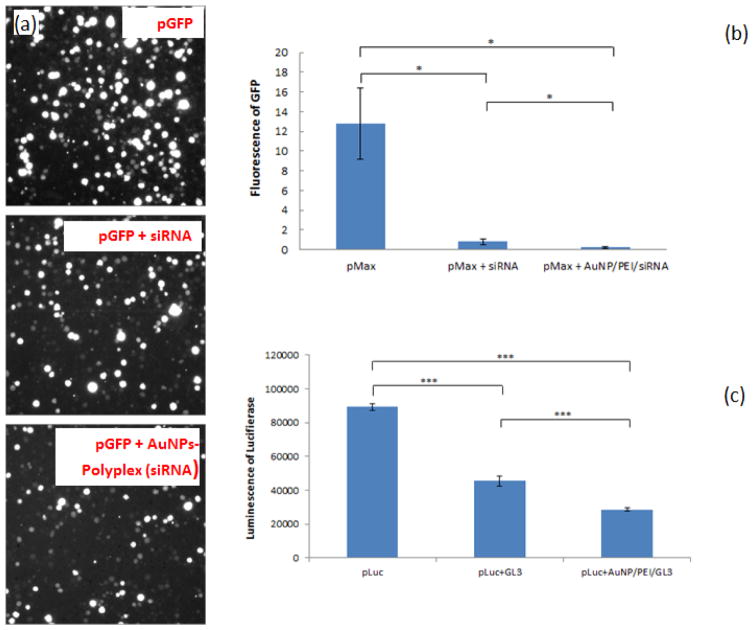
Small interfering RNA (siRNA) is now widely used to down regulate specific gene expression in cells with their complementary nucleotide sequence. To demonstrate the effectiveness of AuNPs-polyplex electroporation to siRNA delivery, we chose siRNA with specific sequences to silence the expression of GFP and Luciferase when co-transfecting with pGFP and pLuc by electroporation. As shown in Figure 6a, clearly less GFP expression was seen when co-delivering pmaxGFP and the corresponding siRNA. AuNPs-polyplex (siRNA) helped turn off more GFP expression than that using free siRNA (Figure 6b). Similar down regulation performance was also found when co-transfecting pLuc and the corresponding siRNA GL3, as shown in Figure 6c. Compared to the interference result of free siRNA GL3, additional ~15% further drop of Luciferase signal was found when siRNA molecules were conjugated in AuNPs-polyplex. Because siRNA have much smaller size than plasmid DNA, neither the delivery of free siRNA nor AuNPs-polyplex to cell cytosol through electroporation is very challenging. However, their quick denature feature makes the delivery focus of siRNA delivery often on their protection from enzyme degradation. Therefore, siRNA delivery with AuNPs-polyplex electroporation could be more beneficial when used for in vivo delivery.
Figure 6.
AuNPs-polyplex electroporation enhanced siRNA delivery on K562 cells: (a) fluorescence images of and (b) intensity measurement on GFP expression level, and (c) the luminescence measurement on Luciferase expression level for free siRNA (“pMax+siRNA” and “pLuc+GL3”) and siRNA from AuNPs-polyplex (“pMax+AuNP/PEI/siRNA” and “pLuc+AuNP/PEI/GL3”). (*P<0.05, ***P<0.001).
It is also worth to point out that the enhancement of AuNPs-polyplex to siRNA interference performance should be better than what was shown in Figure 6. As co-transfection of DNA plasmids and siRNA approach was adopted here, the interference of siRNA to the expression of the targeting reporter gene occurs simultaneously with that particular transgene expression in cells. The early presence of copious siRNA probes could silence the targeting proteins more efficiently than those that already maintain a sustained concentration level in cells. Therefore, both free siRNA and siRNA from AuNPs-polyplex showed efficient down regulation performance here, which allows only limited room to further enhance the interference with AuNPs-polyplex. Moreover, the presence of AuNPs during polyplex (siRNA) delivery simultaneously enhanced the actual expression level of GFP or Luciferase with the same mechanism demonstrated in Figure 5. This means siRNA molecules in AuNPs-polyplex must shut off more GPF or Luciferase proteins than that using free siRNA to reach the similar protein expression level. In another word, the enhancement of AuNPs-polyplex to siRNA down regulation is actually better than what was shown in Figure 6 for their higher starting protein level than that using free siRNA probes.
4. Conclusions
In summary, we immobilized polyplex on gold nanoparticles and delivered them into cells through electroporation. Conjugating with AuNPs helps minimize the cytotoxicity concerns from polyplex after cytoplasmic release while still retains good probe protection. It also avoids poor nanoparticle quality existing in traditional polyplex synthesis, namely large size and broad size variations, by managing molecule interactions and assembly on the surface of AuNPs. Combining with electroporation, conjugated polyplex (AuNPs-Polyplex) showed quick delivery and significant enhancement on the transfection efficiency with no obvious increase of toxicity. Such a combination of physical and chemical delivery concept may stimulate further exploration in the delivery of various therapeutic materials for both in vitro and in vivo applications.
The choice of AuNPs in the enhancement of polyplex delivery lies on their high conductivity and excellent biocompatibility. Their other potential advantages, such as sensing signal enhancement via localized surface plasmon resonance (LSPR) or surface enhanced Raman spectrum (SERS), have not yet been investigated with this gene delivery approach. But there is promising potential of our approach to integrate both diagnostic and therapeutic functions in one nanosystem (namely nanotheranostics) to accomplish both noninvasively tracking the targeting therapeutic probes and measuring their deliver performance simultaneously. This surely will help increase our understanding on the regulation mechanism of many therapeutic probes and quick establishment of appropriate strategies to improve their delivery or treatment performance. Other forms of gold nanostructures, such as nanorod, nanoshell, or nanowires, in principle, could also be used for the similar purposes. Its success will accelerate and broaden the applications of these nanomaterials in drug discovery, cancer diagnosis and treatment, and/or regenerative medicine where quick and precise diagnosis and therapeutics is urgently needed.
Supplementary Material
Acknowledgments
This work was supported by NIH/National Cancer Institute (NCI) Grant (R15CA156146).
References
- 1.Zwaka TP, Thomson JA. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 2.Eiges R, Schuldineer M, Drukker M, Yanuka O, Itskovitz-Eldor J, Benvenisty N. Curr Biol. 2001;11:514–518. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 3.Templeton NS, editor. Gene and cell therapy: therapeutic mechanism and strategies. 2. New York: Marcel Dekker; 2004. [Google Scholar]
- 4.Yin LH, Fu SQ, Nanakorn T, Garcia-Sanchez F, Chung I, Cote R, Pissorno G, Hanania E, Heimfeld S, Crystal R, Deisseroth A. Stem Cells. 1998;16S1:247–250. doi: 10.1002/stem.5530160830. [DOI] [PubMed] [Google Scholar]
- 5.Wu SC, Huang GYL, Liu JH. Biotechnol Progr. 2002;18:617–622. doi: 10.1021/bp020026p. [DOI] [PubMed] [Google Scholar]
- 6.Strair RK, Miller JS, Roberts BE. J Virol. 1988;62:2143–2149. doi: 10.1128/jvi.62.6.2143-2149.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gore ME. Gene Ther. 2003;10:4–4. [Google Scholar]
- 8.Marshall E. Science. 1999;286:2244–2245. doi: 10.1126/science.286.5448.2244. [DOI] [PubMed] [Google Scholar]
- 9.Wagner E, Zenke M, Cotten M, Beug H, Birnstiel ML. Proc Natl Acad Sci USA. 1990;87:3410–3414. doi: 10.1073/pnas.87.9.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang MX, Szoka FC. Gene Ther. 1997;4:823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 12.Leong KW, Mao HQ, Truong-Le VL, Roy K, Walsh SM, August JT. J Control Release. 1998;53:183–193. doi: 10.1016/s0168-3659(97)00252-6. [DOI] [PubMed] [Google Scholar]
- 13.Roy K, Mao HQ, Huang SK, Leong KW. Nat Med. 1999;5:387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- 14.Abraham SA, Waterhouse DN, Mayer LD, Cullis PR, Madden TD, Bally MB. Method Enzymol. 2005;391:71–97. doi: 10.1016/S0076-6879(05)91004-5. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami S, Higuchi Y, Hashida M. J Pharm Sci. 2008;97:726–745. doi: 10.1002/jps.21024. [DOI] [PubMed] [Google Scholar]
- 16.Liu D, Mori A, Huang L. Biochim Biophys Acta. 1992;1104:95–101. doi: 10.1016/0005-2736(92)90136-a. [DOI] [PubMed] [Google Scholar]
- 17.Pannier AK, Shea LD. Mol Ther. 2004;10:19–26. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Even-Chen S, Barenholz Y. Biochim Biophys Acta. 2000;1509:176–188. doi: 10.1016/s0005-2736(00)00292-3. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, Huang L. Biochem Biophys Res Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 21.Leong K. BioMEMS and Biomedical Nanotechnology. Springer; New York: 2006. pp. 239–263. [Google Scholar]
- 22.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Nano Lett. 2009;9:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, Huang Y, Jiang Q, Sun Y, Deng L, Liang Z, Du Q, Zing J, Zhao Y, Wang PC, Dong AX, Liang J. ACS Nano. 2010;4:5505–5511. doi: 10.1021/nn101638u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patra C, Bhattacharya R, Mukhopadhyay D, Mukherjee P. J Biomed Nanotechnol. 2008;4:99–132. [Google Scholar]
- 25.Cai C, Gao T, Hong H, Sun J. Nanotechnol Sci App. 2008;1:17–32. doi: 10.2147/NSA.S3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viator JA, Gupta S, Goldschmidt BS, Bhattacharyya K, Kannan R, Shukla R, Dale PS, Boote E, Katti KV. J Biomed Nanotechnol. 2010;6:1–5. doi: 10.1166/jbn.2010.1105. [DOI] [PubMed] [Google Scholar]
- 27.Cutler JI, Auyeung E, Mirkin CA. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 28.Brown PB, Qureshi AT, Hayes D, Monroe WT. ACS Nano. 2013;7:2948–2959. doi: 10.1021/nn304868y. [DOI] [PubMed] [Google Scholar]
- 29.Chang DC, Reese TS. Biophysical Journal. 1990;58:1–12. doi: 10.1016/S0006-3495(90)82348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann E, Kakorin S. Radiol Oncol. 1998;32:7–17. [Google Scholar]
- 31.Gehl J. Acta Physiol Scand. 2003;177:437–447. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Rubinsky B. Sens Actuators A: Phys. 2003;104:205–212. [Google Scholar]
- 33.Lin YC, Li M, Wu CC. Lab Chip. 2004;4:104–108. doi: 10.1039/b312804k. [DOI] [PubMed] [Google Scholar]
- 34.Lu H, Schmidt MA, Jensen KF. Lab Chip. 2005;5:23–29. doi: 10.1039/b406205a. [DOI] [PubMed] [Google Scholar]
- 35.Khine M, Lau A, Ionescu-Zanetti C, Seo J, Lee LP. Lab Chip. 2005;5:38–43. doi: 10.1039/b408352k. [DOI] [PubMed] [Google Scholar]
- 36.Fei Z, Wang S, Xie Y, Henslee BE, Koh CG, Lee LJ. Anal Chem. 2007;79:5719–5722. doi: 10.1021/ac070482y. [DOI] [PubMed] [Google Scholar]
- 37.Kim JA, Cho K, Shin YS, Jung N, Chung C, Chang JK. Biosens Bioelectron. 2007;22:3273–3277. doi: 10.1016/j.bios.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Valero A, Post JN, Nieuwkasteele JW, Ter Braak PM, Kruijer W, van den Berg A. Lab Chip. 2008;8:62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 39.Wang HY, Lu C. Biotechnol Bioeng. 2008;100:579–586. doi: 10.1002/bit.21784. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Zhang X, Wang W, Lee LJ. Anal Chem. 2009;81:4414–4421. doi: 10.1021/ac9002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fei Z, Hu X, Choi HW, Wang S, Farson D, Lee LJ. Anal Chem. 2010;82:353–358. doi: 10.1021/ac902041h. [DOI] [PubMed] [Google Scholar]
- 42.Geng T, Zhan Y, Wang HY, Witting SR, Cornetta KG, Lu C. J Control Release. 2010;144:91–100. doi: 10.1016/j.jconrel.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 43.Boukany PE, Morss A, Liao WC, Henslee B, Jung HC, Zhang XL, Yu B, Wang XM, Wu Y, Li L, Gao KL, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nat Nanotechnol. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Lee LJ. Biomicrofluidics. 2013;7:011301/1–14. doi: 10.1063/1.4774071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas-Chapana JA, Correa-Duarte MA, Ren Z, Kempa K, Giersig M. NanoLetters. 2004;4:958–988. [Google Scholar]
- 46.Raffa V, Ciofani G, Vittorio O, Pensabene V, Cuschieri A. Bioelectrochemistry. 2010;79:136–141. doi: 10.1016/j.bioelechem.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Weecharangsan W, Opanasopit P, Lee RJ. Anticancer Research. 2007;27:309–314. [PubMed] [Google Scholar]
- 48.Wang S, Zhang X, Yu B, Lee RJ, Lee LJ. Biosen Bioelectron. 2010;81:4414–4421. [Google Scholar]
- 49.Zu Y, Huang S, Liao W, Lu Y, Wang S. J Biomed Nanotech. 2014;10:982–992. doi: 10.1166/jbn.2014.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshmukh H. Enhance the transfection of mammalian cells using polyethylenimine (PEI) coated gold nanoparticles. Louisiana Tech University; 2011. MS thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.