Abstract
Introduction: Current gene therapy involves replacement of defective gene by delivery of healthy genetic material to precede normal function. Virus-mediated gene delivery is the most successful and efficient method for gene therapy, but it has been challenged due to serious safety concerns. Conversely, gene delivery using plasmid DNA (pDNA) is considered safer, but its transfection efficiency is much lower than virus-mediated gene transfer. Recently, mRNA has been suggested as an alternative option to avoid undesired insertion of delivered DNA sequences with higher transfection efficiency and stability.
Area covered: In this review, we summarize the currently available strategies of mRNA modification to increase the therapeutic efficacy; we also highlight the recent improvements of mRNA delivery for in vivo applications of gene therapy.
Expert opinion: The use of mRNA-based gene transfer could indeed be a promising new strategy for gene therapy. Notable advantages include no risk of integration into the genomic DNA, adjustable gene expression and easier modulation of the immune system. By reducing or utilizing the immunogenic properties, mRNA offers a promising tool for gene/or transcript replacement.
Keywords: chemical modification, delivery, gene therapy, mRNA
1. Introduction
Strategies of current gene therapy include replacement of defective gene by delivery of healthy genetic material to precede normal function. For gene therapy, virus-mediated gene delivery has been considered to be the most successful gene replacement method with high efficiency. However, this method has been challenged due to serious safety concerns, including insertion mutagenesis and triggering of the innate immune response [1]. Conversely, non-viral vector-mediated gene delivery using pDNA is safer but has lower transfection efficiency compared to viral vectors because of insufficient nuclear transport. Modification of pDNA for nuclear localization and enhanced transcription by using a strong constitutive promoter increases the expression of therapeutic genes, but integration of delivered sequences into the nuclear DNA also induces unexpected genetic changes [1,2].
To overcome undesired integration of transferred pDNA, mRNA has been suggested as an alternative [3,4]. As sufficient amounts of in vitro transcribed mRNA can be easily obtained by commercially available kits, therapeutic gene delivery using mRNA is now an effective replacement for pDNA in gene therapy. Gene delivery using mRNA has several advantages compared to pDNA. First, unexpected insertion mutation and promoter dependency can be excluded because mRNA is works as a gene/or transcript replacement in the cytoplasm. Therefore, nuclear translocation and transcription is not required. Second, mRNA-mediated gene transfer occurs in non-dividing cells, while pDNA-mediated gene transfer is mostly effective in dividing cells. Third, immunogenicity can be easily modulated by chemical modification.
Although there are many advantages of mRNA-mediated gene delivery, mRNA was previously considered too unstable to be used as a therapeutic molecule. However, transfection efficiency of mRNA has been greatly improved [5,6] and the half-life of mRNA has been dramatically increased, ranging from a few minutes to several hours by chemical modifications [7-9], which facilitates the use of mRNA for therapeutic gene transfer. By combining various mRNA modification and delivery methods, the efficacy of mRNA gene therapy could be greatly improved.
2. Modified mRNA
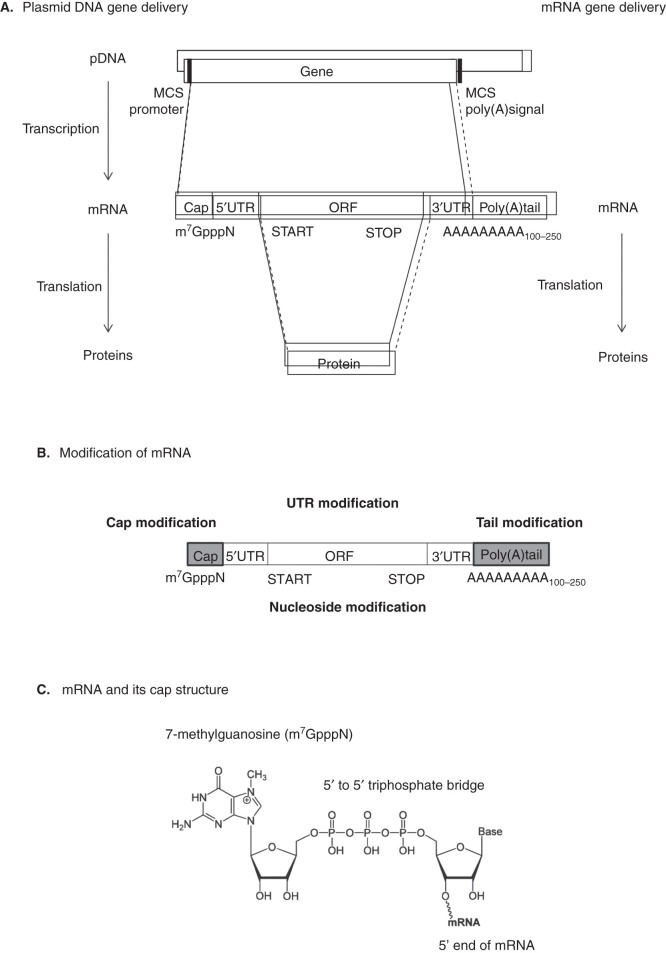
The main reason for mRNA instability is the presence of a hydroxyl group on the second carbon atom of the sugar moiety, which facilitates hydrolytic degradation. Either cis-acting or trans-acting factors can influence mRNA degradation [10]. Mature eukaryotic mRNA consists of five significant portions, including the cap structure ([m7GpppN or m7Gp3N (N: any nucleotide)], the 5’ untranslated region (5’UTR), an open reading frame (ORF), the 3’ untranslated region (3’UTR) and a tail of 100 – 250 adenosine residues (Poly(A) tails) (Figure 1A).
Figure 1.
Typical gene deliveries for therapeutic application. A. pDNA or mRNA-mediated gene transfer is illustrated. pDNA contains the multiple cloning site (MCS), which is used for restriction endonuclease recognition to insert transgene. Mature eukaryotic mRNA consists of five significant portions, including the cap structure ([m7GpppN or m7Gp3N (N: any nucleotide)], the 5’ untranslated region (5’UTR), an open reading frame (ORF), the 3’ untranslated region (3’UTR) and a tail of 100 – 250 adenosine residues (Poly(A) tails). B. Regions of mRNA modifications for increasing their stability. C. Chemical structure of mRNA CAP. D. Standard dinucleotide cap analog. E. Anti-reverse cap analogs (ARCA). F. mRNA degradation pathways. Both major pathways of mRNA decay are initiated by deadenylation.
The cap structure is post-transcriptionally modified with methylated m7GpppN in the nucleus at the 5’ ends of mRNA [11] and plays an important role in normal mRNA function, for example, mRNA splicing [12], stabilization [13], transport [14], recruiting ribosomes [15,16] and translational repression via microRNA [17,18]. This structure contains an uncommon nucleoside, 7-methylguanosine (m7G) and is connected with the 5’-5’ triphosphate bridge to the first transcribed nucleotide (Figure 1C).
To increase the efficiency of mRNA translation, an anti-reverse-cap analogue (ARCA), which contains a modified cap structure containing a 5’-5’ triphosphate bridge, has been suggested (Figure 1D and E) [7]. In vitro transcription performed in the presence of a cap analog may be initiated by an RNA polymerase from either guanosine (G) or m7G to produce correctly-capped (m7GpppG) or reversely-capped (Gpppm7G) mRNA, respectively [19]. The mRNAs bearing reversely-capped structures are poorly translated and more readily degraded. Only the 5’-5’ triphosphate linkage yields a translatable mRNA molecule. Introducing a chemical modification at the 3’- (or 2’-) position of the cap analogs prevents the reverse incorporation and improves both mRNA quality and translation efficiency. ARCA results in attachment in the correct direction only, which is recognized by eukaryotic initiation factor 4E (eIF4E), leading to ribosome recruitment and translation [9]. In addition, it has been reported that a high number of cap modifications and elongated 5’-5’ phosphate bridges in the ARCA improves translation efficiency and stability of mRNA [8].
The length of the poly(A) tail is also crucial for efficient translation and enhancing mRNA stability [20]. In mammalian cells, most actively translated mRNAs contain 100 – 250 poly(A)s [21]. For exogenous application, at least 20 poly(A)s are required for effective translation [22]. Generally, translation efficiency is dependent on the number of poly(A)s [23]. The poly(A) tail binds to numerous polyadenosyl-binding proteins (PABP), which recruit eukaryotic initiation factor 4G (eIF4G), leading to circular mRNA by increasing affinity to the mRNA cap [24]. This synergistic effect of the cap structure and the poly(A) tail has been explained by a cap-eIF4E-eIF4G-PABP-poly(A) closed loop structure, which could facilitate the recycling of ribosomes [25] and protect from nucleolytic degradation [26].
RNA degradation is initiated by shortening of the poly(A) tail and includes a de-capping process (Figure 1F). The cap structure and the poly(A) tail in the mRNA cooperatively function to maintain the stability of the mRNA [27,28]. The mRNA cap binds to the cap-binding protein (CBP) complex, which regulates mRNA transport from the cytoplasm to the nucleus. Degradation of mRNAs takes place in the cytoplasm at sites called P-bodies, which contain 5’-3’ exonucleases, de-capping and de-adenylating enzymes [28-30]. Once the poly(A) tail has been shortened to less than 12 residues, mRNA degradation then occurs by de-capping, along with 5’→3’ exo or 3’→5’ cleavage [31].
Further optimization of the mRNA structure can be achieved by replacing unstable non-coding sequences with non-coding sequences of mRNAs that are known to be stable. Most eukaryotic mRNAs contain mRNA decay signals in their 3’ untranslated regions (3’ UTRs), which affect mRNA stability. Various AU-rich sequences in the 3’ UTR of mRNA have been reported to be involved in the removal of the poly(A) tail [32]. The half-life of mRNA increases when AU-rich sequences are replaced with sequences of 3’ UTR from stable mRNA [10]. Other specific sequences in the 3’ UTR are the so-called iron responsive elements (IREs), which regulate mRNA stability in the 3’ UTR, and affect translation in the 5’ UTR [33]. 3’ UTRs of the human globin gene have also known as stable and insertion of two sequential 3’ UTR and poly (A) tail reported to show enhancement of mRNA stability in DC cells [34].
The coding region of mRNA sometimes facilitates mRNA degradation. Natural coding sequences often show less efficient codon usage for translation in specific organisms. Since different organisms usually show particular preferences for one of the several codons that encode the same amino acid [35], optimized codon usage improves translational efficiency [36]. As ribosomal traffic is modified by change of synonymous codon, mRNA turn-over and stability can be modified by altering RNases accessibility to target sites.
For in vivo applications that use mRNA as an alternative to pDNA, the immune response should be considered. Both DNA and RNA stimulate the mammalian innate immune system through activation of Toll-like receptor (TLRs). Thirteen TLRs have been identified and four of them (TLR3 for dsRNA, TLR7 and 8 for U-rich ssRNA, TLR9 for CpG DNA motif) are involved in nucleic acid recognition. It has been reported that mRNA can be recognized by TLR3 [37] and in vitro transcribed RNAs induce a strong TNF-α response in dendritic cells (DC) [38]. There are other receptors for TLR-independent immune response. RIG-I like receptor (RLR) such as retinoic acid inducible gene 1 (RIG-I) and melanoma differentiation-associated protein 5 (MDA-5) trigger type I IFN by IFN-regulatory factor 3 (IRF3)-dependent pathway, and they can be stimulated by ds/mRNA.
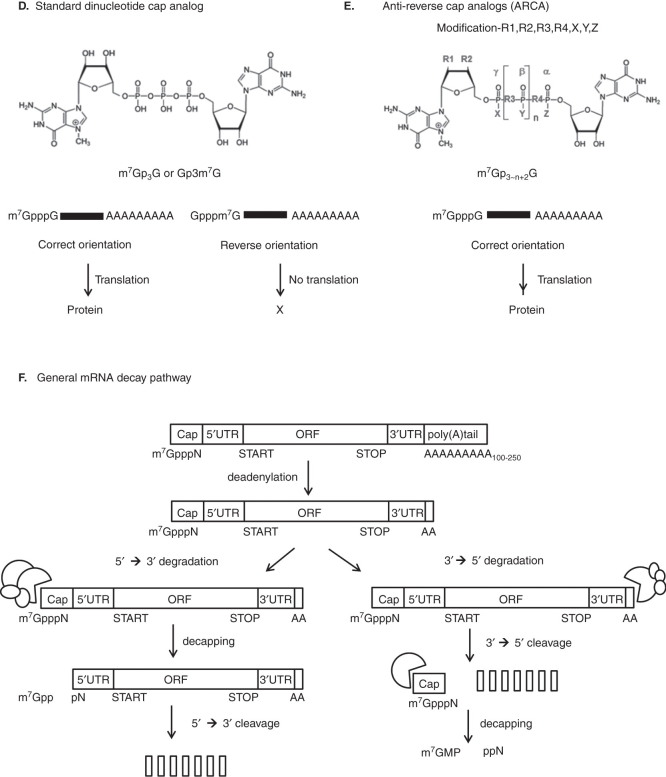
The use of modified nucleosides (Figure 2), such as 2’-O methyl nucleoside for in vitro transcription, shows dramatic suppression of TLR-mediated DC activation [38], but the effects of modified nucleosides on the TLR-independent immune response are still unknown. As many modified nucleosides are present in mammalian RNAs, such as pseudouridine, 2-thiouridine, 5-methylcytidine, 6-methyladenosine, inosine and many 2’-O-methylated nucleosides at the 5’-terminal cap, these nucleosides can be used for modified mRNA to reduce the immune reaction [3].
Figure 2.
Nucleoside modification of mRNA for immune escape.
On the other hand, the strong immune-stimulatory effect of mRNA can in fact be exploited for use in vaccination. Targeting antigen-presenting cells (APCs) with mRNA has been reported to induce tumor or antitumor immunity by activating T and B cells [39,40]. Although mRNAs from organelles like mitochondria are less modified than mRNAs from nuclear and bring stronger immune response, several reports have shown that mRNA-based gene transfer has a higher antigen (e.g., melan A or influenza matrix protein M1) loading efficiency to stimulate cytotoxic T cells [6,41].
3. Delivery systems for modified mRNA
Because the spontaneous uptake of naked nucleic acids by cells is very inefficient, nucleic acid delivery has been developed, adopting both viral and non-viral systems [42]. Replication-deficient recombinant viruses are commonly used vehicles for gene transfer. Use of viral vectors for pDNA has been intensively studied, but only a few cases of mRNA delivery using RNA viruses have been investigated. Generally, viruses which carry therapeutic DNA or RNA can copy to DNA in the cytosol after transfection, and enter into the nucleus. Some RNA viruses have shown localized replication and expression in the cytosol. For mRNA delivery, positive-strand viruses can be directly used and translated into therapeutic proteins [43] but negative-sense RNA should be coupled with its RNA-dependent RNA polymerase so that the positive-sense virus is translated because the negative-sense virus is not infectious [44].
In addition, various transfection reagents have been developed for non-viral delivery, which can be classified into two different groups. One is mRNA delivery across the cellular membrane by physical disturbances, and the other is generation of mRNA endocytosis by cationic carriers. Various physical manipulations have also been used to improve efficiency for direct transfection [45]. Methods utilizing electroporation, gene guns, ultrasound or high-pressure injection can be applied for direct delivery of nucleotides to the cells. These methods can also be used for in vivo as well as in vitro applications. In particular, the use of electroporation is one of the most efficient methods for mRNA delivery [6,46,47]. As the mRNA does not have to enter the nucleus, soft electrical pulses may be applied to reduce cellular toxicity. Another advantage of electroporation is the direct delivery of mRNA into the cytosol, which could surpass the unwanted immune response. For therapeutic vaccination, mRNA-pulsed DCs can be used as APCs and modulate tumor immunity. Compared to transfection of pDNA, higher tumor antigen loading of DCs was observed in the gene transfer by mRNA in human [6,41,46]. The gene gun method uses high-velocity heavy metal particles such as gold, and this particle-mediated mRNA delivery is applicable to most mammalian tissues [48]. Gene gun-mediated GFP mRNA delivery was reported to show detectable expression for a week, and mRNA encoding for the human epidermal growth factor (hEGF) showed a wound healing effect.
Self-assembled lipo- or poly-plexes are well-known transfection vehicles that are spontaneously generated by charge-to-charge interactions between the complexations of negatively charged mRNA and cationic lipids or polymers, such as lipoplexes, polyplexes, polycations and dendrimers [49]. Although various polycations, such as DEAE (diethylaminoethyl)-dextran [50], DOTMA/DOPE (1,2-dioleyl-3-trimethylammoniumchloride/1,2-dioleoyl-3-phosphoethanolamine) [5], poly l-lysine [51] and PEI (polyethylene imine) [49], showed an ability to transfect mRNA, the most efficient was demonstrated to be DOTAP (1,2-dioleoyl-3-trimethylammonium propane) [9]. Single-stranded mRNA strongly binds to cationic polymers compared to pDNA, and cytosolic mRNA is involved in pDNA release from the cationic polymer [52]. As efficient cationic polymers that are used for pDNA transfection may not suitable for efficient mRNA transfection [3], the design of novel cationic polymers for mRNA delivery in human cells has to be carefully considered.
4. Systemic delivery of modified mRNA for gene therapy
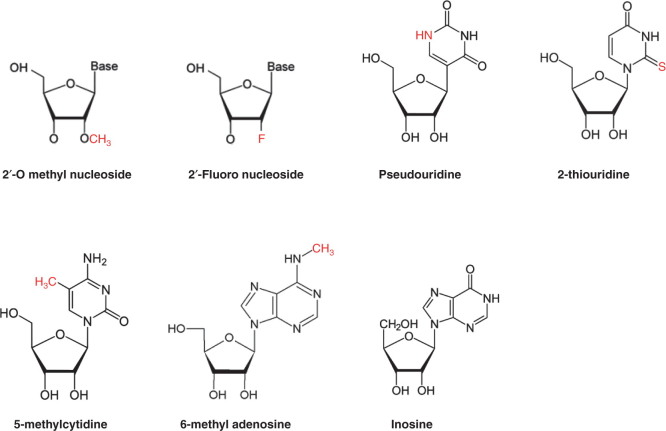
In order to use mRNA as an efficient therapeutic agent for in vivo application, delivery routes also have to be considered. Direct delivery with naked mRNA at the targeted site was usually administrated by local injection, but indirect delivery using carrier-mediated mRNA with targeting moiety was recommended for systemic injection (Figure 3) [53]. For systemic delivery, the increased stability and the prolonged circulation of mRNA is important to improve therapeutic efficacy. In addition to chemical modification to prevent mRNA degradation, cationic lipid or polymer complexes showed successful protection against nucleases and increased stability of mRNA for systemic delivery in a mouse model. Net positively charged complexes showed increased stability of mRNA in vitro, although they sometimes interact with negatively charged serum proteins to form aggregates resulting in rapid clearance or creating clots [54]. To acquire the characteristic of prolonged circulation and to inhibit non-specific uptake by the reticuloendothelial system (RES), conjugation with polyethylene glycol (PEG) has been frequently suggested to play a role [55]. PEGylated lipids or polymers were reported to have higher stability by inhibiting attachment to serum proteins. PEGylation is very important to preserve the integrity of mRNA and to target tumor sites after prolonged circulation.
Figure 3.
Different injection routes of gene delivery are outlined.
Adapted with permission from [53].
On the other hand, immune modulation by mRNA should also be considered for in vivo application. Altough single-stranded RNA is reported to stimulate the innate immune system [56], therapeutic mRNA can be disguised by chemical modification to reduce the innate immune response by resembling the highly modified RNAs from organelles in mammalian cells [57-59]. In addition, the secondary structure of exogenous mRNA is reported to activate IFN-inducible protein kinase R, a global repressor of protein translation, resulting in a low transgene expression [60].
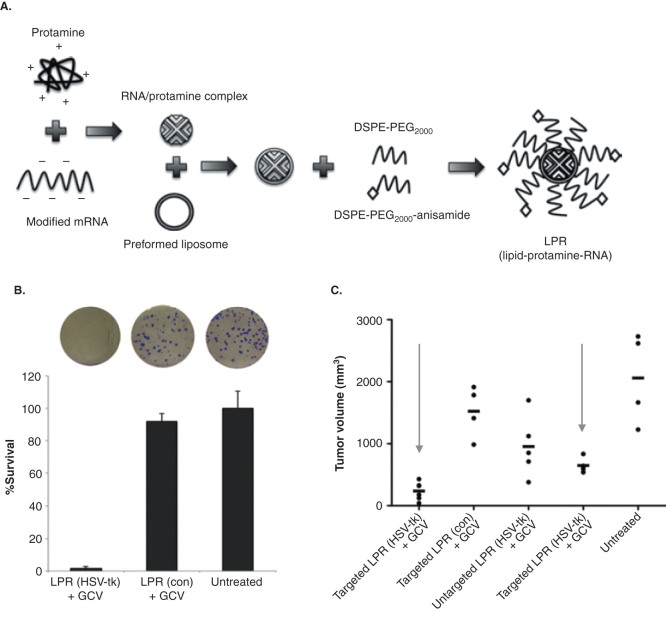
In an effort to improve efficiency of systemic delivery of modified mRNA for gene therapy, specific nanoparticle formulation using liposome-protamine-RNA (LPR) was suggested [61]. To suppress immune activation, Wang et al. modified cytidine triphosphate and uridine triphosphate with 5’-methylcytidine and pseudouridine triphosphate (Figure 4) during the in vitro transcription of mRNA. Modified anionic mRNA was then mixed with polycation (protamine, 4 kDa) to generate an mRNA/protamine complex. Cationic lipid DOTAP/cholesterol was used to generate a liposome, which was then mixed with the mRNA/protamine complex. To reduce the attachment of serum protein and minimize uptake of RES for systemic injection, extensive PEGylation was applied to generate LPR. As sigma receptor overexpressed tumor cells (H460) were used, anisamide was also added to the distal end of PEG for specific targeting (Figure 4A). LPR is small in size (< 100 nm) for easy internalization. Herpes simplex virus 1-thymidine kinase/ganciclovir (HSV-tk/GCV) therapy, one of the most widely used suicidal gene/prodrug coupling for gene therapy, was used in this research. Tumor cells were transfected with LPR, which contains modified mRNA encoding HSV1-tk. After HSV-tk/GCV therapy, tumor cells showed reduced survival in vitro (Figure 4B), and tumor reduction was observed in an in vivo mouse xenograft model (Figure 4C). Interestingly, mRNA (LPR)-transfected tumors showed a greater reduction than pDNA (LPD, liposome-protamine-DNA)-transfected tumors. The delivery of mRNA (LPR) was found to be very rapid (expression within 4 h after transfection), transient and cell cycle independent.
Figure 4.
Systemic delivery of modified mRNA for gene therapy. A. Diagram of formulating modified mRNA into a liposome-protamine-RNA (LPR) complex. B. Proliferation capacity of the surviving H460 cells after HSV-tk/GCV therapy, determined by clonogenic assay. C. Tumor growth inhibition of H460 non-small cell lung cancer after HSV-tk/GCV therapy by systemic delivery of modified mRNA in the LPR.
Adapted with permission from [61].
A. DOTAP: 1,2-Dioleoyl-3-trimethylammonium-propane; DSPE: 1,2-Distearoyl-phosphatidylethanolamine; LPR: Lipid/protamine/mRNA. C. Con: Control; GCV: Ganciclovir; HSV-tk: Herpes simplex virus-thymidine kinase; LPD: lipid/protamine/DNA; LPR: lipid/protamine/mRNA.
The classical vaccination strategy for cancer starts from transfection of DC with tumor antigens using whole cell extracts, but whole cell extracts contain many irrelevant antigens, which can cause autoimmune responses. For this reason, fine-tuned nucleic acid-based vaccination is a promising alternative. A liposome/mRNA vaccine encoding various tumor antigens, such as human CEA (carcinoembryonic antigen) [62], OVA (chicken ovalbumin) [63], hTERT (human telomerase catalytic subunit) [64], AFP (α-fetoprotein, a protein specifically expressed by hepatocellular carcinoma cells) [65] and RHAMM (the receptor for hyaluronan-mediated motility frequently overexpressed in brain tumors) [66] were loaded to DC, and their efficacy was monitored in an in vivo mouse model.
For mRNA vaccination, the administration route was reported to be a critical component [67,68]. There was a marked difference in the cytotoxic T lymphocyte (CTL) response to kill tumor cells after intravenous (i.v.), subcutaneous (s.c.), intramuscular (i.m.) or intradermal (i.d.) injection of liposome encapsulated protamine-condensed mRNA at the base of the ear pinna [67]. Only i.d. administration showed a significant CTL response. Interestingly, mRNA functions not only as an antigen-encoding molecule, but also as an adjuvant by enhancing immunological responses and antigen presentation. In addition, naked RNA vaccine administered into the lymph node was reported to show rapid selective uptake by lymph node DCs driven by micropinocytosis [68].
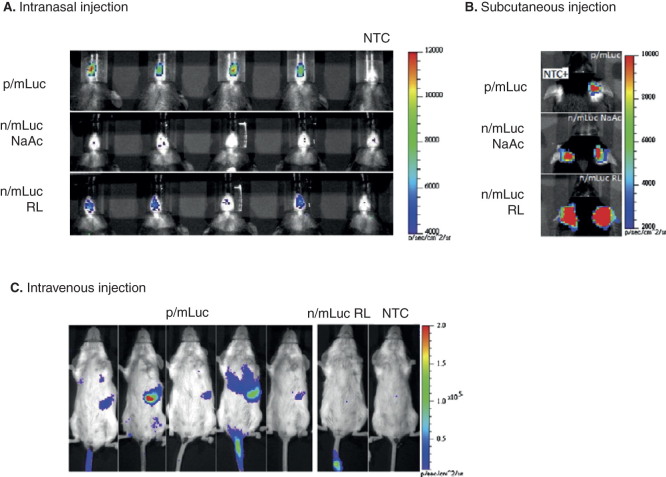
Recently, transfection efficiency and transgene expression using mRNA by various injection routes have also been intensively investigated [69,70]. Figure 5 shows bioluminescence imaging to evaluate in vivo transection efficiency of naked or nanoparticle (ovalbumin)-based mRNA with chemical modification (ARCA) mRNA delivery. Transfection was increased by OVA nanoparticle-mediated mRNA transfer compared to naked mRNA both in intranasal and i.v. injections.
Figure 5.
Evaluating in vivo transfection efficiency of naked or nanoparticle-based modified (ARCA) mRNA according to the administration routes. A. Bioluminescence in C57BL/6 mice transfected intranasally with 4 μg of p/mLuc and n/mLuc over a 4-h time period. B. Bioluminescence in C57BL/6 mice transfected subcutaneously at the base of the ear pinna with p/mLuc and n/mLuc (in NaAc and RL) at 4 h. C. Bioluminescence signal in BALB/c mice intravenously administrated with 26 μg of p/mLuc and m/Luc at 8 h time points with respective color scales.
Adapted with permission from [4].
mLuc: Luciferase mRNA; n: Naked; NaAc: Sodium Acetate; NTC: Non-transfected control; p: mOVA nanoparticle; RL: Ringer’s lactate solution.
5. Conclusions
The use of mRNA-based gene transfer could indeed be a promising new strategy for gene therapy. Notable advantages include no risk of integration into the genomic DNA, adjustable gene expression and easier modulation of the immune system. Various chemical modifications increase the stability of mRNA and provide more chances for the possible use of mRNA-mediated gene transfer for gene therapy. However, one major challenge for the potential use of mRNA in gene therapy is to try to reduce inflammatory reactions after repeated treatment. Although repeated mRNA application seems to be feasible in principle, knowledge about application frequencies for long-term treatment is currently unknown. By reducing or utilizing the immunogenic properties, mRNA offers a promising tool for both gene therapy and vaccination approaches, respectively.
6. Expert opinion
Previously, the stability and transfection efficiency of mRNA was considered too low for it to be used as a therapeutic molecule. However, the efficacy of mRNA transfer has been greatly improved by combining various chemical modification and delivery methods. Since mRNA degradation is initiated by shortening of the poly(A) tail and includes a de-capping process, mRNA structure has been optimized by replacing unstable non-coding sequences with stable non-coding sequence. Various physical manipulations and nanoparticle-mediated mRNA delivery have also been used to improve transfection efficiency. In addition, modification of the cap structure and optimization of codon usages for specific organisms have been frequently used to increase the efficiency of mRNA translation.
Use of mRNA as an alternative to pDNA for therapy has major advantages of its cellular localization; i) risk of insertion mutagenesis can be avoided because mRNA exerts its function in the cytoplasm; ii) promoter-dependent modulation of gene expression is not required because amount of delivered mRNA into cytosol is directly related to its function; iii) effective delivery of mRNA into non-dividing cells is possible because transfection efficiency of mRNA is independent from cell cycle and nuclear transport; iv) vector-induced immunogenicity can be avoidable; v) repeated application is possible.
Although mRNA-based gene transfer was considered as a safe application, induction of unwanted immune response by repeated treatment remains still critical for their in vivo application. By utilizing the immunogenic properties, mRNA-based gene transfer offers a promising tool for both transcript replacement and vaccination approaches, respectively. However, control of immunogenic properties of mRNA to reduce inflammation for mRNA based-transcript replacement therapy has not been carefully investigated to date. Especially, most of researches have been focused on short-term application because mRNA can only render a transient expression, but long-term treatment is required for the treatment of inherited disease. Repeated mRNA treatment seems to be feasible but knowledge about frequency, interval and duration for long-term application is currently unknown. It would be interesting to investigate how to control a protein expression pattern after mRNA delivery under the different pathologic situation.
Recently, mathematical models that predict kinetics and efficiency of mRNA delivery in vitro and in vivo were suggested by several reports [71-73]. Kinetics related to endosomal uptake and cytosolic release of exogenous mRNA in the cellular level should be studied to establish a compartment model with rate equation. These studies established a great co-relation between theories and efficiency of delivered mRNA; thereby provide useful information to optimize mRNA-based therapeutic application. Particularly, a more detailed and quantitative understanding of artificial mRNA delivery is important for in vivo application. The degree of predictive power describing synthetic mRNA expression level and timing will depend on the degree of accuracy with the transfection efficiency and kinetics for the experimental planning. Although mRNA delivery is inherently stochastic and the expression level of every single cell is different, measurement at the single cell level and analysis of the corresponding distribution of functions, in terms of successfully delivered and translated mRNA, is still necessary to acquire the true population response in the mRNA delivery. We are convinced that predictive modeling of mRNA delivery will provide dramatic advances in the theoretical as well as practical aspects of mRNA-based therapy.
Article highlights.
The use of mRNA-based gene transfer could be a new promising strategy for transcript replacement and vaccination therapy.
Various chemical modifications increase the stability of mRNA and provide more chances for the possible use of mRNA-mediated gene transfer for gene therapy.
Recent advances of mRNA-based gene transfer provide notable advantages including no risk of integration into the genomic DNA, adjustable gene expression and easier modulation of the immune system.
Specific nanoparticle formulation using modified mRNA improves the therapeutic efficacy both in transcript replacement and vaccination.
This box summarizes key points contained in the article.
Declaration of interest
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No. HI14C1072, HI13C0826, HI14C0311020014), a grant from the National Research Foundation of Korea (NRF) for the Global Core Research Center (GCRC) funded by the Korea government (MSIP: Ministry of Science, ICT & Future Planning) (No.2011-0030001) and Seoul National University Hospital Research fund (No.042011150). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- Sheridan C. Gene therapy finds its niche. Nat Biotechnol. 2011;29(2):121–8. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- Das SK, Menezes ME, Bhatia S, et al. Gene therapies for cancer: strategies, challenges and successes. J Cell Physiol. 2015;230:259–71. doi: 10.1002/jcp.24791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • .Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current prospects for mRNA gene delivery. Eur J Pharm Biopharm. 2009;71(3):484–9. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]; • This review summarizes various applications of mRNA-mediated gene delivery in detail and suggested future usages.
- Phua KK, Leong KW, Nair SK. Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J Control Release. 2013;166(3):227–33. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci USA. 1989;86(16):6077–81. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tendeloo VF, Ponsaerts P, Lardon F, et al. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98(1):49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- Stepinski J, Waddell C, Stolarski R, et al. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3’-O-methyl)GpppG and 7-methyl (3’-deoxy)GpppG. RNA. 2001;7(10):1486–95. [PMC free article] [PubMed] [Google Scholar]
- Strenkowska M, Kowalska J, Lukaszewicz M, et al. Towards mRNA with superior translational activity: synthesis and properties of ARCA tetraphosphates with single phosphorothioate modifications. New J Chem. 2010;34(5):993–1007. doi: 10.1039/b9nj00644c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohra FT, Chowdhury EH, Tada S, et al. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem Biophysic Res Comm. 2007;358(1):373–8. doi: 10.1016/j.bbrc.2007.04.059. [DOI] [PubMed] [Google Scholar]
- Eberhardt W, Doller A, Akool E-S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol Ther. 2007;114(1):56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Banerjee AK. 5’-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44(2):175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78(4):657–68. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Coutts M, Krowczynska A, Brawerman G. Protection of mRNA against nucleases in cytoplasmic extracts of mouse sarcoma ascites cells. Biochim Biophys Acta. 1993;1173(1):49–56. doi: 10.1016/0167-4781(93)90241-5. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj IW. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J Cell Biol. 1992;118(6):1287–95. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5(11):2108–16. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Ann Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–7. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, et al. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009;35(6):881–8. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5’ caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1(9):957–67. [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. Genome-wide Determination of Poly(A) Tail Length and 3’ End Modifications. Mol Cell. 2014;53(6):1044–52. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Brawerman G. The Role of the poly(A) sequence in mammalian messenger RNA. CRC Crit Rev Biochem. 1981;10(1):1–38. doi: 10.3109/10409238109114634. [DOI] [PubMed] [Google Scholar]
- Elango N, Elango S, Shivshankar P, Katz MS. Optimized transfection of mRNA transcribed from a d(A/T)100 tail-containing vector. Biochem Biophys Res Comm. 2005;330(3):958–66. doi: 10.1016/j.bbrc.2005.03.067. [DOI] [PubMed] [Google Scholar]
- Peng J, Murray EL, Schoenberg DR. In vivo and in vitro analysis of poly(A) length effects on mRNA translation. Methods Mol Biol. 2008;419:215–30. doi: 10.1007/978-1-59745-033-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216(1):1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Michel YM, Poncet D, Piron M, et al. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem. 2000;275(41):32268–76. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- Newbury SF. Control of mRNA stability in eukaryotes. Biochem Soc Trans. 2006;34(Pt 1):30–4. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Ann Rev Biochem. 2004;73:861–90. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25(5):635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32(5):605–15. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • .Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]; • This review explains the mechanism of mRNA decay and pointed out the key components to control its stability.
- Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9(2):239–51. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, et al. Identification of a common nucleotide sequence in the 3’-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83(6):1670–4. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72(1):19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Seimi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–17. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–95. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, et al. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Ni H, Capodici J, et al. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–50. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Palucka AK. Dendritic cells as therapeutic vaccines against cancer. Nat Rev Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- Grunebach F, Muller MR, Brossart P. RNA transfection of dendritic cells. Methods Mol Med. 2005;109:47–54. doi: 10.1385/1-59259-862-5:047. [DOI] [PubMed] [Google Scholar]
- Strobel I, Berchtold S, Gotze A, et al. Human dendritic cells transfected with either RNA or DNA encoding influenza matrix protein M1 differ in their ability to stimulate cytotoxic T lymphocytes. Gene Ther. 2000;7:2028–35. doi: 10.1038/sj.gt.3301326. [DOI] [PubMed] [Google Scholar]
- •• .Tavernier G, Andries O, Demeester J, et al. mRNA as gene therapeutic: how to control protein expression. J Control Release. 2011;150:238–47. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]; •• This review suggested the mRNA as an alternative of pDNA for gene therapy with comprehensive overview.
- Agapov EV, Frolov I, Lindenbach BD, et al. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–94. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzer M, Armeanu S, Lauer UM, Neubert WJ. Sendai virus vectors as an emerging negative-strand RNA viral vector system. J Gene Med. 2003;5:543–53. doi: 10.1002/jgm.426. [DOI] [PubMed] [Google Scholar]
- Li S, Ma Z. Nonviral gene therapy. Curr Gene Therapy. 2001;1:201–26. doi: 10.2174/1566523013348814. [DOI] [PubMed] [Google Scholar]
- Ponsaerts P, van der Sar S, Van Tendeloo VF, et al. Highly efficient mRNA-based gene transfer in feeder-free cultured H9 human embryonic stem cells. Cloning Stem Cells. 2004;6:211–16. doi: 10.1089/clo.2004.6.211. [DOI] [PubMed] [Google Scholar]
- Ponsaerts P, Van Tendeloo VF, Berneman ZN. Cancer immunotherapy using RNA-loaded dendritic cells. Clin Exp Immunol. 2003;134:378–84. doi: 10.1046/j.1365-2249.2003.02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RL, Murray MT, Schwarz K, et al. In-vivo particle mediated delivery of mRNA to mammalian tissues: ballistic and biologic effects. Wound Repair Regen. 2001;9:287–96. doi: 10.1046/j.1524-475x.2001.00287.x. [DOI] [PubMed] [Google Scholar]
- Bettinger T, Carlisle RC, Read ML, et al. Peptide-mediated RNA delivery: a novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 2001;29:3882–91. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G. Interaction of poliovirus-specific RNAs with HeLa cells and E. coli. Curr Top Microbiol Immunol. 1973;62:89–138. doi: 10.1007/978-3-642-65772-6_4. [DOI] [PubMed] [Google Scholar]
- Fisher KJ, Wilson JM. The transmembrane domain of diphtheria toxin improves molecular conjugate gene transfer. Biochem J. 1997;321(Pt 1):49–58. doi: 10.1042/bj3210049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth S, Hoffmann F, von Gersdorff K, et al. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med. 2006;8:1416–24. doi: 10.1002/jgm.975. [DOI] [PubMed] [Google Scholar]
- Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647–52. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- Tros de Ilarduya C, Arangoa MA, Duzgunes N. Transferrin-lipoplexes with protamine-condensed DNA for serum-resistant gene delivery. Methods Enzymol. 2003;373:342–56. doi: 10.1016/s0076-6879(03)73022-5. [DOI] [PubMed] [Google Scholar]
- Noble GT, Stefanick JF, Ashley JD, et al. Ligand-targeted liposome design: challenges and fundamental considerations. Trends Biotech. 2014;32:32–45. doi: 10.1016/j.tibtech.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• .Kormann MS, Hasenpusch G, Aneja MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature Biotechnol. 2011;29:154–7. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]; •• This research article successfully showed therapeutic potential of chemically modified mRNA as an alternative to DNA-based gene therapy in mice.
- Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5’-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–4. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su HH, Yang Y, et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther. 2013;21:358–67. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conry RM, LoBuglio AF, Wright M, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–400. [PubMed] [Google Scholar]
- Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465–72. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeboe-Larssen S, Fossberg E, Gaudernack G. mRNA-based electrotransfection of human dendritic cells and induction of cytotoxic T lymphocyte responses against the telomerase catalytic subunit (hTERT) J Immunol Methods. 2002;259:191–203. doi: 10.1016/s0022-1759(01)00506-3. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Zhang LW, Ren J, et al. Induction of alpha-fetoprotein-specific CD4- and CD8-mediated T-cell response using RNA-transfected dendritic cells. Cell Immunol. 2006;239:144–50. doi: 10.1016/j.cellimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Amano T, Kajiwara K, Yoshikawa K, et al. Antitumor effects of vaccination with dendritic cells transfected with modified receptor for hyaluronan-mediated motility mRNA in a mouse glioma model. J Neurosurg. 2007;106:638–45. doi: 10.3171/jns.2007.106.4.638. [DOI] [PubMed] [Google Scholar]
- Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Diken M, Kreiter S, Seimi A, et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene ther. 2011;18:702–8. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- Van Lint S, Goyvaerts C, Maenhout S, et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 2012;72:1661–71. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- Phua KK, Staats HF, Leong KW, Nair SK. Intranasal mRNA nanoparticle vaccination induces prophylactic and therapeutic anti-tumor immunity. Sci Rep. 2014;4:5128. doi: 10.1038/srep05128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua KK, Leong KW, Nair SK. Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J Control Release. 2013;166:227–33. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon TS, Leonhardt C, Rädler JO. Multi-level kinetic model of mRNA delivery via transfection of lipoplexes. PLoS One. 2014;9:e107148. doi: 10.1371/journal.pone.0107148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt C, Schwake G, Stögbauer TR, et al. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine. 2014;10:679–88. doi: 10.1016/j.nano.2013.11.008. [DOI] [PubMed] [Google Scholar]